Abstract
It has been known since the early 1970s that nuclear receptor complexes bind DNA in association with coregulatory proteins. Characterization of these nuclear receptor coregulators has revealed diverse enzymatic activities that temporally and spatially coordinate nuclear receptor activity within the context of local chromatin in response to diverse hormone signals. Chromatin-modifying proteins, which dictate the higher-order chromatin structure in which DNA is packaged, in turn orchestrate orderly recruitment of nuclear receptor complexes. Modifications of histones include acetylation, methylation, phosphorylation, ubiquitylation, sumoylation, ADP ribosylation, deimination, and proline isomerization. At this time, we understand how a subset of these modifications regulates nuclear receptor signaling. However, the effects, particularly of acetylation and demethylation, are profound. The finding that nuclear receptors are directly acetylated and that acetylation in turn directly regulates contact-independent growth has broad therapeutic implications. Studies over the past 7 yr have led to the understanding that nuclear receptor acetylation is a conserved function, regulating diverse nuclear receptor activity. Furthermore, we now know that acetylation of multiple and distinct substrates within nuclear receptor signaling pathways, form an acetylation signaling network from the cell surface to the nucleus. The finding that nicotinamide adenine dinucleotide (NAD)-dependent histone deacetylases, the sirtuins, are capable of deacetylating nuclear receptors provides a new level of complexity in the control of nuclear receptor activity in which local intracellular concentrations of NAD may regulate nuclear receptor physiology.
IN THE EARLY 1970s, initial attempts at purifying nuclear receptors were confounded by the large number of coassociated proteins. The O’Malley laboratory had characterized the nuclear progesterone receptor/DNA complex and the thyroid hormone receptor associated with a heterogeneous group of proteins that was regulated in a ligand-dependent manner (2,3). It was apparent that transcription factors contained transactivation domains that functioned as modular surfaces to regulate transcription independently of direct binding to DNA (4). The laboratory of Tjian and others (5) characterized the TATA box binding protein-associated factors termed TAFs. Several cell-type-specific activities were characterized and shown to regulate transcription factor activity. In this regard, a B cell-specific activity designated Oct coactivator from B cells (OCA-B) regulated Oct-dependent B-cell-specific transcription (6).
Cross-squelching experiments by the Chambon laboratory (7) suggested distinct classes of transcriptional activation domains existed within nuclear receptors. Consistent with the notion that nuclear receptors were capable of repressing transcription, formal evidence that nuclear receptors contain specific repression domains was provided by studies of the progesterone receptor and retinoic acid receptor (8,9). These studies provided the rational basis for the identification of proteins mediating transcriptional activation and repression of nuclear receptors. Yamamoto and colleagues (10) identified the SWI protein as a key activator of the glucocorticoid receptor in yeast. In 1994, cAMP response element-binding protein-binding protein (CBP) was cloned as a coactivator of cAMP response element-binding protein (CREB) (11) and p300 as an E1A-interacting protein (12,13). Of fundamental importance was the identification of histone acetyltransferase enzymatic activity within the p300 activation domain. These proteins were shown to function as rate-limiting coactivators of nuclear receptor activity partially dependent upon their intrinsic histone acetyltransferase activity.
A dynamic and rapidly evolving field has characterized diverse types of enzymes (14). Furthermore, the assembly of these enzymes was shown to be temporally coordinated. The histone acetyltransferase, p300, enhanced the efficiency of transcriptional initiation from an estrogen-regulated template assembled within chromatin. The reassembly of active complexes during subsequent rounds of reinitiation did not require p300 (14). Indeed, consistent with these findings, chromatin immunoprecipitation experiments identified temporarily coordinated multiprotein complexes associated with estrogen receptor-α (ERα) and with endogenous ERα DNA-binding sites. These studies showed coactivators were recruited in a cyclical manner in association with local chromatin. p300 was recruited to the promoter region of the ERα-responsive genes in the first phase of ERα binding but not in subsequent cycles of ERα recruitment (15).
