Abstract
Uterine leiomyomas (ULs) are benign monoclonal tumors originating from myometrial tissue in the uterus. Genetic pathways that lead to myometrial transformation into leiomyomas are largely unknown. Approximately 40% of ULs are karyotypically abnormal by G-banding; however, the remaining 60% of leiomyomas do not contain cytogenetically visible genomic rearrangements. Recent technological advances such as array based comparative genomic hybridization (array CGH) and dense single nucleotide polymorphism (SNP) arrays have enabled genome-wide scanning for genomic rearrangements missed by karyotype banding analysis. In the current study, we employed a high resolution SNP microarray on 16 randomly selected ULs and normal myometrium samples to detect submicroscopic (<5 Mb) chromosomal aberrations. The SNP array identified gene dosage changes in 56% of the fibroids (9/16), 25% of which (4/16) had aberrations >5 Mb, whereas 31% of which (5/16) contained only submicroscopic copy number changes (<5 Mb). We corroborated 3/5 submicroscopic changes using quantitative PCR, meaning that ultimately, 19% of our samples (3/16) were found to contain only submicroscopic changes. Novel submicroscopic aberrations on chromosomal segments 1q42.13, 11q13.1 and 13q12.13 and large, previously unreported deletions on 15q11.2–q23, 17p–q21.31 and 22q12.2-q12.3 were identified. Previously reported deletions on 1p, 3q, 7q, 13, and chromosome 14q were also noted. RHOU, MAP3K11 and WASF3 gene copy numbers were changed in the subset of leiomyomas with submicroscopic aberrations, and these genes have previously been implicated in tumorigenesis. Our findings support the hypothesis that a significant fraction of ULs without visible cytogenetic changes harbor submicroscopic genomic rearrangements which may in turn contribute to transformation of normal myometrial tissue into leiomyomas.
Keywords: copy number variation, leiomyoma and fibroid, microdeletion and microduplication, SNP microarray, uterus
Introduction
Uterine leiomyomas (ULs), or fibroids, are benign monoclonal smooth muscle tumors which arise from the underlying myometrial tissue in the uterus. It has been estimated that between 25% (Buttram and Reiter, 1981) and 77% (Cramer and Patel, 1990) of women will develop fibroids in their reproductive lifetime, and fibroids are a leading indication for hysterectomies performed in the USA. Many causes for leiomyomas have been proposed, and it is clear that a hereditary predisposition to fibroid development exists. The genetic causes of these tumors is largely unknown, although their formation has been linked to hormone levels (Wei et al., 2007), enzymes (Ishikawa et al., 2009), and cytogenetically visible genomic alterations. The cytogenetic alterations that have been associated with leiomyomas are varied and include large deletions on chromosome 7q (Ishwad et al., 1997, Xing et al., 1997), 12q, 14q (Pandis et al., 1991) and 13q (Meloni et al., 1991). Breakpoints in the more common cytogenetic rearrangements have exposed several candidate genes including ORC5L, LHFPL3 and PCOLCE (Ligon et al., 2002; Ptacek et al., 2007). Genomic rearrangements observed in leiomyomas may be causative of myometrial transformation and these large genomic rearrangements may result from mitotic errors secondary to smaller genomic rearrangements. Recent results suggest that submicroscopic genomic duplications and/or deletions, also termed copy number variants (CNVs), are also a significant cause of genetic pathology. With the development of single nucleotide polymorphism (SNP)/CNV array platforms, previously undetectable genetic alterations (typically those <3–5 Mb) can be found. We hypothesized that a substantial number of karyotypically ‘normal’ fibroids contained submicroscopic deletions and/or duplications. In this study we utilized a 370 K Illumina SNP/CNV array to study the full extent of CNV occurrences in a set of human ULs.
Materials and Methods
Tissue collection and genomic DNA isolation
The current study was approved by Baylor's College of Medicine Institutional Review Board. Fibroid samples were collected from 17 women who were diagnosed with ULs and underwent medically indicated abdominal hysterectomy. Patients were 34–65 years of age (mean age = 41) and were predominantly of Hispanic origin. The ULs underwent histological examination and were classified as typical leiomyomas, with low mitotic activity. All of the leiomyomas were >3 cm in size. A paired sample of adjacent normal myometrium was collected from each patient at the same time for array reference and downstream analyses. All samples were immediately frozen in a mixture of liquid nitrogen/100% ethanol and stored at −80°C until genomic DNA extraction.
