Abstract
The Target Of Rapamycin (TOR) kinase belongs to the highly conserved eukaryotic family of phosphatidylinositol-3-kinase-related kinases (PIKKs). TOR proteins are found at the core of two distinct evolutionarily conserved complexes, TORC1 and TORC2. Disruption of TORC1 or TORC2 results in characteristically dissimilar phenotypes. TORC1 is a major cell growth regulator, while the cellular roles of TORC2 are not well understood. In the fission yeast Schizosaccharomyces pombe, Tor1 is a component of the TORC2 complex, which is particularly required during starvation and various stress conditions. Our genome-wide gene expression analysis of Δtor1 mutants indicates an extensive similarity with chromatin structure mutants. Consistently, TORC2 regulates several chromatin-mediated functions, including gene silencing, telomere length maintenance, and tolerance to DNA damage. These novel cellular roles of TORC2 are rapamycin insensitive. Cells lacking Tor1 are highly sensitive to the DNA-damaging drugs hydroxyurea (HU) and methyl methanesulfonate, similar to mutants of the checkpoint kinase Rad3 (ATR). Unlike Rad3, Tor1 is not required for the cell cycle arrest in the presence of damaged DNA. Instead, Tor1 becomes essential for dephosphorylation and reactivation of the cyclin-dependent kinase Cdc2, thus allowing reentry into mitosis following recovery from DNA replication arrest. Taken together, our data highlight critical roles for TORC2 in chromatin metabolism and in promoting mitotic entry, most notably after recovery from DNA-damaging conditions. These data place TOR proteins in line with other PIKK members, such as ATM and ATR, as guardians of genome stability.
The TOR protein kinase is a major cell growth regulator that links cellular growth with cell divisions (18, 42, 64, 65). TOR is an atypical protein kinase conserved from yeast to humans that was isolated as the target of the immunosuppressive and anticancer drug rapamycin (28). TOR proteins can be found in two distinct complexes, known as TORC1 and TORC2 (27, 64). These complexes mediate their distinct cellular functions via phosphorylation and activation of different sets of AGC-like kinases, including mammalian p70S6K, downstream of TORC1, and AKT/protein kinase B (PKB) downstream of TORC2 (18). TORC1 in mammals contains mTOR (Tor1 or Tor2 in Saccharomyces cerevisiae; Tor2 in Schizosaccharomyces pombe) and the Raptor protein (Kog1 in S. cerevisiae; Mip1 in S. pombe). TORC1 in many different eukaryotes plays a central role in the control of growth (mass accumulation) in response to external stimuli, particularly nutrient availability. Disruption of TORC1, either by mutating its components or by rapamycin treatment, can lead to a starvation-like phenotype (64). The cellular roles of TORC2, on the other hand, are less well defined. TORC2 in mammals contains mTOR (Tor2 in S. cerevisiae; Tor1 in S. pombe) together with Rictor (Avo3 in S. cerevisiae; Ste20 in S. pombe) and mSin1 (Avo1 in S. cerevisiae; Sin1 in S. pombe). TORC2 plays a role in regulating the actin cytoskeleton and cell wall integrity pathway in S. cerevisiae (3, 15, 27), a function that is at least partially conserved in human cells (17, 47).
Fission yeast contains two TOR homologues, Tor1 and Tor2 (59), which form the TORC2 and TORC1 complexes, respectively (14, 32). Disruption tor2+ (TORC1) mimics nitrogen starvation responses (1, 14, 32, 56, 57, 62), while disruption of tor1+ (TORC2) results in pleiotropic defects, including elongated cells, sensitivity to osmotic and oxidative stress, inability to execute developmental processes in response to nutrient depletion, and a decrease in amino acid uptake (16, 22, 59). Tor1 regulates cell survival under stress conditions and starvation responses via the AGC protein kinase Gad8, a putative homologue of mammalian AKT/PKB (16).
In budding yeast and mammalian cells, TORC1 mediates the rapamycin-sensitive signaling branch while TORC2 is far less sensitive to inhibition by this drug (27, 48). Curiously, rapamycin does not inhibit growth of S. pombe cells but partially inhibits sexual development and amino acid uptake (60-62). Inhibition of amino acid uptake is likely a result of inhibiting Tor1 (61, 62). Accordingly, a tor1 rapamycin-defective allele (tor1S1834E) confers rapamycin resistance to strains that are dependent on amino acid uptake for their growth (61). Yet rapamycin also induces a response similar to that for a shift from rich to poor nitrogen conditions, an effect that may involve inhibition of both Tor1 and Tor2 (41).
While other members of the phosphatidylinositol-3-kinase-related kinase (PIKK) family of proteins, such as ATM and ATR, have been shown to play central roles in the DNA damage response, little is known about roles that TOR proteins might play in such processes. Recently it was shown that the rapamycin-sensitive TORC1 complex participates in regulating cell survival under DNA-damaging conditions (24, 42, 49). Currently, no such role has been attributed to TORC2.
Here we show that Tor1 (TORC2) is critical for cell survival under DNA-damaging conditions, gene silencing at heterochromatic regions, and telomere length maintenance and for regulation of cell cycle progression. Since the TOR complexes are highly conserved in evolution, this novel TORC2 function may also be conserved in other organisms.
MATERIALS AND METHODS
Yeast techniques.
S. pombe strains are described in Table S1 in the supplemental material. All experiments were performed by using standard genetic and molecular yeast techniques as described in reference 35. Growth medium was prepared as described previously (59). Rapamycin (R0395; Sigma) was used at a final concentration of 100 ng/ml. For cell killing assays, HU (H8627; Sigma) or methyl methanesulfonate (MMS) (129925; Sigma) was added at appropriate concentrations. UV irradiation was performed using a UV Stratalinker 1800 crosslinker (Stratagene). Cells were visualized using a Nikon eclipse E600 fluorescence microscope and photographed using a Nikon digital camera (DXM1200) and the ACT1 software. Cell length was determined at septation and measured using Scion Image software. For fluorescence-activated cell sorting (FACS) analysis, nuclei were isolated as described previously (10), stained with propidium iodide, and analyzed by a Becton Dickinson FACSort cell sorter. Data were analyzed by Cell Quest software for Macintosh.
Telomere gels.
DNA was isolated from logarithmically growing cells, digested with EcoRI, and subjected to Southern blotting (33). A DNA probe corresponding to the telomere repeats was generated from pIRT2-TELO29 (33).
RNA and protein manipulations.
RNA for microarray hybridization and Northern blots was prepared using the hot phenol method. Northern blot analysis was carried out as described previously (62). Gene-specific probes were labeled with [α-32P]dCTP using the Random Primer DNA labeling kit (20-101-25A; Biological Industries). Transcripts were quantified using Gelquant software. For Western blot analysis, 50 ml of logarithmically growing cells were harvested, resuspended in protein extraction buffer (20% glycerol, 20 mM HEPES [pH 7.9], 50 mM NH2SO4, 5 mM EDTA [pH 8.0]) in the presence of protease inhibitor and broken with glass beads. Immunoblotting was performed as described previously (59).
Microarray experiments and data evaluation.
We used DNA microarrays displaying probes for >99% of all known and predicted genes of S. pombe spotted in duplicate onto glass slides. RNA extraction, hybridization, and initial data processing and normalization were performed as described previously (29). Three independent biological experiments were performed, including a dye swap. The data were visualized and analyzed using the GeneSpring software program (Agilent). The significance of overlaps between different gene lists was calculated in GeneSpring using a standard Fisher exact test, and P values were adjusted with a Bonferroni multiple testing correction. Cutoff values of 1.5-fold change in all biological repeats were used. Gene annotations were downloaded from S. pombe GeneDB (http://www.genedb.org/genedb/pombe/). The data can be obtained from the ArrayExpress account at www.ebi.ac.uk/aerep/login, accession number E-MTAB-39.
