Summary
Single domain response regulators (SD-RRs) are signaling components of two-component phosphorylation pathways that harbor a phosphoryl receiver domain but lack a dedicated output domain. The E. coli protein CheY, the paradigm member of this family, regulates chemotaxis by relaying information between chemoreceptors and the flagellar switch. New data provide a more complex picture of CheY-mediated motility control in several bacteria and suggest diverging mechanisms in control of cellular motors. Moreover, advances have been made in understanding cellular functions of SD-RRs beyond chemotaxis. We review recent reports indicating that SD-RRs constitute a family of versatile molecular switches that contribute to cellular organization and dynamics as spatial organizers and/or as allosteric regulators of histidine protein kinases.
Keywords: two-component systems, single-domain response regulators, receiver domain, chemotaxis, Caulobacter
Introduction
For bacteria, growth and survival critically relies on the ability to sense and respond to environmental information in order to adapt to an ever-changing environment. To shuttle information through the membrane, most bacteria make use of conserved signal transduction architectures, including one- and two-component systems. In two-component systems (TCSs) information is relayed through an autophosphorylation reaction of the histidine protein kinase (HPK) and subsequent phosphotransfer to the cognate response regulator (RR) [1] (Fig. 1). RRs usually consist of an N-terminal (phosphoryl-) receiver domain and a wide range of different C-terminal output domains that generate a cellular response [2]. The level of RR phosphorylation ultimately determines the output response.
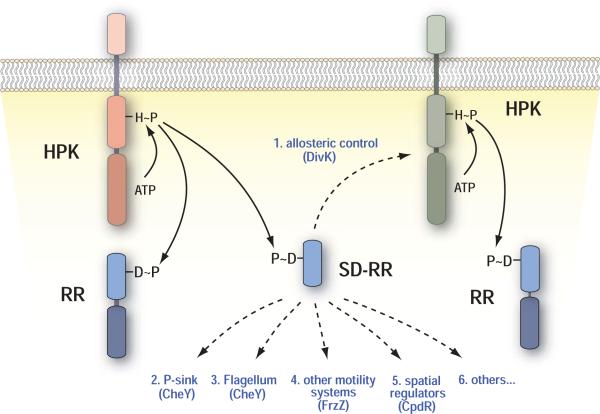
Figure 1. Functional properties of single-domain response regulators.
The domain structure of histidine protein kinases and response regulators are shown schematically with continuous lines indicating phosphorylation reactions and dashed lines summarizing the known regulatory mechanisms for SD-RRs.
The RR receiver domain with its structurally conserved (βα)5 fold represents the central regulatory switch of TCSs (Fig. 2). This domain catalyzes the transfer of a phosphoryl group from the conserved histidine of the HPK to one of its own aspartic acid residues localized inside a surface exposed acidic pocket. Receiver domains undergo dynamic structural changes with phosphorylation shifting the distribution from the inactive to the active conformation [3-5] (Fig. 2). Activation results in subtle conformational changes that are localized primarily to the α4-β5-α5 face of the receiver domain surface [3-7]. Consequently, interactions of the receiver domain with downstream elements are largely mediated by different parts of the α4-β5-α5 interface (summarized in: [2]). This surface of the receiver domain influences the associated effector domains either through direct interaction [8-10] or by the formation of receiver domain dimers [6,7,11-14]. Similarly, isolated receiver domains, so-called single-domain response regulators (SD-RRs), can engage in intermolecular interactions with downstream effector proteins [15]. Although SD-RRs form the second largest class of response regulators [2,16] little is known about their specific functions in bacterial signaling. Here we review some recent progress in understanding the molecular and cellular role of members of this regulatory family. This review does not cover SD-RRs of the Spo0F type that are involved in phosphotransfer reactions as part of phosphorelays [17,18].