Genomic DNA (gDNA) was extracted from 100 mg of leiomyomatous and matched myometrial tissue (according to Qiagen's Puregene protocol, Valencia, CA, USA). The gDNA was precipitated by centrifugation and subsequently washed with 70% ethanol. The DNA was then rehydrated overnight in DNA Hydration Solution (Qiagen, Cat # D-5004). DNA concentrations were determined on the NanoDrop instrument prior to freezing of gDNA aliquots at −20°C.
SNP array and data analysis
We utilized Illumina's (San Diego, CA, USA) 370 Duo SNP beaded array technology on the Beadstation 500GX to genotype both fibroid and myometrial gDNA. The fibroid and matched myometrial samples were run separately; however, two samples selected at random (F03 and F13) were run in both instances to serve as a control for array consistency and comparison.
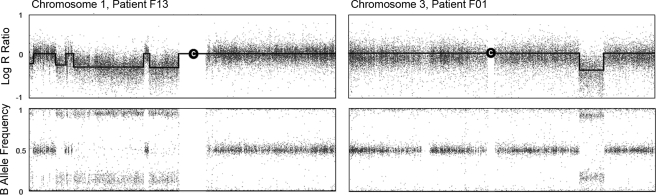
The SNP array data was analyzed using BeadStudio Genotyping Module software (Illumina) for the presence of CNVs. Fibroid and myometrial sample data were pooled into a single project and all SNP's from both myometrium and fibroid gDNA were clustered together. The ‘call rate’ algorithm, which measures the percent of probes which successfully identified a genotype, was used as an index for the validity of identified CNVs. We excluded one sample pair which returned a call rate of <96% (F06-F, F06-M) from the array and from any downstream analyses. To determine copy number changes between the paired fibroid and myometrium samples, we used the cnvPartition v1.0.2 analysis in BeadStudio with the confidence threshold set at 75 (default is 35). CNVs were discernable in two ways using the BeadStudio software. By mapping the log of the signal intensity (log R ratio) across a given chromosome, abnormalities could be visualized as an increase or a decrease from the baseline which was standardized to zero. Additionally, mapping the frequency of the genotyped ‘B’ allele versus the ‘A’ allele (B-allele frequency) across a given chromosome allowed the detection of regions of homozygosity. These two techniques used in concert permitted the identification of regions with high probability for true CNV changes although differentiating them from regions of copy-neutral loss of heterozygosity (i.e. diploid regions of DNA which contain long stretches of homozygosity). Figure 1 shows how two large deletions identified in this study cause changes in these two parameters.
Figure 1.
Log R ratio and B allele frequency plots for large copy number deletions (indicated in the graphs by a depression in the black line) on chromosomes 1 and 3 in samples F13 and F01, respectively.
A decrease in intensity, measured by log R ratio, is seen coupled with a loss of heterozygosity, measured by B-allele frequency. Notice how these aberrations can affect a single area (F01, right) or multiple areas (F13, left) on a chromosome. Chromosomal centromere is depicted by the ‘C’.
Screening and selection of CNVs
From the results of the CNV partition, a list of probable CNVs was exported and overlapping regions were grouped together by their respective samples. CNV regions shared between fibroid and myometrial tissues were excluded from the downstream analyses, as these areas likely represent non-pathologic polymorphisms. The remaining CNVs were screened against known polymorphisms deposited in the Database of Genomic Variants online (http://projects.tcag.ca/variation). If the CNVs identified in leiomyomas were present in the Database of Genomic Variants, they were presumed to represent benign polymorphisms and were excluded from the downstream analyses. A list of novel variants specific to the leiomyomatous tissues are displayed in Table I. Out of the 17 UL-specific CNVs described in the table, we selected five CNVs which were <500 kb in size, and five larger CNVs ranging from 14 to 50 Mb for real-time PCR analysis. All selected CNV regions were hemizygous deletions.
Table I.