Clustering along chromosomes of genes with induced expression in Δtor1 cells was analyzed using an in-house Perl script which compares clustered genes to a random distribution (31). P values were adjusted for multiple testing using the Benjamini-Hochberg false discovery rate.
RESULTS
tor1+ deletion leads to upregulation of repeated elements and subtelomeric genes.
In order to uncover the underlying mechanism for the pleiotropic defects in cells lacking tor1+ (Δtor1), we performed genome-wide gene expression profiling. This analysis revealed that in growing cells, 117 and 48 genes were at least 1.5-fold upregulated or downregulated, respectively, in Δtor1 cells compared to results for wild-type cells (see Fig. S1 and S2 in the supplemental material and data submitted at ArrayExpress [www.ebi.ac.uk/aerep/login]). Comparison of these transcriptional profiles with profiles of other mutants showed an extensive overlap among upregulated genes with either clr6-1, Δclr3 clr6-1 (12), or Δrsc58 mutants (34) (Fig. 1A). Clr3 and Clr6 are histone deacetylases (HDACs), while Rsc58 is part of the conserved RSC complex, a member of the SWI/SNF chromatin-remodeling family. Genes that are upregulated in the absence of Tor1 include repeated genes, such as the wtf elements, and several noncoding RNA telomeric duplications, suggesting that the Δtor1 mutation leads to a derepression of gene transcription mediated by heterochromatin. Accordingly, upregulated genes were significantly clustered at subtelomeric regions compared to a random distribution (P < 0.05). Using Northern blot analysis, we verified that genes that are upregulated in clr3 or clr6-1 cells (12) are also upregulated in Δtor1 mutants (Fig. 1B). One of these genes, C186.05c, is located close (∼30 kbp) to the telomeric region (12).
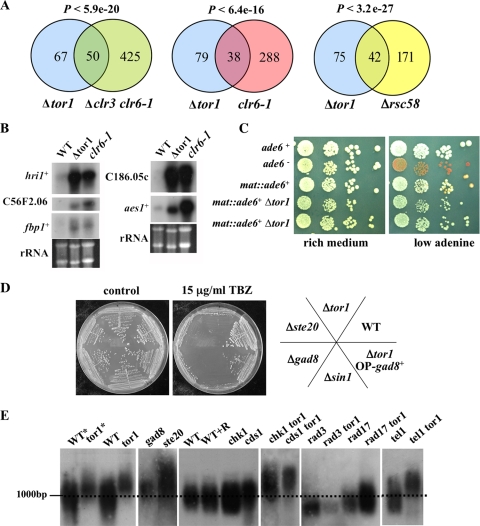
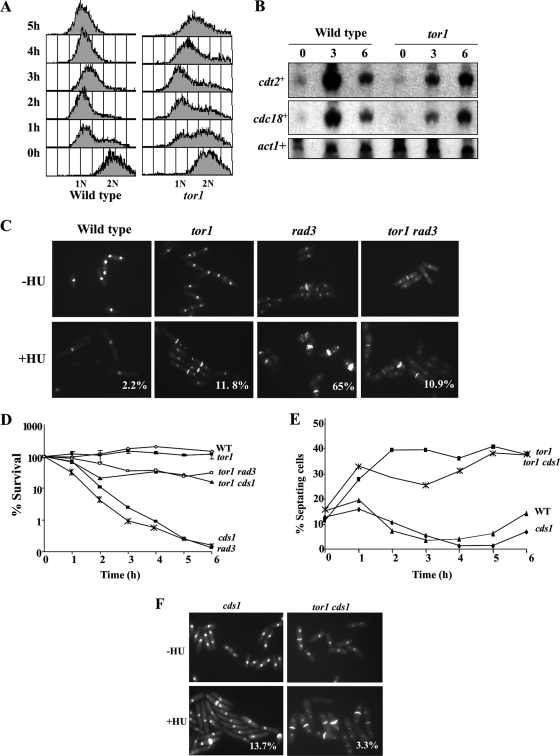
FIG. 1.
Tor1 is required for gene silencing and maintenance of telomere length. (A) The set of genes upregulated by loss of Tor1 significantly overlaps with the set of genes upregulated in histone deacetylase mutants. The number of genes that were upregulated 1.5-fold in the indicated mutants is presented in Venn diagrams, along with corresponding P values. (B) Northern blot analysis. Total RNA was prepared from wild-type (WT), Δtor1, and clr6-1 mutants grown to mid-log phase in rich medium. Northern blots were probed with the indicated genes. (C) Tor1 promotes silencing at the mating type locus. Strains containing an ade6+ cassette at the mating type locus were spotted onto the indicated plates. In an otherwise wild-type background, the ade6+ gene insertion produced a typical position variegation effect, since only a portion of the colonies is white (express the ade6+ gene) while others are red due to a decreased level of ade6+ transcript and accumulation of a red pigment. Only white colonies are present in cells carrying the Δtor1 mutation. (D) Tor1 is required for tolerance to microtubule destabilizing agents. Cells were streaked on plates containing the indicated levels of TBZ. (E) Tor1 is required for the maintenance of telomere length regulation. DNA was extracted from cells grown in rich medium (or minimal medium; asterisk). When rapamycin was added (R), the cells were grown in the presence of 100 ng/ml rapamycin. Genomic DNA was digested with EcoRI, which in wild-type cuts about 1 kb from the terminus, and analyzed by Southern blotting. The resulting filter was probed with α32-P-labeled telomere repeat DNA.
S. pombe contains heterochromatin in centromeric and telomeric regions and at the mating type locus. Since our microarray experiments suggested upregulation of genes at heterochromatic regions, we examined the expression of a reporter gene, ade6+, inserted at the mating type locus (2). We found that a loss of Tor1 relieved the repression of ade6+ inserted at the mating type locus (Fig. 1C), further supporting a role for Tor1 in chromatin-mediated gene silencing.
Inhibition of HDACs caused hyperacetylation at centromeres and defective chromosome segregation (52). Accordingly, clr6-1 mutants exhibit sensitivity to thiabendazole (TBZ), a drug that destabilizes microtubules and thus aggravates chromosome loss in strains with compromised centromeres (52). Similarly, we found that Δtor1 mutants are highly sensitive to TBZ (Fig. 1D), raising the possibility that tor1 mutants are also defective in accurate chromosome segregation.
Among the genes that were downregulated in Δtor1 mutants, we noted several transporters, including str1+, encoding a component of the iron-siderophore system. The transcription of str1+ is also downregulated in clr1, clr3, and clr4 mutants (12). The findings that transporters and stress-responsive genes are aberrantly expressed in clr mutants led Hansen et al. (12) to examine the sensitivity of clr mutants to osmotic stress. Indeed, the clr6-1 Δclr3 double mutant was highly sensitive to 1 M KCl (12), showing an osmotic sensitivity similar to that observed in Δtor1 mutants (59). Thus, Δtor1 mutants share with HDAC mutants their gene expression pattern, derepression of genes at heterochromatic regions, and sensitivity to TBZ and KCl.
Tor1 is required for telomere length maintenance.