NUCLEAR RECEPTOR ACETYLATION GOVERNS CELLULAR GROWTH POTENTIAL
Histone acetyltransferases have been shown to acetylate diverse substrates. The first evidence that nuclear receptors served as direct substrates for histone acetyltransferases were studies by Fu et al. (16). The residues of androgen receptor (AR) acetylated by p300 in vitro were conserved between species. Point substitution mutations of the acetylation sites identified in vitro regulated ligand-dependent transactivation. Subsequent studies demonstrated that the nuclear receptor acetylation site is conserved between a subset of nuclear receptors, including the ERα, thyroid hormone receptor-β (17), progesterone receptor, and the glucocorticoid receptor (18). With each of the nuclear receptors characterized to date, the acetylation sites regulate a subset of nuclear receptor functions with the AR currently being the best characterized. The addition of ligand, dihydrotestosterone, or other agonists such as bombesin enhances AR acetylation (19). When reintroduced into AR-deficient human prostate cancer cells, gain of function point substitution of the AR acetylation site resulted in receptors that promote prostate tumor growth, both in vitro and in vivo (20). Characterization of the mechanism by which the AR acetylation site regulated contact-independent growth, indicated both enhancement of cellular proliferation and a reduction in cellular apoptosis (20,21). The charge of lysine residues in the AR acetylation site regulated recruitment of p300 and, in a reciprocal manner, disengagement of corepressor complexes including nuclear receptor corepressor (NCoR), histone deacetylase (HDAC), and mothers against decapentaplegic homolog 3 (20).
The AR acetylation site regulates prostate cellular apoptosis. The Jun kinase kinase signaling pathway (MAPK kinase kinase 1) induces cellular apoptosis in the presence of the AR. The AR acetylation site governs TRAIL-induced cellular apoptosis (21). Cells transduced with AR acetylation site gain-of-function mutants enhanced proliferation, associated with the induction of cell cycle markers such as Ki67 and cyclin D1. The relative abundance of cyclin D1 was increased in cells transduced with the AR activating mutants.
Chromatin immunoprecipitation analysis demonstrated that in addition to the enhanced recruitment of the AR to an androgen-responsive element binding site, the HDAC inhibitor, trichostatin A (TSA), enhanced AR recruitment in an AR acetylation site-dependent manner (22). AR acetylation site dead mutants showed reduced recruitment to the PSA promoter androgen-responsive element and reduced TSA-dependent recruitment. This suggests that the acetylation site of the nuclear receptor regulated access in the context of local chromatin (22).
The ERα was shown to be acetylated at the same motif as the AR (23). Deletion analysis identified the primary site acetylated by p300, and a minimal peptide sufficient to serve as a substrate for p300 was characterized. The peptide encoding the ERα acetylation site conveyed similar substrate specificity as histone H3. Proteomic analysis including liquid chromatography/mass spectrometry and Edman degradation sequencing, demonstrated K302 and K303 were preferential sites of acetylation. Lys K303 is frequently mutated in human breast cancer (24). The ERα acetylation mutants function as hyperactive mutants in the presence of low concentrations on its ligand estradiol (23,24). The ERα coactivator steroid receptor coactivator 1 (SRC1) is acetylated, and point mutation of the SRC1 acetylation site results in an activator of thyroid and retinoic acid receptor (ACTR) hyperactive mutant that fails to be transcriptionally down-regulated (25). As mutation of the ERα and the ERα coactivator ACTR acetylation site, enhances ligand-dependent acetylation may serve to repress hormone signaling by recruiting corepressor activity as shown for peroxisome proliferator-activated receptor-γ (PPARγ) coactivator-1α (PGC1α) (26) or through docking other as-yet-to-be-defined complexes. Consistent with this model, ERα repression by BRCA1 (27,28) is abolished by mutation of the ERα K302/303 acetylation site (Pestell, R. G., and C. Wang, unpublished data). Like PGC1α and p53, in which multiple acetylation sites have been identified (29), ERα has recently been shown to be acetylated at the additional residue K266/K268 (30). In these studies, SRC1/p300 was used as the source of histone acetyltransferase. The relative importance of the K302/303 vs. the K266/K268 site in the diverse physiological roles of the ERα remains to be determined.