A list of novel CNVs identified by SNP array in fibroid tissue DNA
| Patient | Deletions (loci) | Size (Mb) | Duplications (loci) | Size (Mb) |
|---|---|---|---|---|
| F01 | 3q25.1–q26.1 | 14.5 | ||
| F05 | 20q11.21* | 0.03 | ||
| F07 | 12q13.13* | 0.11 | ||
| F10 | 7q21.12* | 0.35 | ||
| 7q21.2–q31.31 | 26 | |||
| 7q32.3 | 1.6 | |||
| F12 | 11q13.1 | 0.07 | ||
| F13 | 1p | Multiple deletions (>5 Mb) | ||
| 2q37.3 | 2.3 | |||
| 13 | Multiple deletions (>5 Mb) | |||
| 14q23.3-qter | 42 | |||
| 15q11.2–q23 | 53 | |||
| 17p–q21.31 | 39 | |||
| 22q12.2–q12.3 | 5.9 | |||
| F14 | 7q21.3–q31.1 | 16 | ||
| F15 | 13q12.13 | 0.08 | ||
| F17 | 1q42.13 | 0.26 |
*Not corroborated by quantitative PCR.
Real-time PCR
For each of the ten CNVs selected for testing, we chose a relevant protein coding gene lying near the center of the purported CNV and designed 100 bp amplicons to run a 5′ nuclease real-time PCR analysis. Primers and probes were custom designed using Primer3Plus online (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). Primers were purchased from Integrated DNA Technologies (San Diego, CA, USA) and dual labeled probes (5′ 6-FAM; 3′ BHQ-1) were purchased from Sigma (St. Louis, MO, USA). The following primer sets selected for each CNV locus were utilized: 1q42.13, forward primer (F) 5′-GATGGCTTTCAGGGCAGTAG-3′, reverse primer (R) 5′-CACAGGAAGCAGCCAAGTATG-3′, internal probe (P) 5′-CATGCACCTGGCTGGCTACACG-3′; 7q21.1, (F) 5′-GCATTGGCAATAGCCAGAATAG-3′, (R) 5′-TGCACTGTGTGCTCAGGTTAG-3′, (P) 5′-CCCTTTCCTCTGTTACCCTCTGGGAATT-3′; 11q13.1, (F) 5′-AGCTCCAATGCTCTCGTAGTG-3′, (R) 5′-TGACCCTCTGGCTTCCTAAG-3′, (P) 5′-CTTTCACTTGAAGAAAGGGCTTCTCACG-3′; 12q13.13, (F) 5′-TTCCGTTGTGCCAATTAGTG-3′, (R) 5′-GCTGGACAAAGAGGGGTAAC-3′, (P) 5′-TTGTCAAATTTGCAATGTCAGGTTTCTTCC-3′; 20q11.21, (F) 5′-ACCCACGTCTGGCTCATTAC-3′, (R) 5′-GCTGTGGCCAGAGACATTAAC-3′, (P) 5′-TCCATGGAGTCATTGAAGCATGCTC-3′; 1p, (F) 5′-CCCTTGGAGTCGTGGGTTAG-3′, (R) 5′-CGTGTGCACGTGTGTGTAATC-3′, P: 5′-AACCCCACAGCACAGGTTTTCCAC-3′; 3q25.1-26.1, (F) 5′-TCCTGCCCATCCGATACTC-3′, (R) 5′-ACTTTTGCTTGGTGGGTGTC-3′, (P) 5′-CTGCGCCCTAACGGCTACTTAAACG-3′; 5q11.2-23, (F) 5′-TGGGGTGGAACATTGAAATTAG-3′, (R) 5′-GGACGGATGCAGACAGTACAC-3′, (P) 5′-CCTTTACCAAGGTTAGGTGCCCAGGA-3′; 7q21.2-31.31, (F) 5′-AACAGTTTCCCGGGAGTTAGTC-3′, (R) 5′-TGGAGCAAGAGTGGGGAGTAG-3′, (P) 5′-CCAAGCAGACATCTCGCCGGTTACT-3′.
Real-time PCR was conducted using 50 ng of gDNA, 0.5 μM forward and reverse primers, 0.2 μM dual labeled probe, 1× TaqMan Fast Universal PCR Master Mix (Applied Biosystems, Austin, TX, USA) and water to a total volume of 25 μl per well. Equal gDNA concentrations of myometrial and fibroid samples were run in five replicates amplifying a segment within the reported CNV and within a control segment located in a region outside the reported CNVs. The Biorad (Richmond, CA, USA) iQ™ 5 Real-Time PCR Detection System for real-time quantification was utilized with an annealing temperature of 57°C for 30 s followed by denaturing at 95°C for 10 s, for 40 cycles. Ct values were calculated and averaged for each replicate in the final analysis and means were used in the calculation of error.