Mutations in chromatin modifiers can affect telomere length (12). Thus, we examined telomere length in different TORC2 mutants. We found that telomeres of Δtor1, Δste20, or Δgad8 mutants were elongated by ∼150 bp compared to those of the wild type, similar to the elongation observed in clr6-1 mutants. In contrast, for wild-type cells grown in the presence of 100 ng/ml rapamycin, the length of telomeres was not affected (Fig. 1E). We conclude that TORC2-Gad8 regulates telomere length in a rapamycin-insensitive manner. Overexpression of Gad8 did not suppress telomere overelongation in Δtor1 mutants (data not shown). However, since Gad8 is a substrate for phosphorylation by Tor1, it is likely to be poorly active in Δtor1 mutants.
DNA checkpoint proteins play a central role in telomere maintenance. Mutations in Rad3, the primary DNA damage checkpoint kinase, or in any of the subunits of the heterotrimeric checkpoint clamp complex Rad9-Rad1-Hus1 (9-1-1) or its clamp loader Rad17 result in short telomeres (38). We found that the telomeres of the Δtor1 Δrad3 or Δtor1 Δrad17 double mutant are as short as those of single Δrad3 or Δrad17 mutants, respectively. Thus, Tor1 may induce telomere overelongation via Rad3 and Rad17 (Fig. 1E). Chk1 and Cds1, the downstream effectors of Rad3 in the DNA damage and DNA replication checkpoints, respectively, play little or no role in regulating telomere length (38). Consistently, deletion of tor1+ in either a Δchk1 or Δcds1 background resulted in a telomere elongation similar to that in single Δtor1 mutants (Fig. 1E). We also tested for involvement of Tel1, a PIKK kinase similar to ATM that works together with Rad3 to regulate telomere length (38). Telomere overelongation in Δtor1 mutants did not require the presence of Tel1 (Fig. 1E). We thus suggest that Tor1 acts in a Rad3-dependent pathway to maintain proper telomere length and this function is independent of Tel1.
TORC2 is required under DNA-damaging conditions.
Defects either in the RSC complex or in HDAC complexes can lead to sensitivity to DNA damage and replication stress conditions (26, 34, 39). We examined the sensitivity of Δtor1 mutants to the drug hydroxyurea (HU), which halts DNA replication by inhibiting nucleotide synthesis from the ribonucleotide reductase (4). Deletion of each of the genes encoding specific TORC2 components, Tor1, Ste20, or Sin1, or the downstream effector Gad8 resulted in strong sensitivity to HU (Fig. 2A). HU sensitivity in Δtor1 mutants has been observed previously (57), with no further analysis of the underlying mechanism.
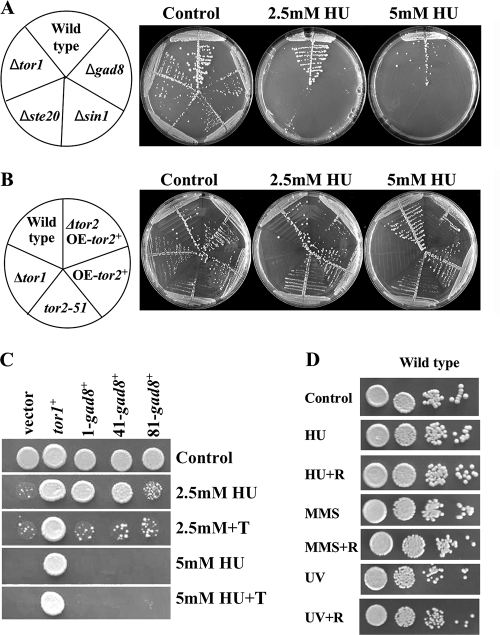
FIG. 2.
Mutations in TORC2 but not TORC1 confer sensitivity to DNA replication stress in a rapamycin-independent manner. (A and B) TORC2 but not TORC1 components are required for HU tolerance. Strains were streaked on plates with or without the indicated amounts of HU. (C) Overexpression of gad8+ partially rescues the lethal phenotype of Δtor1 cells on HU. gad8+ is expressed from the thiamine (T)-repressible nmt1+ promoter from the plasmids pREP1, -41, and -81, which allow strong, moderate, and weak expression, respectively. tor1+ is expressed from a plasmid under the regulation of its own promoter. (D) Rapamycin does not affect tolerance to DNA-damaging conditions. Serial dilutions of wild-type cells in the presence of 2.5 mM HU or 0.0025% MMS or UV irradiated at 75 J/m2 with or without 100 ng/ml rapamycin (R).
In contrast, reduction of Tor2 (TORC1) activity, overexpression of Tor2, or deletion of tsc1+ or tsc2+ did not markedly affect HU sensitivity (Fig. 2B and data not shown). Thus, it appears that cells carrying mutations in TORC2 but not TORC1 are sensitive to HU. Overexpression of gad8+ partially suppressed the HU sensitivity of Δtor1 mutants, further suggesting that Tor1 acts via Gad8 in tolerating replication stress (Fig. 2C).
We also found that Δtor1 cells were strongly sensitive to the DNA-alkylating agent MMS (Fig. 3B) and slightly sensitive to UV irradiation (data not shown). Rapamycin did not affect sensitivity to these drugs (Fig. 2D), indicating that the functions of TORC2 under DNA-damaging conditions are rapamycin insensitive.
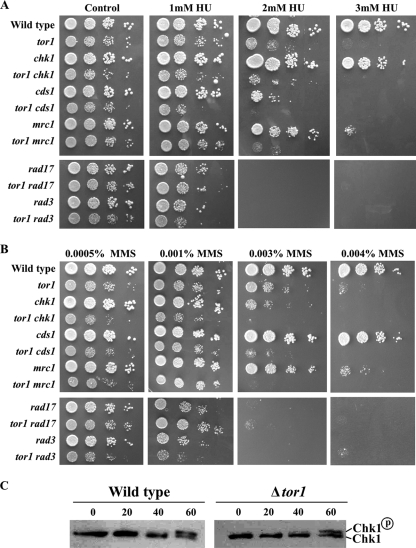
FIG. 3.
Mutations in TORC2 confer sensitivity to DNA-damaging conditions independently of Cds1 or Chk1. (A and B) Tor1 functions independently of Chk1 or Cds1. Serial dilutions of mutant cells were plated with or without the indicated amounts of HU or MMS. (C) Tor1 is not required for phosphorylation of Chk1. Western blot analysis of HA-tagged Chk1. Wild-type or Δtor1 cells containing HA-tagged Chk1 were grown to log phase. Protein was extracted from untreated cells or treated with 0.2% MMS for the indicated times (minutes).
Cells lacking Tor1 are almost as sensitive to HU or MMS as mutants lacking the main checkpoint kinase Rad3 or mutants lacking the RFC-like protein Rad17 (Fig. 3A and B). Combining the Δtor1 mutation with Δrad3 or Δrad17 did not result in further sensitivity to the DNA-damaging conditions (Fig. 3A and B). Thus, the function of Tor1 in the DNA damage response, as in telomere length control, may depend on the functions of Rad3 and Rad17. In fission yeast, the Rad3 kinase controls two checkpoint pathways: one responds to the DNA replication block, mainly through the Cds1 kinase (mammalian Chk2), while the other responds to DNA damage through activation of the Chk1 kinase (4). Cells lacking Tor1 exhibited HU sensitivity comparable to that of cells lacking Cds1, the main effector of the DNA replication stress response pathway (Fig. 3A). The sensitivity of Δtor1 mutants to HU was further augmented in combination with loss of function of either cds1+ or its specific mediator, mrc1+, encoding a Claspin homologue (50) (Fig. 3A). Thus, it appears that Tor1 acts in a Cds1-Mrc1-independent pathway.