A body of evidence has contributed to the current understanding that PGC1α, a transcriptional coactivator of nuclear receptors, is acetylated, and the acetylation of PGC1α regulates cellular metabolism. The metabolic coactivator PGC1α modulates the gluconeogenic pathway in fasting and diabetic states through interaction with several transcription factors (31,32). PGC1α forms part of a multiprotein complex that includes the tartrate-resistant acid phosphatase (TRAP)/mediator complex, including TRAP220 binding to its C terminus (33). The endogenous PGC1α complex includes the histone acetyltransferase general control of amino acid synthesis 5 (GCN5). p300 and SRC1 bind the N-terminal activation domain of PGC1α (34). PGC1α is directly acetylated by GCN5, resulting in a transcriptionally inactive protein that relocalizes from promoter regions to nuclear foci (26).
ACETYLATION OF COMPONENTS WITHIN HORMONE SIGNALING PATHWAYS
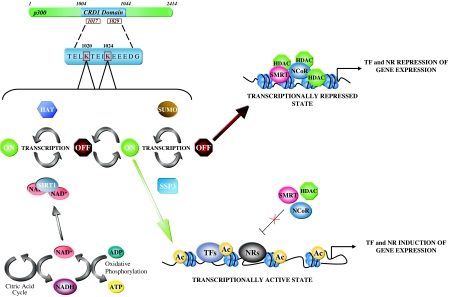
The addition of liganded nuclear receptor to a cell induces multiple downstream signaling cascades. A growing body of evidence has demonstrated that several additional components that regulate nuclear receptor signaling pathways are themselves substrates for histone acetyltransferases (Fig. 1). Additional components of the nuclear receptor signaling cascade that are directly acetylated include MAPK kinase kinase 1, heat-shock protein 90 (Hsp90) (35,36), and IκB kinase (37). The acetylation of these nuclear receptor-binding proteins, in turn, affects nuclear receptor function. Thus, for example, Hsp90-mediated glucocorticoid receptor maturation is regulated by acetylation of Hsp90 (36). Within the nucleus, components of the nuclear receptor coactivator or corepressor complexes are themselves substrates for histone acetyltransferases. Thus, ACTR and GCN5 are histone acetylases, and ACTR and PGC1α serve as substrates for histone acetyltransferases (25,26). Other transcription factors known to work in trans to regulate nuclear receptor signaling, such as the forked protein, are acetylated, providing an additional level of complexity for cross-talk between forkhead homolog in rhabdomyosarcoma (FKHR) and ERα signaling (38).
Figure 1.
The Histone Acetyltransferase (HAT) Protein, p300, Is Capable of Regulating Gene Expression through Transcription Factors (TFs) and Nuclear Receptors (NRs) Binding to DNA-Specific Sequences within Euchromatin
This effect is governed by K1020 and K1024 acetylation within p300’s cell cycle regulatory domain (CRD1). SIRT1, the function of which is induced by NAD+ produced as a byproduct of oxidative phosphorylation, serves as the master regulatory protein in this process. Ac, Acetylase; SMRT, silencing mediator of retinoid and thyroid receptor; SUMO, small ubiquitin-related modifier.
THE NICOTINAMIDE ADENINE DINUCLEOTIDE (NAD)-DEPENDENT HISTONE DEACETYLASES REGULATE HORMONE SIGNALING
The HDACs are divided into three groups. The class 1 and 2 HDACs share homology to the yeast Rpd3p and Hda1p proteins. The class 3 HDACs are homologs to the yeast transcriptional repressor Sir2p. The class 3 HDACs are NAD dependent. Mammalian sirtuins are conserved with seven genes homologous to the yeast Sir2 gene. The nuclear sirtuins (SIRT1, SIRT6, and SIRT7), the mitochondrial sirtuin (SIRT3, SIRT4, and SIRT5) and cytosolic sirtuin (SIRT2) regulate diverse metabolic functions. The distinct substrates and subcellular localization of the mammalian sirtuins suggests significant diversity of function (39,40).