We utilized the 2−ΔΔCt method to derive any significant fold changes in copy number between myometrial and fibroid tissue.
Results
We chose to compare CNVs between leiomyomas and matched myometrial tissue to investigate whether leiomyomas harbor submicroscopic genomic rearrangements. Because ULs are monoclonal, tumor heterogeneity is not a significant problem. A total of 325 genomic rearrangements were identified by the SNP array in the gDNA of 16 fibroid and 16 myometrial samples (∼10 per sample). Of these genomic rearrangements, 113 (35%) were excluded because they were found in both leiomyomas and surrounding myometrial tissue, and another 195 genomic rearrangements (60%) were excluded after being identified as known polymorphisms in the Database of Genomic Variants (http://projects.tcag.ca/variation). Table I lists 17 unique genomic rearrangements that were neither common polymorphisms nor present in the matched myometrial tissue. These genomic rearrangements were considered specific for the leiomyomas. The total number of novel abnormalities identified by the microarray in all samples included eight submicroscopic CNVs (<5 Mb), and nine large genomic rearrangements (>5 Mb). DNA from patient F13 alone contained seven, or 41%, of the total leiomyoma specific genomic rearrangements. Control myometrial tissue did not show any novel genomic rearrangements, which argues that leiomyomatous genomic rearrangements are unlikely to represent benign polymorphisms.
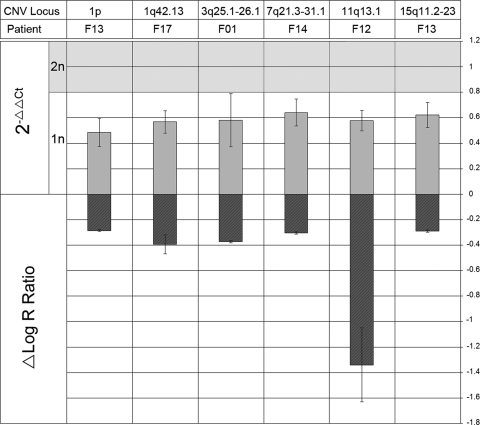
We verified SNP array findings by real-time PCR analysis on a subset of genomic rearrangements. Threshold cycle (Ct) shifts were indeed seen in the exponential phase of the amplification in 7 out of 10 quantified CNVs. No significant change between tissue types was seen in the amplification of three of our CNVs; those on 7q21.12, 12q13.13 and 20q11.21 in patients F10, F07 and F05, respectively. No significant Ct shifts were detected in any of the control amplifications where we performed real-time PCR on genomic regions that did not show copy number variation. We also performed a 2−ΔΔCt analysis (Fig. 2), with the standard error of the mean difference given as the error bars. This is plotted along side the mean shift in intensity (log R ratio) as determined by our array, so that both microarray and qPCR data can be viewed in concert. Significant fold reduction is indicated by values lying below the shaded (diploid) region, and was seen on 1p in F13, 1q42.13 in F17, 3q25.1–26.1 in F01, 7q21.3–31.1 in F14, 11q13.1 in F12 and 15q11.2–q23 in F13.
Figure 2.
Data from SNP array and quantitative real-time PCR plotted as ΔLog R Ratio and 2−ΔΔCt, respectively, for six regions of interest.
Regions of interest were first determined based on log R ratio shifts, where the reported copy number reductions in fibroid tissue are indicated above by a negative shift in the ΔLog R Ratio (fibroid − myometrium). 2−ΔΔCt values were then obtained by real-time PCR on the target regions. Values lying within the shaded region are indicative of a normal diploid copy number of 2, and those below are indicative of a copy number reduction of 1, a fold change of approximately 0.5 (1/2). Hemizygous deletions were corroborated on 1p in patient F13, 1q42.13 in patient F17, 3q25.1–26.1 in patient F01, 7q21.3–31.1 in patient F14, 11q13.1 in patient F12 and 15q11.2–23 in patient F13.