Cells lacking Tor1 show sensitivity to MMS comparable to that of cells lacking Chk1, the main effector of the DNA damage response pathway (Fig. 3B). Yet the Δtor1 mutation showed additive effects with Δchk1 cells with respect to MMS sensitivity (Fig. 3A). Thus, Tor1 appears to act independently of Chk1. Consistently, Tor1 was not required for activation of Chk1 by phosphorylation in response to MMS treatment (Fig. 3C). Taken together, our genetic analysis is consistent with the possibility that Tor1 lies on the same pathway as Rad3 but acts independently of either Chk1 or Cds1 (see also below and our model in Fig. 6B).
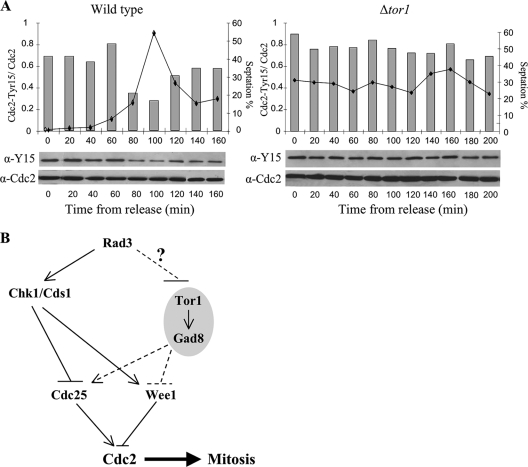
FIG. 6.
Tor1 is required for activation of Cdc2 after release from HU arrest. (A) Wild-type and Δtor1 cells were treated with 12 mM HU for 3.5 h, washed, and resuspended in fresh yeast extract. Samples from the indicated time points were taken for septation index measurement and Western blot analysis. (B) A working model. In response to DNA damage or DNA replication stress, Rad3 activates Chk1 or Cds1, respectively, leading to a delay in mitotic entry. In parallel, Rad3 keeps Tor1 inactive until DNA replication is completed. Regulation of Tor1 activity is not essential to prevent premature entry into mitosis but is required for reentry upon recovery from checkpoint arrest.
Aberrant response of Δtor1 cells to DNA replication stress induced by HU.
A nonsynchronized wild-type population of fission yeast cells contains mainly G2 cells. Addition of HU to such a population results in a doubling in cell number, since cells proceed through the first mitosis and then arrest in the subsequent S phase (8, 9). While HU induces a cell cycle arrest in wild-type cells, cellular growth continues, resulting in elongated cells (8, 9).
FACS analysis of Δtor1 cells indicated that cells accumulated with 1N DNA content in response to HU, although with delayed kinetics compared to wild-type cells (Fig. 4A). Note that the FACS analysis presented in Fig. 4A is of isolated nuclei. A “drift” of the DNA content toward a content of 1.5N DNA is observed at 4 to 5 h in HU in Δtor1 nuclei. The meaning of this drift is not clear. However, since Δtor1 cells maintain full viability following incubation for 4 to 5 h in HU (see below), we suggest that this “drift” reflects changes in the structure or size of Δtor1 nuclei rather than the inability of Δtor1 cells to properly arrest in G1.
FIG. 4.
Tor1 is required for a normal response to HU. (A and B) The response of tor1 mutants to HU is delayed. Wild-type and Δtor1 cells were grown to log phase and shifted to medium containing 12 mM HU. (A) Samples were taken every hour, and nuclei were isolated and subjected to FACS analysis. (B) Total RNA was prepared from samples taken at the indicated time points (hours) after a shift to 12 mM HU. Northern blots were probed with cdt2+ and cdc18+ (MBF targets) and with act1+ (loading control). (C) Loss of Tor1 rescues the mitotic catastrophe of Δrad3 mutants. Cells were incubated with or without 12 mM HU for 6 h at 30°C and then stained with DAPI and calcofluor to visualize nuclear DNA and septa, respectively. Percentages indicate abnormal mitosis, scoring for the “cut” phenotype in which the septum is formed despite the absence of chromosome replication. (D) The rapid loss of viability of the Δrad3 or Δcds1 mutant strain is rescued by Δtor1. Cells were grown to log phase and shifted to 12 mM HU for 6 h, and samples were taken every hour to determine cell viability by assessing plating efficiency on rich medium. WT, wild type. (E and F) Loss of Tor1 is epistatic over loss of Cds1. Strains were grown to log phase and shifted to 12 mM HU. The percentage of cells with septa was measured at the indicated times by staining with calcofluor and DAPI and visualized by fluorescence microscopy.
Consistent with the slower kinetics with which Δtor1 nuclei accumulated with 1N DNA content in response to HU, we detected a slow and reduced accumulation of Cdc10/MBF-dependent S-phase-specific transcripts (e.g., cdt2+ and cdc18+) (63) in Δtor1 mutants compared to results in wild-type cells (Fig. 4B). This finding could reflect either a defect in cell cycle progression in Δtor1 mutants or a more direct defect in activating the transcriptional response to HU (5-7, 46).
Although Δtor1 cells arrested with nuclei of 1N DNA content and showed induction of S-phase-specific transcripts, these cells did not show elongation in response to HU (Fig. 4C). Staining of the cells with 4′,6′-diamidino-2-phenylindole (DAPI) and calcofluor in order to view nuclei and septa, respectively, revealed that exposure of Δtor1 mutants to HU resulted in a ∼40% increase in the number of septated cells. In contrast, addition of HU to wild-type cells resulted in a sharp reduction in the number of cells containing a septum (Fig. 4C and E), as shown previously (8). The septated HU-arrested Δtor1 cells contained two condensed 1C nuclei (Fig. 4C) and maintained a high level of viability (Fig. 4D). These findings suggest that in response to HU, Δtor1 cells are arrested with 1C nuclei content but cytokinesis of the previous cell cycle is delayed. A similar delay in septation in the presence of HU has been reported for mutants lacking Liz1, a pantothenate transporter (37, 53); this delay results from an indirect effect of HU on pantothenate biosynthesis (53). Unlike the case with Δliz1 mutants, addition of pantothenate to the medium did not rescue the HU sensitivity of Δtor1 mutants (data not shown); thus, the aberrant response to HU in Δtor1 mutants occurs via a distinct mechanism. Importantly, however, our observation that Δtor1 cells are highly sensitive to MMS suggests that Δtor1 cells have a general defect in coping with DNA damage, rather than a specific defect concerning the response to HU.
The viability of Δtor1 mutants in response to a short exposure to HU is in sharp contrast to the rapid drop in viability observed in checkpoint-deficient Δrad3 or Δcds1 mutants (4). As previously described, Δrad3 mutants do not elongate but continue to divide in the presence of unreplicated DNA, leading to a lethal phenotype known as “cut.” This phenotype is characterized by anucleate cells or cells with <1C DNA (8, 9) and can be observed by staining both nuclei and septa (Fig. 4C). The response of the Δtor1 Δrad3 double mutant to HU was similar to that of single Δtor1 mutants, and very few cells with a “cut” phenotype were observed (Fig. 4C). Consistently, the Δtor1 mutation partially rescued the lethality of Δrad3 mutants in response to acute exposure to HU (Fig. 4D). Δcds1 mutants do not show the lethal “cut” phenotype in the presence of HU, yet they die rapidly in HU (4). The Δtor1 Δcds1 double mutants displayed phenotypes similar to those of single Δtor1 mutants (Fig. 4E and F), and like the interaction with Δrad3 mutants, Δtor1 partially rescued the rapid loss of viability of Δcds1 mutants in response to HU (Fig. 4D). Notably, Δtor1 rescued only the lethality of Δrad3 or Δcds1 upon short but not constant exposure to HU. We suggest that in the absence of Tor1, the death that occurs in the presence of HU in Δcds1 or Δrad3 cells is postponed due to slow progression during the first mitosis, before cells halt in early S phase. However, when cells eventually enter S phase, the Δtor1 mutation cannot rescue the lethal events that occur in Δcds1 or Δrad3 mutants.