SIRT1 has been characterized in its interactions with nuclear receptors and nuclear receptor coactivators. The functional interactions between SIRT1 and nuclear receptor signaling has been examined for the cointegrators p300 (41) and PGC1α (42,43,44) and for the nuclear receptor AR (45), PPARγ (46), and ERα (47). SIRT1 has been shown to deacetylate coactivators PGC1α and p300 and the nuclear receptors ERα and AR. SIRT1 regulates repression of the coactivator p300, providing a mechanism by which the NAD-dependent HAT deacetylases may regulate diverse nuclear receptor functions (41). The p300 coactivator functions as a rate-limiting protein in regulating diverse nuclear receptor functions. A direct link between the NAD-dependent HDACs and the histone acetyltransferases was first demonstrated by Bouras et al. (41) (Fig. 1). SIRT1 repressed p300 in an NAD-dependent manner. The lysine residues within the cell cycle regulatory domain (CRD1) of p300 were essential for SIRT1-mediated repression. Importantly, Bouras et al. (41) demonstrated that lysine residues within p300 function as substrates for SIRT2. SIRT1 also functions as a substrate for sumoylation (41). SUMO-1-specific protease 3 antagonized SIRT1-mediated repression of p300. Importantly these studies suggest a link between local metabolic changes that are anticipated to alter the NAD/NADH ratio and p300 activity. Because sirtuins are regulated by NAD, endogenous levels of nicotinamide may limit SIRT1 function and thereby alter activity of many transcription factors and nuclear receptors that are regulated by p300.
Sirtuins have well characterized functions in regulating responses to caloric restriction and glucose metabolism, cellular growth, and DNA repair. SIRT1 regulates gluconeogenesis by promoting the repression of glycolytic gene function (40). Importantly, recent studies have linked SIRT1-dependent deacetylation of the nuclear receptor coactivator PGC1α to glucose homeostasis. PGC1α is acetylated in vivo (43). Acetylation of PGC1α was augmented by treatment with the SIRT1 inhibitor nicotinamide or by expression of the transcriptional coactivator p300 (28). The finding that SIRT1 associates with PGC1α (28) led to studies examining the biological significance of PGC1α acetylation. Fasting was shown to induce PGC1α deacetylation in skeletal muscle. This important study, identifying SIRT1 as a functional regulator of PGC1α to induce mitochondrial fatty acid oxidation in vivo, has broad implications for the understanding of adaptations to altered availability of nutrients. SIRT1 is increased in the fasted liver, thereby deacetylating and activating PGC1α, enhancing hepatic glucose output (43).
These studies imply that PGC1α abundance may in turn govern metabolic diseases such as obesity and diabetes. Recent studies using resveratrol, a known activator of SIRT1 (48), demonstrated an increased aerobic capacity and induction of oxidative phosphorylation and mitochondrial biogenesis, primarily mediated by resveratrol-mediated inhibition of PGC1α acetylation and an increase in PGC1α activity (49). Resveratrol was also shown to oppose the effects of a high-calorie diet producing changes associated with longer lifespan (50). Caloric restriction enhances SIRT1 binding to and repression of PPARγ-responsive gene promoters. SIRT1 repressed PPARγ activity through binding the corepressors NCoR and silencing mediator of retinoid and thyroid receptor (46). Consistent with the physiological significance of SIRT1-mediated repression of PPARγ activity, SIRT1+/− mice demonstrated defective mobilization of fatty acids from white adipocytes upon fasting (46). Whether PPARγ or components within a PPARγ complex are direct substrates of acetylation remains to be determined. Collectively, these studies demonstrate the important physiological role for PGC1α acetylation and a key role for the NAD-dependent HDAC SIRT1 in regulating fat metabolism.