Our SNP array identified that 9/16 (56%) of the samples contained genomic rearrangements (Table I), two of which (F05 and F07) were not corroborated. Out of the remaining 7/16 samples, 4/16 (25%) of the samples had large genomic rearrangements that should be visible using G-banding analysis (F01, F10, F13 and F14), although three leiomyomas (F12, F15 and F17) contained only submicroscopic genomic variations, such that an additional 19% of the total was found to have genetic alterations when high resolution scanning was utilized. Among these three submicroscopic CNVs, two are predicted to cause haploinsufficiency in RHOU (1q42.13) in sample F17, and MAP3K11, SSSCA1, FAM89B, EHBP1L1, KCNK7 and PCNXL3 genes (11q13.1) in sample F12. We have corroborated the haploinsufficiency of these genes by real-time quantitative PCR (Fig. 2). The third CNV is predicted to cause over-expression of WASF3 from a gain in 13q12.13 in sample F15. RHOU has been implicated in WNT signaling, regulation of the cytoskeleton and cell proliferation (Tao et al., 2001). RHOU has not been previously implicated in the etiology of leiomyomas. Little is known about the role of MAP3K11 in leiomyomatous growth, although MAP3K11 plays a significant role in solid tumor invasiveness and proliferation (Yu et al., 2008). WASF3 is another gene that has been implicated in tumorigenesis, and regulates actin polymerization, cytoskeleton organization, and cell motility (Sossey-Alaoui et al., 2007) but has not been previously implicated in the etiology of leiomyomatous growth.
Discussion
Leiomyomas are genetically heterogeneous tumors that display cytogenetically visible deletions in 40–50% of cases. Little is known about submicroscopic changes that may be present in the remaining 50–60% of leiomyomas which appear normal. Gene CNVs may represent biologically more relevant pathologic lesions whose impact may be underestimated. CNVs may lead to aberrant mitosis, and subsequently cause large genomic rearrangements, and aneuploidy.
In order to identify genuine submicroscopic lesions, we utilized both whole-genome SNP array and real-time quantification. It is important to note the strengths and weaknesses of these methods. The advantage of using whole genome SNP arrays rather than oligonucleotide arrays lies in their ability to detect uniparental disomy and loss of heterozygosity, although oligonucleotide arrays have also been extensively utilized to determine CNVs (Levy et al., 2000, Nishio et al., 2004, Toruner et al., 2007). Although modern microarrays provide near unparalleled resolution on a genome-wide scale, even the most robust microarrays are estimated to be reliable only down to ∼50 Kb (Albertson and Pinkel, 2003, Coe et al., 2007). As the fallibility of the array data increases as the window approaches the SNP array's maximum resolution, other approaches are often used in concert to corroborate the results, such as real-time PCR and fluorescent in-situ hybridization. Additionally, even high resolution arrays may tag only about 50% of the CNVs in a given sample (Cooper et al., 2008), which suggests that the extent of CNVs in UL may be generally underestimated.
The majority of CNVs that we detected in UL were deletions rather than duplications. Our study differs from previous studies in this respect, in which duplications were more prevalent (Packenham et al., 1997; Sarlomo-Rikala et al., 1998; Levy et al., 2000). Secondly, with the exclusion of F13 leiomyoma, we found less de novo variations per tissue sample when compared with these previous studies. This could be due either to the inherent variability of the disorder, or the stringency of our selection criteria in which we excluded CNVs which were shared in normal tissue or known to be polymorphic.
Real-time analysis of the regions identified by SNP microarray corroborated the existence of various CNVs in our fibroid samples. Macrodeletions (>5 Mb) which have been reported previously for UL were verified on 1p, 3q, 7q, 13 and 14q. We also found novel macrodeletions on chromosomes 15q11.2–q23, 17p–q21.31 and 22q12.2–q12.3 all of which were present in patient F13, indicating that this particular tissue underwent massive genetic rearrangements. Additionally, we identified novel submicroscopic deletions in 1q42.13, 11q13.1 and a novel submicroscopic duplication in 13q12.13.
The various CNVs presented in this study may contribute to the formation of ULs. Macrodeletions, which we discovered on multiple chromosomes, would have the effect of altering genetic expression patterns of thousands of genes with diverse functions that may contribute to fibroid development. In contrast, the microdeletions revealed in this study would have an effect on only a handful of genes but their implications in UL development may be as significant. RHOU (ras homolog gene family, member U, 1q42.13) encodes a GTPase of the Rho family expressed in uterus and is associated with a variety of cancers (Tao et al., 2001). Furthermore, the RHOU gene is responsive to estradiol (Kirikoshi and Katoh, 2002). Fluctuations in hormone levels may consequently alter the expression of RHOU in myometrium tissue and contribute to UL growth. MAP3K11 (mitogen-activated protein kinase 11, 11q13.1) and WASF3 (WAS protein family member 3, 13q12.13) are two additional genes that have been associated with metastasis, survival and cell motility (Yu et al., 2008, Sossey-Alaoui et al., 2007) and were altered in a subset of our fibroid tissue. A variation in copy number in these regions could alter gene dosage and consequently abet alterations in downstream cellular functions.