Tor1 promotes mitotic entry via Cdc2.
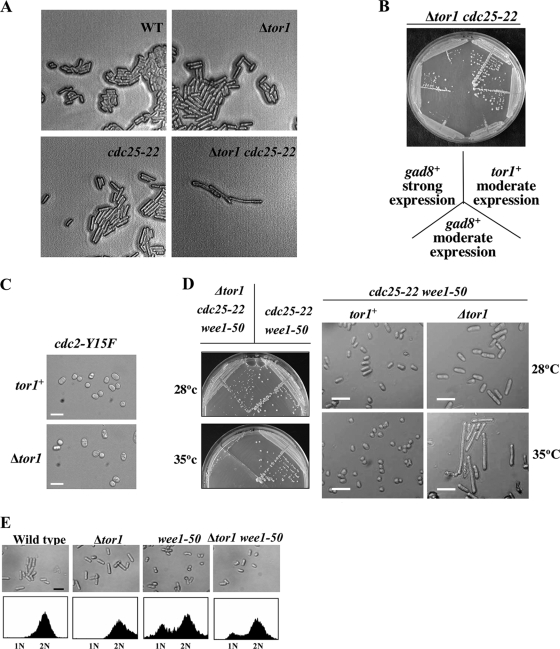
Disruption of tor1+ generates moderately elongated cells, indicative of a delay in entry into mitosis (59). Accordingly, we found that Δtor1 is synthetic lethal with the temperature-sensitive mutation in cdc25-22 (Fig. 5A). Cdc25 is a phosphatase that activates Cdc2, the cyclin-dependent kinase that controls mitotic entry (12). Overexpression of Gad8 partially rescued the synthetic lethality between Δtor1 and cdc25-22 (Fig. 5B), suggesting that Tor1 affects entrance into mitosis via Gad8. This is also in concert with recent studies that reported lethality between Δgad8 and cdc25-22 (16), supporting a positive role for TORC2-Gad8 in regulating mitotic entry.
FIG. 5.
Tor1 positively regulates mitosis. (A) The Δtor1 mutation is synthetic lethal with cdc25-22. A diploid strain heterozygous for Δtor1 and cdc25-22 was subjected to meiosis and tetrad analysis. Plates were incubated at 25°C. (B) Overexpression of gad8+ rescues the synthetic lethality of tor1 cdc25-22. The same diploid strain as above was transformed with pIRT2-tor1+, pREP41-gad8+ (moderate overexpression), and pREP1-gad8+ (strong overexpression). Two double mutant spores containing each of the plasmids were isolated and streaked onto plates at 28°C (no viable spores were obtained with an empty vector). (C) The cdc2-Y15F mutation suppresses the elongated phenotype of cells lacking Tor1. Bars: 20 μm. (D) The Δtor1 mutation reverses the suppression of cdc25-22 by wee1-50. Cells from the indicated genotypes were streaked onto plates either at 28°C or 35°C (left panel), and cells were visualized by light microscopy (right panel). Bars: 20 μm. (E) The wee1-50 mutation partially suppresses the elongated phenotype of cells lacking Tor1. Cells were grown to mid-log phase at 30°C, photographed, and subjected to FACS analysis.
Two major antagonistic branches, the Cdc25- and Wee1-dependent pathways, regulate the status of Cdc2 phosphorylation on its tyrosine-15 residue (36). The cdc2-Y15F mutation, expressing an unphosphorylatable and constitutively active form of Cdc2 (11), completely reversed the elongated morphology of Δtor1 mutants, and the Δtor1 cdc2-Y15F double mutant strain looked indistinguishable from the single cdc2-Y15F mutant (Fig. 5C). Thus, it appears that Tor1 controls entrance into mitosis by regulating the status of Cdc2 phosphorylation.
Introduction of the Δtor1 mutation into the genetic background of Δcdc25 cdc2-3w cells resulted in cell cycle elongation (Table 1), indicating that Tor1 can regulate cell size in the absence of Cdc25. However, Tor1 is also capable of affecting cell size in the absence of Wee1. Combining the Δtor1 mutation with the wee1-50 mutation resulted in a slight elongation of the “wee” (very short) phenotype (Table 1; Fig. 5E). Similarly, Δwee1 Δtor1 double mutants were slightly more elongated than single Δwee1 mutants (our unpublished observation). Cells lacking Wee1 show a G1 delay, since they are “born” at a cell size shorter than the threshold required for the G1-S transition (see reference 36). Our FACS analysis indicated that double mutant wee1-50 Δtor1 cells are also delayed in G1, albeit slightly less so than single wee1-50 mutants (Fig. 5E). In addition, we also found that the elongation of Δtor1 cells was highly augmented in combination with deletion of cdr2+, encoding a negative regulator of Wee1 (20) (Table 1), suggesting that Tor1 does not require Cdr2 for its cell cycle effect.
TABLE 1.
Effects of mutations on cell size
| Genotype | Temp (°C) | Mean cell length (μm)± SDa
|
|
|---|---|---|---|
| tor1+ | Δtor1 | ||
| Wild type | 30 | 15.1 ± 0.9 | 17.5 ± 2.3 |
| cdc25-22 | 28 | 20.4 ± 1.4 | >35 (synthetic lethal) |
| wee1-50 | 35 | 8.0 ± 1.3 | 8.9 ± 1.7 |
| cdc2-1w | 30 | 8.0 ± 1.6 | 11.2 ± 1.8 |
| cdc2-3w | 30 | 8.3 ± 1.7 | 12.6 ± 2.3 |
| Δnim1 | 30 | 15.1 ± 1.5 | 16.0 ± 1.6 |
| Δcdr2 | 30 | 17.2 ± 1.6 | >30 (synthetic sick) |
| Δcdc25 cdc2-3w | 30 | 14.0 ± 2/9 | 21.1 ± 1.8 |
| Δsty1 | 30 | 24.3 ± 2.9 | 19.0 ± 2.3 |
Cell length at division (n = 200). Cell lengths of double or triple mutant cells are presented in bold.
The elongation conferred by the Δtor1 mutation was suppressed by two different activated alleles of cdc2, cdc2-3w or cdc2-1w (Table 1), which are largely insensitive to Wee1 or Cdc25, respectively (45). This finding is consistent with the idea that Tor1 does not act solely via either the Wee1 or Cdc25 function.
The Δtor1 mutation caused lethality when combined with the genetic background of wee1-50 cdc25-22 and resulted in extreme cell size elongation at the restrictive temperature (Fig. 5D). The wee1-50 cdc25-22 double mutant represents a genetic background in which the activity of Cdc2 is poorly regulated since both negative and positive effectors are missing. Another mutation which reverses the suppression of cdc25-22 by the wee1-50 mutation is the deletion of the stress-activated mitogen-activated protein kinase Spc1/StyI (equivalent to p38 in mammalian cells). Moreover, deletion of spc1+/sty1+ or its upstream effector wis1+ resulted in a set of genetic interactions with cell cycle mutants highly similar to that recorded by us for Δtor1, including synthetic lethality with the cdc25-22 mutation (51, 58). It has previously been suggested that Spc1/Sty1 regulates Polo kinase (Plo1) via its phosphorylation and localization to the spindle pole body, which in turn affects the balance between the activities of Wee1 and Cdc25 and determines mitotic progression (30, 40). It is possible that Tor1 also acts by affecting both Wee1 and Cdc25 (see our model, Fig. 6B). Recently it was also demonstrated that Tor1 acts upstream of Spc1/Sty1 (41). Combining Δtor1 with Δspc1/sty1 resulted in an intermediate cell elongation compared to single Δtor1 or Δspc1/sty1 mutants, in concert with the possibility that Tor1 and Spc1/StyI act in the same pathway (Table 1; also see Discussion).