Recent studies have provided indirect evidence for an additional potential functional interaction between nuclear receptors and the sirtuins. Studies from the laboratories of Rosenfeld and Glass (51) demonstrated the induction of transient double-stranded DNA breaks upon the addition of estradiol, and the recruitment of ERα, poly-(ADP-ribose) polymerase 1 (PARP-1), and DNA topoisomerase IIβ. The BRCA1 gene, which plays an essential role in DNA repair, is known to bind the ERα in the context of local chromatin in a temporally coordinated manner (28). Although the role of ERα acetylation in recruitment to double-stranded DNA breaks is unknown, sirtuins, in particular SIRT6, regulate double-stranded DNA break repair. SIRT6 plays a key role in resistance to DNA damage and suppression of genomic instability (52). The fibroblasts of SIRT1−/− mice showed enhanced sensitivity to DNA-damaging agents. Although the sirtuins and the ERα have both been independently implicated in double-stranded DNA break repair, it remains to be determined whether sirtuins regulate this nuclear receptor function.
The use of SIRT1 inhibitors has strongly implicated endogenous SIRT1 in regulating AR activity in prostate cancer cells (37). SIRT1 colocalizes with the AR in nuclear subdomains (37). SIRT1 inhibits cellular proliferation in an AR-dependent manner. SIRT1 antagonists induce endogenous AR expression and enhance dihydrotestosterone-mediated AR-dependent gene expression. SIRT1 physically binds to and deacetylates the AR at a conserved lysine motif. The AR served as a substrate for the SIRT1 orthologs of several species including the mouse mSir2a, hSirt1, and two other enzymes, Cerevisiae (SIR2p) and Archaeoglobus fulgidus (Sir2af2). Each of these enzymes was examined for deacetylation activity against a synthetic AR peptide known to be acetylated by p300. The deacetylation of the AR by the sirtuins of several species was characterized by liquid chromatography/mass spectrometry and tandem mass spectrometry fragmentation analysis. Thus, the AR is deacetylated by SIRT1 to inhibit ligand-dependent AR activity to thereby inhibit dihydrotestosterone-dependent prostate cancer cellular proliferation.
What might be the physiological role of sirtuin-dependent regulation of nuclear receptor function? Sirtuin expression and activity is itself under complex control and may impact nuclear receptor function. The importance of NAD in Sir2 activity implies Sir2 activity may be regulated by intracellular NAD, by the NAD/NADH ratio, or nicotinamide (53,54). Elevated lactate, which reduces the NAD/NADH ratio, is predicted to inhibit SIRT1 (55). Inhibition of SIRT1 may enhance AR function (45) with consequent anabolic repair effects within muscle (56,57). During prostate cancer progression, a shift toward cytosolic glycolysis occurs (58,59). Increased local lactate concentration correlates with prostate cancer progression (1,60), which may be predicted to inhibit sirtuin activity and activate the AR (45). Sirtuins are located in diverse subcellular locations, and it will be of interest to determine whether receptors in nonnuclear compartments are regulated by sirtuins.
CONCLUSION
Nuclear receptors serve as substrates for sirtuins and play a role in glucose metabolism, cell growth, and DNA repair. Evidence suggests sirtuins regulate nuclear receptors and nuclear receptor coactivators to coordinate diverse physiological functions. Histones are modified by at least eight distinct types of enzyme. Posttranslational modification of histones by specific enzymes determines their subsequent types of enzymatic modification. Acetylation of lysine residues provides docking sites for other enzymes that coordinate sequential enzymatic reactions. Distinct modifications of lysine residues are mutually exclusive. It has been proposed that posttranslational modification of histones imparts specific genetic output and signaling specificity. Thus, posttranslational modification of lysines within histone tails results in specific types of signaling. Nuclear receptors undergo acetylation, phosphorylation, sumoylation, and methylation. It is likely that these posttranslational modifications are part of a similar coordinated signaling cascade that occurs within the receptor. The mechanisms by which posttranslational modification within nuclear receptors contributes to the specificity of hormone signaling has broad implications for health and disease, in particular hormone-responsive cancers.
Footnotes
This work was supported by National Institutes of Health Grants R01CA70896, R01CA75503, R01CA86072, and R01CA86071 (to R.G.P.) and the Susan Komen Breast Cancer Foundation Grant BCTR0504227 (to C.W.). This project is funded in part from the Dr. Ralph and Marian C. Falk Medical Research Trust (to R.G.P.) and grants from the Pennsylvania Department of Health (to R.G.P. and C.W.).
Disclosure Statement: The authors have nothing to disclose.