The genetics of UL pathogeneses is complex. Leiomyomas likely arise spontaneously and genetic rearrangements therein arise independently from person to person, and from tumor to tumor. This has been suggested in studies observing the clonality of multiple ULs from single patients (Mashal et al., 1994; Zhang et al., 2006). However, it is evident that certain genomic rearrangements, particularly those on 1p, 7q and 13q, occur with significant frequency in many studies including our own. The etiology of such genomic rearrangement is unknown. Future studies will examine genomic rearrangements in a larger population of leiomyomatous samples to determine better the prevalence of submicroscopic CNVs in ULs and to determine whether gene expression patterns in such leiomyomas differ from leiomyomas that show more extensive and cytogenetically visible genomic rearrangements. It is possible that microdeletions play an important role in the pathogenesis of leiomyomas and that a subset of the observed complex and large genomic rearrangements are the result of initial submicroscopic genomic rearrangements.
Funding
This work was supported by the National Institutes of Health [HD058125].
References
- Albertson DG, Pinkel D. Genomic microarrays in human genetic disease and cancer. Hum Mol Genet. 2003;12:R145–R152. doi: 10.1093/hmg/ddg261. [DOI] [PubMed] [Google Scholar]
- Buttram VC, Jr, Reiter RC. Uterine leiomyomata: etiology, symptomatology, and management. Fertil Steril. 1981;36:433–445. doi: 10.1016/s0015-0282(16)45789-4. [DOI] [PubMed] [Google Scholar]
- Coe BP, Ylstra B, Carvalho B, Meijer GA, Macaulay C, Lam WL. Resolving the resolution of array CGH. Genomics. 2007;89:647–653. doi: 10.1016/j.ygeno.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet. 2008;40:1199–1203. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer SF, Patel A. The frequency of uterine leiomyomas. Am J Clin Pathol. 1990;94:435–438. doi: 10.1093/ajcp/94.4.435. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Reierstad S, Demura M, Rademaker AW, Kasai T, Inoue M, Usui H, Shozu M, Bulun SE. High aromatase expression in uterine leiomyoma tissues of African-American women. J Clin Endocrinol Metab. 2009;94:1752–1756. doi: 10.1210/jc.2008-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishwad CS, Ferrell RE, Hanley K, Davare J, Meloni AM, Sandberg AA, Surti U. Two discrete regions of deletion at 7q in uterine leiomyomas. Genes Chromosomes Cancer. 1997;19:156–160. doi: 10.1002/(sici)1098-2264(199707)19:3<156::aid-gcc4>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Kirikoshi H, Katoh M. Expression of WRCH1 in human cancer and down-regulation of WRCH1 by beta-estradiol in MCF-7 cells. Int J Oncol. 2002;20:777–783. [PubMed] [Google Scholar]
- Levy B, Mukherjee T, Hirschhorn K. Molecular cytogenetic analysis of uterine leiomyoma and leiomyosarcoma by comparative genomic hybridization. Cancer Genet Cytogenet. 2000;121:1–8. doi: 10.1016/s0165-4608(00)00225-9. [DOI] [PubMed] [Google Scholar]
- Ligon AH, Scott IC, Takahara K, Greenspan DS, Morton CC. PCOLCE deletion and expression analyses in uterine leiomyomata. Cancer Genet Cytogenet. 2002;137:133–137. doi: 10.1016/s0165-4608(02)00547-2. [DOI] [PubMed] [Google Scholar]
- Mashal RD, Fejzo ML, Friedman AJ, Mitchner N, Nowak RA, Rein MS, Morton CC, Sklar J. Analysis of androgen receptor DNA reveals the independent clonal origins of uterine leiomyomata and the secondary nature of cytogenetic aberrations in the development of leiomyomata. Genes Chromosomes Cancer. 1994;11:1–6. doi: 10.1002/gcc.2870110102. [DOI] [PubMed] [Google Scholar]