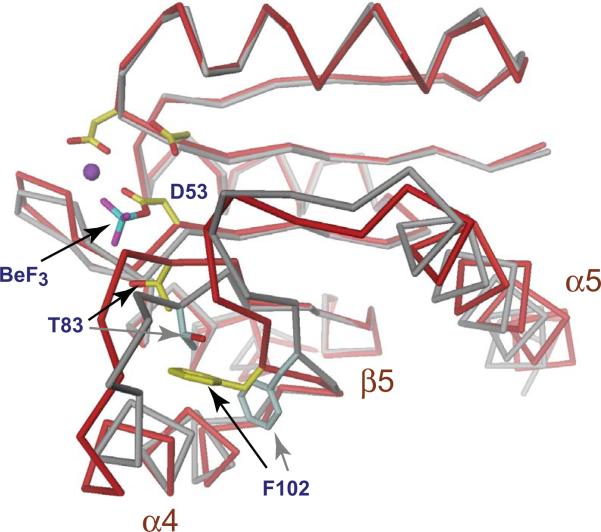
Figure 2. Structural changes upon activation of the response regulator receiver domain.
The structure of the phosphorylated (red) and unphosphorylated (grey) Rec1 domain of PleD is shown to demonstrate the conformational changes imposed upon activation of receiver domain switches [7,66]. The acidic pocket is shown with the phosphoryl-acceptor residue Asp53, a Mg2+ ion (magenta) and a BeF3 moiety. Two additional highly conserved sites, a hydroxyl-containing residue (Thr83) at the C-terminus of β4 and an aromatic residue (Phe102) in β5 are indicated. Upon phosphorylation of the active site aspartate the relative orientations of these residues change from an inactive “outward” conformation (grey) to the active “inward” conformation (yellow). Movement of this molecular lever results in subtle conformational changes that are localized primarily to the α4-β5-α5 face of the receiver domain surface.
Distribution of single-domain response regulators
Single-domain response regulators are found in all three domains of life, Bacteria, Archaea and Eukaryotes. Among Bacteria, they are encoded in the sequenced genomes of all bacterial phyla except for Chlamydia, Fusobacteria and Tenericutes (Mollicutes). Among Archaea, SD-RRs are widespread in members of Euryarchaeota but only poorly represented in Crenarchaeota. SD-RRs are also found in certain eukaryotic microorganisms, such as ciliates, diatoms, some yeasts, as well as in various green algae.
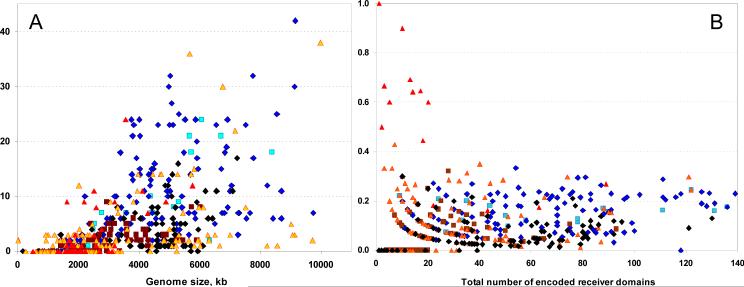
The number of SD-RRs per cell correlates with the genome size and the total number of the encoded RRs (Fig. 3) and varies from none in many bacterial parasites to more than 30 in certain α- and δ-proteobacteria [16]. The fraction of SD-RRs among all RRs encoded in a given genome reaches 90-100% in such archaea as Archaeoglobus fulgidus, Methanococcus aeolicus and N. maritimus but is under 30% in most other bacteria and archaea. Some important pathogens, such as Shigella dysenteria Sd197 and Mycobacterium leprae, do not encode any SD-RRs, despite their relatively large genome sizes (4552 and 3270 kb, respectively). Many other bacteria encode just one or two, usually the chemotaxis response regulators of the CheY family. Several SD-RRs have been experimentally characterized (Tables 1 and 2, supplemental material); selected examples are discussed in detail below.
Figure 3. Distribution of SD-RRs.
(A) The number of SD-RRs per cell in various bacteria and archaea. Five organisms encoding more than 30 SD-RRs are Acidobacteria bacterium Ellin345, Anaeromyxobacter dehalogenans, Methylobacterium sp. 4-46, Myxococcus xanthus and Solibacter usitatus. (B) The fraction of SD-RRs among all RRs. Blue diamonds, α-, β-, δ- and ε-proteobacteria; black diamonds, γ-proteobacteria; cyan squares, cyanobacteria; brown squares, firmicutes; yellow triangles, other bacteria; red triangles, archaea. For details, see supplemental material and the RR census web site, http://www.ncbi.nlm.nih.gov/Complete Genomes/RRcensus.html.