Tor1 is required for Cdc2 activation following recovery from HU-induced arrest.
In wild-type cells, Tyr15 phosphorylation on Cdc2 is required for the replication checkpoint arrest, and removal of the phosphate residue is critical to allow mitotic entry following recovery (43). We therefore examined the role of Tor1 in dephosphorylation of Cdc2-Tyr15 following release from HU arrest. To this aim, we incubated wild-type and Δtor1 cells in the presence of 12 mM HU for 3.5 h before release into fresh medium that does not contain HU. Following treatment with HU, both wild-type and Δtor1 cells arrested with highly phosphorylated Cdc2 on Tyr15 (Fig. 6A). In wild-type cells, dephosphorylation of Cdc2 occurred at 100 min following release from HU, consistent with results in previous studies (43), while in Δtor1 cells, Cdc2 remained phosphorylated on Tyr15 for at least 200 min following release from HU (Fig. 6A). We conclude that Tor1 is required for activation of Cdc2 by Tyr15 dephosphorylation following recovery from HU treatment.
DISCUSSION
Our data reveal novel and unexpected roles for TORC2 in regulating gene silencing, telomere length, and survival under DNA-damaging conditions. These TORC2-dependent functions are rapamycin insensitive and thus could easily be overlooked in studies of mammalian cells, which are largely based on the use of rapamycin as a specific inhibitor of TOR. Global gene expression analysis of Δtor1 mutants revealed an extensive overlap with expression signatures in mutants in histone deacetylase genes (clr3 and clr6) or in the gene encoding the RSC58 subunit of the RSC complex. Like these chromatin structure mutants, Δtor1 cells derepressed gene expression at heterochromatic regions, exhibited elongated telomeres, and were sensitive to osmotic stress, DNA damage, and the microtubule destabilizer TBZ. In budding yeast, TORC1 regulates the chromatin structure in a rapamycin-sensitive manner via Rpd3 (44, 55). Our data suggest that TORC2 may carry out a similar function, although the precise mechanism is yet to be determined.
A striking observation is that Δtor1 cells have longer telomeres than wild-type cells. It will clearly be important to determine whether TOR also affects telomere length in higher eukaryotes. Loss of Tor1 induced overelongation of telomeres in the Δtel1, Δchk1, or Δcds1 checkpoint mutant but not in cells lacking the ATR-like kinase Rad3. Thus, this is although highly speculative at present, Tor1 and Rad3 may work in the same pathway, regulating telomeres in an antagonistic manner. Elongated telomeres have also been observed in several chromatin defect mutants, including the clr6-1 mutant (12) and with loss of set1+, encoding the histone H3-K4 methyltransferase in fission yeast (19), raising the possibility of a mechanistic link between chromatin structure defects and telomere elongation. The TOR signaling pathway may provide a link between nutrient signaling and cellular processes that govern chromatin and telomere structures.
Like Δtor1 mutants, the Δste20 or Δsin1 mutant is also highly sensitive to TBZ and HU and shows highly elongated telomeres (Fig. 1E and 2A). Thus, it appears that TORC2 is the TOR complex required under DNA replication stress and for regulation of telomere length. Gad8 (equivalent to AKT/PKB1) acts downstream of Tor1 (TORC2) (16). Overexpression of Gad8 in the background of Δtor1 cells partially rescued HU or TBZ sensitivity (Fig. 1D and 2C). Thus, most of the newly identified functions of Tor1 (TORC2) appear to be mediated via Gad8. Cells lacking Gad8 also exhibited elongated telomeres, yet overexpression of gad8+ did not suppress telomere overelongation in Δtor1 mutants (data not shown). It is likely that Gad8 is not fully activated in the absence of Tor1 and thus cannot fully rescue defects associated with disruption of tor1+.
Unlike Clr6 and Clr3, Tor1 has also been strongly implicated in regulating cell cycle progression and the response to nitrogen starvation (22, 41, 59; this work). In addition, Δtor1 cells are far more sensitive to DNA-damaging conditions than the clr6 or clr3 mutant cells. For example, the growth of Δtor1 mutant cells is greatly inhibited at the concentrations of 2 mM HU or 0.003% MMS (Fig. 3A and B). In contrast, the growth of clr6-1 mutant cells is inhibited at the concentrations of 10 mM of HU or 0.01% of MMS (12; our unpublished data). We speculate that the cell cycle defects observed in Δtor1 mutants contribute to their sensitivity to DNA-damaging conditions.
How may Tor1 (TORC2) integrate its function in mitotic progression and the response to DNA-damaging conditions? The Rad3 kinase is a major DNA damage sensor that regulates cell cycle progression via activation of the Chk1 or Cds1 kinase in response to DNA damage or replication stress. Activated Chk1 or Cds1 inhibits mitotic entry by regulating Cdc25 and Wee1/Mik1 activity (4). Our data suggest that Tor1 is not required for arresting mitotic entry in the presence of DNA damage or replication stress. Indeed, Tor1 is required for mitotic progression, a function that seems critical upon removal of HU. Thus, if Tor1 acts downstream of Rad3, it would be expected that Rad3 negatively regulates Tor1, keeping Tor1 inactive till DNA replication or repair is completed (Fig. 6B). However, the connection between Rad3 and Tor1 is yet to be determined. Our genetic data showing that the sensitivity of Δtor1 cells to HU or MMS is augmented in combination with Δcds1 or Δchk1, respectively, suggest that Tor1 acts independently of either Cds1 or Chk1. Consistently, Chk1 is normally phosphorylated in response to DNA damage in the absence of Tor1 (Fig. 3C). Yet, like Chk1/Cds1, Tor1 affects mitotic progression via regulation of the phosphorylation status of Cdc2 at the tyrosine-15 residues, possibly by controlling the balance between Cdc25 and Wee1 activity, as depicted in our working model (Fig. 6B).
Our data indicate that Tor1 acts as a positive regulator of mitotic entry under normal growth conditions. Moreover, Tor1 is critical for dephosphorylation of Cdc2 Tyr-15 upon recovery from HU treatment, thus promoting reentry into mitosis and cellular proliferation. It has been reported that reducing the level of Tor1 induced entry into mitosis via regulating the Spc1/StyI pathway (41). Our results are consistent with a role of Tor1 in the same pathway as Spc1/StyI but argue that Tor1 is a positive regulator of mitosis. This apparent discrepancy may be explained by the use of different tor1 mutants in the two studies; while we used a complete disruption of tor1+, Petersen and Nurse (41) based their conclusions on cells expressing low levels of Tor1. Thus, the effect of Tor1 on mitotic entry may rely on its level of activity. Indeed, while we were revising the manuscript, it was reported that Tor1 can act as part of TORC1 in regulating entrance into mitosis (13). It is the inhibition of a Tor1-Mip1 (TORC1) complex that induces entrance into mitosis under poor nitrogen conditions (13). Thus, whether Tor1 acts as an inducer or inhibitor of mitosis may also rely on its partner proteins.