- Meloni AM, Surti U, Sandberg AA. Deletion of chromosome 13 in leiomyomas of the uterus. Cancer Genet Cytogenet. 1991;53:199–203. doi: 10.1016/0165-4608(91)90096-d. [DOI] [PubMed] [Google Scholar]
- Nishio J, Iwasaki H, Ohjimi Y, Ishiguro M, Kobayashi K, Nabeshima K, Naito M, Kikuchi M. Chromosomal imbalances in angioleiomyomas by comparative genomic hybridization. Int J Mol Med. 2004;13:13–16. [PubMed] [Google Scholar]
- Packenham JP, du Manoir S, Schrock E, Risinger JI, Dixon D, Denz DN, Evans JA, Berchuck A, Barrett JC, Devereux TR, et al. Analysis of genetic alterations in uterine leiomyomas and leiomyosarcomas by comparative genomic hybridization. Mol Carcinog. 1997;19:273–279. doi: 10.1002/(sici)1098-2744(199708)19:4<273::aid-mc9>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Pandis N, Heim S, Bardi G, Floderus UM, Willen H, Mandahl N, Mitelman F. Chromosome analysis of 96 uterine leiomyomas. Cancer Genet Cytogenet. 1991;55:11–18. doi: 10.1016/0165-4608(91)90229-n. [DOI] [PubMed] [Google Scholar]
- Ptacek T, Song C, Walker CL, Sell SM. Physical mapping of distinct 7q22 deletions in uterine leiomyoma and analysis of a recently annotated 7q22 candidate gene. Cancer Genet Cytogenet. 2007;174:116–120. doi: 10.1016/j.cancergencyto.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Sarlomo-Rikala M, El-Rifai W, Lahtinen T, Andersson LC, Miettinen M, Knuutila S. Different patterns of DNA copy number changes in gastrointestinal stromal tumors, leiomyomas, and schwannomas. Hum Pathol. 1998;29:476–481. doi: 10.1016/s0046-8177(98)90063-6. [DOI] [PubMed] [Google Scholar]
- Sossey-Alaoui K, Safina A, Li X, Vaughan MM, Hicks DG, Bakin AV, Cowell JK. Down-regulation of WAVE3, a metastasis promoter gene, inhibits invasion and metastasis of breast cancer cells. Am J Pathol. 2007;170:2112–2121. doi: 10.2353/ajpath.2007.060975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Pennica D, Xu L, Kalejta RF, Levine AJ. Wrch-1, a novel member of the Rho gene family that is regulated by Wnt-1. Genes Dev. 2001;15:1796–1807. doi: 10.1101/gad.894301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toruner GA, Streck DL, Schwalb MN, Dermody JJ. An oligonucleotide based array-CGH system for detection of genome wide copy number changes including subtelomeric regions for genetic evaluation of mental retardation. Am J Med Genet A. 2007;143A:824–829. doi: 10.1002/ajmg.a.31656. [DOI] [PubMed] [Google Scholar]
- Wei T, Geiser AG, Qian HR, Su C, Helvering LM, Kulkarini NH, Shou J, N'Cho M, Bryant HU, Onyia JE. DNA microarray data integration by ortholog gene analysis reveals potential molecular mechanisms of estrogen-dependent growth of human uterine fibroids. BMC Womens Health. 2007;7:5. doi: 10.1186/1472-6874-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing YP, Powell WL, Morton CC. The del(7q) subgroup in uterine leiomyomata: genetic and biologic characteristics. Further evidence for the secondary nature of cytogenetic abnormalities in the pathobiology of uterine leiomyomata. Cancer Genet Cytogenet. 1997;98:69–74. doi: 10.1016/s0165-4608(96)00406-2. [DOI] [PubMed] [Google Scholar]
- Yu K, Ganesan K, Tan LK, Laban M, Wu J, Zhao XD, Li H, Leung CH, Zhu Y, Wei CL, et al. A precisely regulated gene expression cassette potently modulates metastasis and survival in multiple solid cancers. PLoS Genet. 2008;4:e1000129. doi: 10.1371/journal.pgen.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhang C, Hao J, Sung CJ, Quddus MR, Steinhoff MM, Lawrence WD. Use of X-chromosome inactivation pattern to determine the clonal origins of uterine leiomyoma and leiomyosarcoma. Hum Pathol. 2006;37:1350–1356. doi: 10.1016/j.humpath.2006.05.005. [DOI] [PubMed] [Google Scholar]