Role of CheY-like SD-RR in bacterial taxis
The concept of a response regulator was put forward by Dan Koshland to explain the mechanisms of bacterial chemotaxis [19]. Accordingly, the prototype and best-characterized single domain response regulator is the chemotaxis protein CheY from E. coli. Chemotaxis refers to the process by which bacteria bias their motility to move towards favorable or away from unfavorable environments. Changes in chemo-effector concentration are detected by a series of ligand-specific MCP chemoreceptor proteins that assemble with the coupling protein CheW and the HPK CheA to form large signal transduction clusters at the cell pole. CheA-mediated phosphorylation of CheY increases its affinity for the FliM component of the flagellar motor switch complex causing a change in flagellar rotation from counter-clockwise to clockwise [20-23]. Binding of CheY~P to FliM is mediated by the same α4-β5-α5 face of the receiver domain surface that normally mediates interaction with RR effector domains [24,25].
Many bacteria have evolved more sophisticated engines and more complex chemo-sensory pathways than E. coli. Some of these have multiple chemosensory systems with CheY molecules having adopted novel roles in controlling motor activity. The flagellar motor of Rhodobacter sphaeroides is modulated by a start-stop mechanism [26]. Two distinct chemosensory clusters, one localized at the cell pole and one in the cytoplasm, control the R. sphaeroides motor. Each cluster has dedicated CheA homologs that phosphorylate distinct CheY molecules; CheA3 and CheA4 of the cytoplasmic cluster activate CheY6, while CheA2 from the polar cluster phosphorylates CheY3 and CheY4. Although all three CheY homologs are able to bind to the FliM switch component of the R. sphaeroides flagellum in a phosphorylation-dependent manner, none of them can mediate chemotaxis alone [27•,28]. While CheY6 can stop the flagellar motor the effect of FliM binding by the other CheYs is unknown. Based on this observation, it has been speculated that some CheYs compete for FliM binding sites with CheYs that can stop motor rotation and by that contribute to signal integration from the cytoplasmic and polar chemoreceptor cluster [27•]. Similarly, Sinorhizobium meliloti requires two CheYs for chemotaxis. In this organism CheY2 is the main regulator of flagellar rotation, while CheY1 modulates CheY2 activity by acting as phosphate sink [29]. CheY1 has lost the ability to interact with the motor but controls the phosphorylation state of CheY2 by catalyzing rapid phosphotransfer from CheY2 via CheA to CheY1 under conditions where CheA is inactive. Since S. meliloti lacks signal-terminating CheY phosphatases it is assumed that this additional CheY protein fulfills the function of terminating the response in the absence of signal input.
In addition to flagella-based motility, CheY homologs control twitching and gliding motility (e.g. in Cytophaga and Flavobacterium), cyanobacterial swimming, and archaeal motility. These systems are not homologous to bacterial flagella [30] and neither of them includes FliM-related proteins. Components of these nanomotors that interact with CheY~P remain to be identified. A fascinating picture of motility control emerges in Myxococcus xanthus. This organism has a complex life cycle that includes surface motility based cell aggregation and cell differentiation. M. xanthus cells lack flagella but are equipped with two distinct engines [31], eight Che-like chemosensory systems [32] and a record number of 42 SD-RRs [16]. Cell movement is powered by the extension and retraction of type IV pili (S-motility) and by the action of a gliding motor (A-motility) (summarized in: [33,34]). By virtue of the two engines, Myxococcus cells can move forward or backwards along their longitudinal axis. Directionality of cell movement is accomplished through modulating the reversal frequency in response to environmental stimuli, a process that requires strict spatial coordination of the two engines. Recent work has indicated that Myxococcus cell reversal is associated with synchronized oscillations of components of both motility systems to the leading and lagging cell pole, respectively [35, 36, 37•,38•]. This dynamic behavior is orchestrated by a small Ras-like GTPase, MglA, the only known protein to be essential for both S- and A-motility [39,40]. MglA acts as central recruitment factor for several key components of A- and S-motility engines and thus appears to direct cell reversal through spatial coordination of the two motility machineries [33].