An intriguing question is how TORC2 may affect nuclear functions. TOR proteins seem to locate primarily in the cytoplasm (54) but have also been reported to shuttle into the nucleus in both mammalian (23) and budding yeast (25) cells. In growing fission yeast cells, Tor2 fused to green fluorescent protein localizes to the cytoplasm and to the perinuclear region, while no localization data exist for Tor1 (14). Thus, whether Tor1 affects nuclear functions directly or by controlling other regulators remains to be resolved in future experiments.
Finally, recent work (14, 21) demonstrated that Tel2, a fission yeast homologue of mammalian Clk2/Rad-2 required for the replication checkpoint, physically interacts with all PIKKs, suggesting a possible functional link among this family of proteins. Our study is consistent with this intriguing observation and argues that TORC2 is a regulator of survival under DNA damage conditions. Together, these findings place the TOR proteins alongside the other PIKKs, ATR and ATM, as regulators of nuclear processes and guardians of genome integrity and stability.
Supplementary Material
Acknowledgments
We thank P. Russell, A. M. Carr, P. Fantes, P. Young, J. Millar, M. Yamamoto, K. Shiozaki, S. Moreno, A. Cohen, O. Rog, J. Cooper, and the Yeast Genetic Resource Center, Japan, for strains and plasmids, F. Schubert for help with the Perl scripting, and members of the Kupiec laboratory for encouragement and support.
This research was supported by grants from the Israel Science Foundation (397/03-16.2), the Association for International Research (08-0070), and the Chief Scientist Office of the Ministry of Health, Israel, to R.W. and from Cancer Research UK to J.B. L.L.-M. is supported by a FEBS postdoctoral fellowship.
Footnotes
Published ahead of print on 22 June 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alvarez, B., and S. Moreno. 2006. Fission yeast Tor2 promotes cell growth and represses cell differentiation. J. Cell Sci. 1194475-4485. [DOI] [PubMed] [Google Scholar]
- 2.Ayoub, N., I. Goldshmidt, and A. Cohen. 1999. Position effect variegation at the mating-type locus of fission yeast: a cis-acting element inhibits covariegated expression of genes in the silent and expressed domains. Genetics 152495-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bickle, M., P. A. Delley, A. Schmidt, and M. N. Hall. 1998. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 172235-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carr, A. M. 2002. DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair (Amsterdam) 1983-994. [DOI] [PubMed] [Google Scholar]
- 5.Chu, Z., J. Li, M. Eshaghi, X. Peng, R. K. Karuturi, and J. Liu. 2007. Modulation of cell cycle-specific gene expressions at the onset of S phase arrest contributes to the robust DNA replication checkpoint response in fission yeast. Mol. Biol. Cell 181756-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Bruin, R. A., T. I. Kalashnikova, C. Chahwan, W. H. McDonald, J. Wohlschlegel, J. Yates III, P. Russell, and C. Wittenberg. 2006. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol. Cell 23483-496. [DOI] [PubMed] [Google Scholar]
- 7.Dutta, C., P. K. Patel, A. Rosebrock, A. Oliva, J. Leatherwood, and N. Rhind. 2008. The DNA replication checkpoint directly regulates MBF-dependent G1/S transcription. Mol. Cell. Biol. 285977-5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enoch, T., A. M. Carr, and P. Nurse. 1992. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 62035-2046. [DOI] [PubMed] [Google Scholar]
- 9.Enoch, T., and P. Nurse. 1990. Mutation of fission yeast cell cycle control genes abolishes dependence of mitosis on DNA replication. Cell 60665-673. [DOI] [PubMed] [Google Scholar]
- 10.Forsburg, S. L., and N. Rhind. 2006. Basic methods for fission yeast. Yeast 23173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gould, K. L., and P. Nurse. 1989. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature 34239-45. [DOI] [PubMed] [Google Scholar]
- 12.Hansen, K. R., G. Burns, J. Mata, T. A. Volpe, R. A. Martienssen, J. Bahler, and G. Thon. 2005. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol. Cell. Biol. 25590-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartmuth, S., and J. Petersen. 2009. Fission yeast Tor1 functions as part of TORC1 to control mitotic entry through the stress MAPK pathway following nutrient stress. J. Cell Sci. 1221737-1746. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi, T., M. Hatanaka, K. Nagao, Y. Nakaseko, J. Kanoh, A. Kokubu, M. Ebe, and M. Yanagida. 2007. Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells 121357-1370. [DOI] [PubMed] [Google Scholar]
- 15.Helliwell, S. B., A. Schmidt, Y. Ohya, and M. N. Hall. 1998. The Rho1 effector Pkc1, but not Bni1, mediates signalling from Tor2 to the actin cytoskeleton. Curr. Biol. 81211-1214. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda, K., S. Morigasaki, H. Tatebe, F. Tamanoi, and K. Shiozaki. 2008. Fission yeast TOR complex 2 activates the AGC-family Gad8 kinase essential for stress resistance and cell cycle control. Cell Cycle 7358-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacinto, E., R. Loewith, A. Schmidt, S. Lin, M. A. Ruegg, A. Hall, and M. N. Hall. 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 61122-1128. [DOI] [PubMed] [Google Scholar]
- 18.Jacinto, E., and A. Lorberg. 2008. TOR regulation of AGC kinases in yeast and mammals. Biochem. J. 41019-37. [DOI] [PubMed] [Google Scholar]
- 19.Kanoh, J., S. Francesconi, A. Collura, V. Schramke, F. Ishikawa, G. Baldacci, and V. Geli. 2003. The fission yeast spSet1p is a histone H3-K4 methyltransferase that functions in telomere maintenance and DNA repair in an ATM kinase Rad3-dependent pathway. J. Mol. Biol. 3261081-1094. [DOI] [PubMed] [Google Scholar]
- 20.Kanoh, J., and P. Russell. 1998. The protein kinase Cdr2, related to Nim1/Cdr1 mitotic inducer, regulates the onset of mitosis in fission yeast. Mol. Biol. Cell 93321-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanoh, J., and M. Yanagida. 2007. Tel2: a common partner of PIK-related kinases and a link between DNA checkpoint and nutritional response? Genes Cells 121301-1304. [DOI] [PubMed] [Google Scholar]
- 22.Kawai, M., A. Nakashima, M. Ueno, T. Ushimaru, K. Aiba, H. Doi, and M. Uritani. 2001. Fission yeast tor1 functions in response to various stresses including nitrogen starvation, high osmolarity, and high temperature. Curr. Genet. 39166-174. [DOI] [PubMed] [Google Scholar]
- 23.Kim, J. E., and J. Chen. 2000. Cytoplasmic-nuclear shuttling of FKBP12-rapamycin-associated protein is involved in rapamycin-sensitive signaling and translation initiation. Proc. Natl. Acad. Sci. USA 9714340-14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, C. H., K. Inoki, M. Karbowniczek, E. Petroulakis, N. Sonenberg, E. P. Henske, and K. L. Guan. 2007. Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J. 264812-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, H., C. K. Tsang, M. Watkins, P. G. Bertram, and X. F. Zheng. 2006. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 4421058-1061. [DOI] [PubMed] [Google Scholar]
- 26.Liang, B., J. Qiu, K. Ratnakumar, and B. C. Laurent. 2007. RSC functions as an early double-strand-break sensor in the cell's response to DNA damage. Curr. Biol. 171432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M. N. Hall. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10457-468. [DOI] [PubMed] [Google Scholar]