First Published Online December 28, 2007
Abbreviations: ACTR, Activator of thyroid and retinoic acid receptor; AR, androgen receptor; ERα, estrogen receptor-α; GCN5, general control of amino acid synthesis 5; HDAC, histone deacetylase; Hsp90, heat-shock protein 90; NAD, nicotinamide adenine dinucleotide; NCoR, nuclear receptor corepressor; PGC1α, peroxisome proliferator-activated receptor-γ coactivator-1α; PPARγ, peroxisome proliferator-activated receptor-γ; SRC1, steroid receptor coactivator 1; TSA, trichostatin A.
References
- Rossi S, Graner E, Febbo P, Weinstein L, Bhattacharya N, Onody T, Bubley G, Balk S, Loda M 2003 Fatty acid synthase expression defines distinct molecular signatures in prostate cancer. Mol Cancer Res 1:707–715 [PubMed] [Google Scholar]
- Spelsberg TC, Steggles AW, O’Malley BW 1971 Progesterone-binding components of chick oviduct. Chromatin acceptor sites. J Biol Chem 246:4188–4197 [PubMed] [Google Scholar]
- Murray MB, Towle HC 1989 Identification of nuclear factors that enhance binding of the thyroid hormone receptor to a thyroid hormone response element. Mol Endocrinol 3:1434–1442 [DOI] [PubMed] [Google Scholar]
- Ma J, Ptashne M 1988 Converting a eukaryotic transcriptional inhibitor into an activator. Cell 55:443–446 [DOI] [PubMed] [Google Scholar]
- Dynlacht BD, Hoey T, Tjian R 1991 Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell 66:563–576 [DOI] [PubMed] [Google Scholar]
- Luo RX, Postigo AA, and Dean DC 1998 Rb interacts with histone deacetylase to repress transcription. Cell 92:463–473 [DOI] [PubMed] [Google Scholar]
- Tasset D, Tora L, Fromental C, Scheer E, Chambon P 1990 Distinct classes of transcriptional activating domains function by different mechanisms. Cell 62:1177–1187 [DOI] [PubMed] [Google Scholar]
- Vegeto E, Allan GF, Schrader WT, Tsai MJ, McDonnell DP, O’Malley BW 1992 The mechanism of RU486 antagonism is dependent on the conformation of the carboxyterminal tail of the human progesterone receptor. Cell 69:703–713 [DOI] [PubMed] [Google Scholar]
- Allan GF, Leng X, Tsai SY, Weigel NL, Edwards DP, Tsai MJ, O’Malley BW 1992 Hormone and antihormone induce distinct conformational changes which are central to steroid receptor activation. J Biol Chem 267:19513–19520 [PubMed] [Google Scholar]
- Yoshinaga SK, Peterson CL, Herskowitz I, Yamamoto KR 1992 Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258:1598–1604 [DOI] [PubMed] [Google Scholar]
- Kwok RPS, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SGE, Green MR, Goodman RH 1994 Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370:223–226 [DOI] [PubMed] [Google Scholar]
- Yee SP, Branton PE 1985 Detection of cellular proteins associated with human adenovirus type 5 early region 1A polypeptides. Virology 147:142–153 [DOI] [PubMed] [Google Scholar]
- Harlow E, Whyte P, Franza Jr BR, Schley C 1986 Association of adenovirus early-region 1A proteins with cellular polypeptides. Mol Cell Biol 6:1579–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, Kadonaga JT 1998 p300 and estrogen receptor cooperatively activate transcription via differential enhancement of intiation and reinitiation. Genes Dev 12:331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M 2000 Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843–852 [DOI] [PubMed] [Google Scholar]
- Fu M, Wang C, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L, Ogryzko V, Avantaggiati ML, Pestell RG 2000 p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J Biol Chem 275:20853–20860 [DOI] [PubMed] [Google Scholar]
- Lin HY, Hopkins R, Cao HJ, Tang HY, Alexander C, Davis FB, Davis PJ 2005 Acetylation of nuclear hormone receptor superfamily members: thyroid hormone causes acetylation of its own receptor by a mitogen-activated protein kinase-dependent mechanism. Steroids 70:444–449 [DOI] [PubMed] [Google Scholar]
- Ito K, Yamamura S, Essilfie-Quaye S, Cosio B, Ito M, Barnes PJ, Adcock IM 2006 Histone deacetylase 2-mediated deacetylation of the glucocorticoid receptor enables NF-κB suppression. J Exp Med 203:7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Zhu J, Goodman OB, Pestell RG, Schlegel PN, Nanus DM, Shen R 2006 Activation of p300 histone acetyltransferase activity and acetylation of the androgen receptor by bombesin in prostate cancer cells. Oncogene 25:2011–2021 [DOI] [PubMed] [Google Scholar]
- Fu M, Wang C, Wang J Sakamaki T, Di Vizio D, Zhang X, Albanese C, Balk S, Chang C, Fan S, Rosen E, Palvimo JJ, Janne OA, Muratoglu S, Avantaggiati M, Pestell RG 2003 Acetylation of the androgen receptor enhances coactivator binding and promotes prostate cancer cell growth. Mol Cell Biol 23:8563–8575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Wang C, Wang J, Sakamaki T, Zhang X, Yeung YG, Chang C, Hopp T, Fuqua SA, Jaffray E, Hay RT, Palvimo JJ, Jänne OA, Pestell RG 2002 The androgen receptor acetylation governs transactivation and MEKK1-induced apoptosis without affecting in vitro sumoylation and transrepression function. Mol Cell Biol 22:3373–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Rao M, Wu K, Wang C, Zhang X, Hessien M, Yeung YG, Gioeli D, Weber MJ, Pestell RG 2004 The androgen receptor acetylation site regulates cAMP and AKT but not ERK-induced activity. J Biol Chem 279:29436–29449 [DOI] [PubMed] [Google Scholar]
- Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, Hopp T, Fuqua SA, Lopez GN, Kushner PJ, Pestell R G 2001 Direct acetylation of the estrogen receptor α hinge region by p300 regulates transactivation and hormone sensitivity. J Biol Chem 276:18375–18383 [DOI] [PubMed] [Google Scholar]
- Fuqua SA, Wiltschke C, Zhang QX, Borg A, Castles CG, Friedrichs WE, Hopp T, Hilsenbeck S, Mohsin S, O’Connell P, Allred DC 2000 A hypersensitive estrogen receptor-α mutation in premalignant breast lesions. Cancer Res 60:4026–4029 [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM 1999 Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98:675–686 [DOI] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P 2006 GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metab 3:429–438 [DOI] [PubMed] [Google Scholar]
- Fan S, Wang J, Yuan R, Ma Y, Meng Q, Erdos MR, Pestell RG, Yuan F, Auborn KJ, Goldberg ID, Rosen EM 1999 BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science 284:1354–1356 [DOI] [PubMed] [Google Scholar]
- Wang C, Fan S, Li Z, Fu M, Rao M, Ma Y, Lisanti MP, Albanese C, Katzenellenbogen BS, Kushner PJ, Weber B, Rosen EM, Pestell RG 2005 Cyclin D1 antagonizes BRCA1 repression of estrogen receptor α activity. Cancer Res 65:6557–6567 [DOI] [PubMed] [Google Scholar]
- Knights CD, Catania J, Giovanni SD, Muratoglu S, Perez R, Swartzbeck A, Quong AA, Zhang X, Beerman T, Pestell RG, Avantaggiati ML 2006 Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J Cell Biol 173:533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Woo EM, Chon YT, Homenko DR, Kraus WL 2006 Acetylation of estrogen receptor a by p300 at lysines 266 and 268 enhances the deoxyribonucleic acid binding and transactivation activities of the receptor. Mol Endocrinol 20:1479–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM 2001 Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413:131–138 [DOI] [PubMed] [Google Scholar]
- Knutti D, Kralli A 2001 PGC-1, a versatile coactivator. Trends Endocrinol Metab 12:360–365 [DOI] [PubMed] [Google Scholar]
- Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG 2003 Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1α. Mol Cell 12:1137–1149 [DOI] [PubMed] [Google Scholar]
- Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O’Malley B, Spiegelman BM 1999 Activation of PPARγ coactivator-1 through transcription factor docking. Science 286:1368–1371 [DOI] [PubMed] [Google Scholar]
- Yu X, Guo ZS, Marcu MG, Neckers L, Nguyen DM, Chen GA, Schrump DS 2002 Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J Natl Cancer Inst 94:504–513 [DOI] [PubMed] [Google Scholar]
- Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP 2005 HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell 18:601–607 [DOI] [PubMed] [Google Scholar]