- 28.Lorberg, A., and M. N. Hall. 2004. TOR: the first 10 years. Curr. Top. Microbiol. Immunol. 2791-18. [DOI] [PubMed] [Google Scholar]
- 29.Lyne, R., G. Burns, J. Mata, C. J. Penkett, G. Rustici, D. Chen, C. Langford, D. Vetrie, and J. Bahler. 2003. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacIver, F. H., K. Tanaka, A. M. Robertson, and I. M. Hagan. 2003. Physical and functional interactions between polo kinase and the spindle pole component Cut12 regulate mitotic commitment in S. pombe. Genes Dev. 171507-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mata, J., R. Lyne, G. Burns, and J. Bahler. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32143-147. [DOI] [PubMed] [Google Scholar]
- 32.Matsuo, T., Y. Otsubo, J. Urano, F. Tamanoi, and M. Yamamoto. 2007. Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol. Cell. Biol. 273154-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, K. M., O. Rog, and J. P. Cooper. 2006. Semi-conservative DNA replication through telomeres requires Taz1. Nature 440824-828. [DOI] [PubMed] [Google Scholar]
- 34.Monahan, B. J., J. Villen, S. Marguerat, J. Bahler, S. P. Gygi, and F. Winston. 2008. Fission yeast SWI/SNF and RSC complexes show compositional and functional differences from budding yeast. Nat. Struct. Mol. Biol. 15873-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194795-823. [DOI] [PubMed] [Google Scholar]
- 36.Moser, B. A., and P. Russell. 2000. Cell cycle regulation in Schizosaccharomyces pombe. Curr. Opin. Microbiol. 3631-636. [DOI] [PubMed] [Google Scholar]
- 37.Moynihan, E. B., and T. Enoch. 1999. Liz1p, a novel fission yeast membrane protein, is required for normal cell division when ribonucleotide reductase is inhibited. Mol. Biol. Cell 10245-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura, T. M., B. A. Moser, and P. Russell. 2002. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics 1611437-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicolas, E., T. Yamada, H. P. Cam, P. C. Fitzgerald, R. Kobayashi, and S. I. Grewal. 2007. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat. Struct. Mol. Biol. 14372-380. [DOI] [PubMed] [Google Scholar]
- 40.Petersen, J., and I. M. Hagan. 2005. Polo kinase links the stress pathway to cell cycle control and tip growth in fission yeast. Nature 435507-512. [DOI] [PubMed] [Google Scholar]
- 41.Petersen, J., and P. Nurse. 2007. TOR signalling regulates mitotic commitment through the stress MAP kinase pathway and the Polo and Cdc2 kinases. Nat. Cell Biol. 91263-1272. [DOI] [PubMed] [Google Scholar]
- 42.Reiling, J. H., and D. M. Sabatini. 2006. Stress and mTORture signaling. Oncogene 256373-6383. [DOI] [PubMed] [Google Scholar]
- 43.Rhind, N., and P. Russell. 1998. Tyrosine phosphorylation of cdc2 is required for the replication checkpoint in Schizosaccharomyces pombe. Mol. Cell. Biol. 183782-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rohde, J., J. Heitman, and M. E. Cardenas. 2001. The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 2769583-9586. [DOI] [PubMed] [Google Scholar]
- 45.Russell, P., and P. Nurse. 1987. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell 49559-567. [DOI] [PubMed] [Google Scholar]
- 46.Rustici, G., J. Mata, K. Kivinen, P. Lio, C. J. Penkett, G. Burns, J. Hayles, A. Brazma, P. Nurse, and J. Bahler. 2004. Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 36809-817. [DOI] [PubMed] [Google Scholar]
- 47.Sarbassov, D. D., S. M. Ali, D. H. Kim, D. A. Guertin, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 141296-1302. [DOI] [PubMed] [Google Scholar]
- 48.Sarbassov, D. D., S. M. Ali, S. Sengupta, J. H. Sheen, P. P. Hsu, A. F. Bagley, A. L. Markhard, and D. M. Sabatini. 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22159-168. [DOI] [PubMed] [Google Scholar]
- 49.Shen, C., C. S. Lancaster, B. Shi, H. Guo, P. Thimmaiah, and M. A. Bjornsti. 2007. TOR signaling is a determinant of cell survival in response to DNA damage. Mol. Cell. Biol. 277007-7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shikata, M., F. Ishikawa, and J. Kanoh. 2007. Tel2 is required for activation of the Mrc1-mediated replication checkpoint. J. Biol. Chem. 2825346-5355. [DOI] [PubMed] [Google Scholar]
- 51.Shiozaki, K., and P. Russell. 1995. Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature 378739-743. [DOI] [PubMed] [Google Scholar]
- 52.Silverstein, R. A., W. Richardson, H. Levin, R. Allshire, and K. Ekwall. 2003. A new role for the transcriptional corepressor SIN3; regulation of centromeres. Curr. Biol. 1368-72. [DOI] [PubMed] [Google Scholar]
- 53.Stolz, J., T. Caspari, A. M. Carr, and N. Sauer. 2004. Cell division defects of Schizosaccharomyces pombe liz1− mutants are caused by defects in pantothenate uptake. Eukaryot. Cell 3406-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sturgill, T. W., A. Cohen, M. Diefenbacher, M. Trautwein, D. Martin, and M. N. Hall. 2008. TOR1 and TOR2 have distinct locations in live cells. Eukaryot. Cell. 71819-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsang, C. K., P. G. Bertram, W. Ai, R. Drenan, and X. F. Zheng. 2003. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 226045-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Urano, J., T. Sato, T. Matsuo, Y. Otsubo, M. Yamamoto, and F. Tamanoi. 2007. Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc. Natl. Acad. Sci. USA 1043514-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uritani, M., H. Hidaka, Y. Hotta, M. Ueno, T. Ushimaru, and T. Toda. 2006. Fission yeast Tor2 links nitrogen signals to cell proliferation and acts downstream of the Rheb GTPase. Genes Cells 111367-1379. [DOI] [PubMed] [Google Scholar]
- 58.Warbrick, E., and P. A. Fantes. 1991. The wis1 protein kinase is a dosage-dependent regulator of mitosis in Schizosaccharomyces pombe. EMBO J. 104291-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weisman, R., and M. Choder. 2001. The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J. Biol. Chem. 2767027-7032. [DOI] [PubMed] [Google Scholar]
- 60.Weisman, R., M. Choder, and Y. Koltin. 1997. Rapamycin specifically interferes with the developmental response of fission yeast to starvation. J. Bacteriol. 1796325-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weisman, R., I. Roitburg, T. Nahari, and M. Kupiec. 2005. Regulation of leucine uptake by tor1+ in Schizosaccharomyces pombe is sensitive to rapamycin. Genetics 169539-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weisman, R., I. Roitburg, M. Schonbrun, R. Harari, and M. Kupiec. 2007. Opposite effects of tor1 and tor2 on nitrogen starvation responses in fission yeast. Genetics 1751153-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White, S., F. Khaliq, S. Sotiriou, and C. J. McInerny. 2001. The role of DSC1 components cdc10+, rep1+ and rep2+ in MCB gene transcription at the mitotic G1-S boundary in fission yeast. Curr. Genet. 40251-259. [DOI] [PubMed] [Google Scholar]
- 64.Wullschleger, S., R. Loewith, and M. N. Hall. 2006. TOR signaling in growth and metabolism. Cell 124471-484. [DOI] [PubMed] [Google Scholar]
- 65.Yang, Q., and K. L. Guan. 2007. Expanding mTOR signaling. Cell Res. 17666-681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.