- Mittal R, Peak-Chew SY, McMahon HT 2006 Acetylation of MEK2 and IκB kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc Natl Acad Sci USA, 103:18574–18579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuur ER, Loktev AV, Sharma M, Sun Z, Roth RA, Weigel RJ 2001 Ligand-dependent interaction of estrogen receptor-α with members of the forkhead transcription factor family. J Biol Chem 276:33554–33560 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Schoonjans K, Auwerx J 2007 Sirtuin functions in health and disease. Mol Endocrinol 21:1745–1755 [DOI] [PubMed] [Google Scholar]
- Yang T, Fu M, Pestell R, Sauve AA 2006 SIRT1 and endocrine signaling. Trends Endocrinol Metab 17:186–191 [DOI] [PubMed] [Google Scholar]
- Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, Hay RT, Gu W, Pestell RG 2005 SIRT1 deacetylation and repression of P300 involves lysine residues 1020/1024 within the cell-cycle regulatory domain 1. J Biol Chem 280:10264–10276 [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P 2007 Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J 26:1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P 2005 Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434:113–118 [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T 2005 SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J Biol Chem 280:16456–16460 [DOI] [PubMed] [Google Scholar]
- Fu M, Liu M, Sauve AA, Jiao X, Zhang X, Wu X, Powell MJ, Yang T, Gu W, Avantaggiati ML, Pattabiraman N, Pestell TG, Wang F, Quong AA, Wang C, Pestell RG 2006 Hormonal control of androgen receptor function through SIRT1. Mol Cell Biol 26:8122–8135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L 2004 Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature 429:771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JR, Powell MJ, Popov VM, Shirley LA, Wang C, Pestell RG 2007 Sirtuins, nuclear hormone receptor acetylation and transcriptional regulation. Trends Endocrinol Metab 18:356–364 [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood J, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA 2003 Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425:191–196 [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J 2006 Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell 127:1109–1122 [DOI] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA 2006 Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444:337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG 2006 A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312:1798–1802 [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW 2006 Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124:315–329 [DOI] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L 2000 Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289:2126–2128 [DOI] [PubMed] [Google Scholar]
- Lin SJ, Ford E, Haigis M, Liszt G, Guarente L 2004 Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev 18:12–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V 2003 Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell 12:51–62 [DOI] [PubMed] [Google Scholar]
- Grinspoon S, Corcoran C, Lee K, Burrows B, Hubbard J, Katznelson L, Walsh M, Guccione A, Cannan J, Heller H, Basgoz N, Klibanski A 1996 Loss of lean body and muscle mass correlates with androgen levels in hypogonadal men with acquired immunodeficiency syndrome and wasting. J Clin Endocrinol Metab, 81:4051–4058 [DOI] [PubMed] [Google Scholar]
- Grool LJ 1995 Anabolic steroids. In DeGroot LJ, ed. Endocrinology. 3rd ed. Philadelphia: Saunders; 2362–2369 [Google Scholar]
- Altenberg B, Greulich KO 2004 Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 84:1014–1020 [DOI] [PubMed] [Google Scholar]
- Chowdhury SK, Gemin A, Singh G 2005 High activity of mitochondrial glycerophosphate dehydrogenase and glycerophosphate-dependent ROS production in prostate cancer cell lines. Biochem Biophys Res Commun 333:1139–1145 [DOI] [PubMed] [Google Scholar]
- Baron A, Migita T, Tang D, Loda M 2004 Fatty acid synthase: a metabolic oncogene in prostate cancer? J Cell Biochem 91:47–53 [DOI] [PubMed] [Google Scholar]