Genetic evidence suggested that one of the eight M. xanthus chemosensory pathways, the Frz system, controls cell reversal by interfering with MglA activity [41]. According to the E. coli chemotaxis paradigm, signaling through the FrzCD chemoreceptor would activate the CheA homolog, FrzE, which in turn would lead to cell reversal through the activation of the CheY homolog FrzZ. However, in contrast to CheY, FrzZ is a “double-faced Janus” with two functional receiver domains arranged in tandem. Both receivers have conserved aspartate phosphoryl acceptor residues, which can be phosphorylated by FrzE in vitro [42•]. Consistent with this observation, both receiver domains are required for FrzZ-mediated motility control [42•]. Just how each receiver domain contributes to Frz signaling and which downstream motility components they interact with, remains to be determined. It is possible that, analogous to CheY2 in S. meliloti, one of the FrzZ receiver domains acts as a phosphate sink to control the Frz output via its neighboring domain. Such a model would be in line with the observation that inactivation of either one of the phosphoryl acceptor sites produces partial, but distinct motility phenotypes [42•]. Alternatively, duplication of the receiver domain might increase the versatility of FrzZ, e.g. by providing additional interaction surface for downstream signaling components. This has recently been suggested for the Caulobacter crescentus RR PleD, which uses tandem receiver domains for dimerization and dynamic sequestration to one cell pole [7,43]. It is plausible that FrzZ, like some of its downstream components, dynamically oscillates between cell poles during cell reversal, where it might interact with spatial regulators as well as downstream signaling components of the Frz system. Analogous to Janus, the double-faced roman god, who according to a legend had received the gift to see both future and past, the double receiver domain protein FrzZ could represent a branching point in the FrzZ pathway. Whether FrzZ directly interacts with MglA to control its activity, remains to be shown.
SD-RRs as allosteric regulators of histidine protein kinases
Preliminary evidence for a novel molecular role of receiver domains again came from studies of the M. xanthus Frz system. The CheA homolog FrzE has an additional receiver domain fused at the C-terminus of its kinase domain. Hybrid HPKs with an additional CheY-like domain are generally believed to be part of phosphorelays with the receiver domain being involved in phosphotransfer from the conserved histidine of the kinase domain to the histidine of the downstream phosphotransfer component [1]. However, recent work by Inclan and coworkers [42•,44•] argues against such a role for the FrzE receiver domain. Both genetic and biochemical data argue that the FrzE receiver domain is not involved in phosphotransfer to FrzZ and is not required for normal co-ordination of A- and S-motility. Rather, the observation that the C-terminal receiver domain inhibits FrzE autophosphorylation suggests that it plays a modulating role in Frz signaling [44•]. Based on this and on additional experiments, Inclan and colleagues speculate that in its unphosphorylated state the receiver domain might interact with the FrzE HPT subdomain, thereby preventing it from interacting with the catalytic domain of the kinase. Phosphorylation of the C-terminal receiver domain by another HPK could provide FrzE with the possibility to crosstalk with other chemosensory pathways involved in M. xanthus motility control. Alternatively, the observation that the C-terminal receiver domain can be phosphorylated by the FrzE kinase itself opens up the possibility that this domain functions as an intramolecular device to allosterically regulate FrzE kinase activity.
More direct evidence for a role of SD-RRs as allosteric effectors of HPK activity has recently been provided by studies on Caulobacter crescentus development [45••]. C. crescentus divides asymmetrically to produce two progeny cells with distinct developmental and cell cycle programs. Whereas the sessile stalked cell immediately enters S-phase, the motile swarmer progeny remains in G1 before it differentiates into a stalked cell and enters S-phase (Fig. 4A). Two HPKs, DivJ and PleC, which are positioned to opposing poles of the Caulobacter predivisional cell, control asymmetry and cell differentiation [46,47] (Fig. 4A). Genetic and biochemical experiments have identified two RRs, which are directly controlled by PleC and DivJ; DivK, a SD-RR, and PleD, an unorthodox RR with a GGDEF diguanylate cyclase output domain [45••,48-51]. Although their molecular roles are distinct, the two RRs are not simply part of diverging readouts but are functionally interconnected.
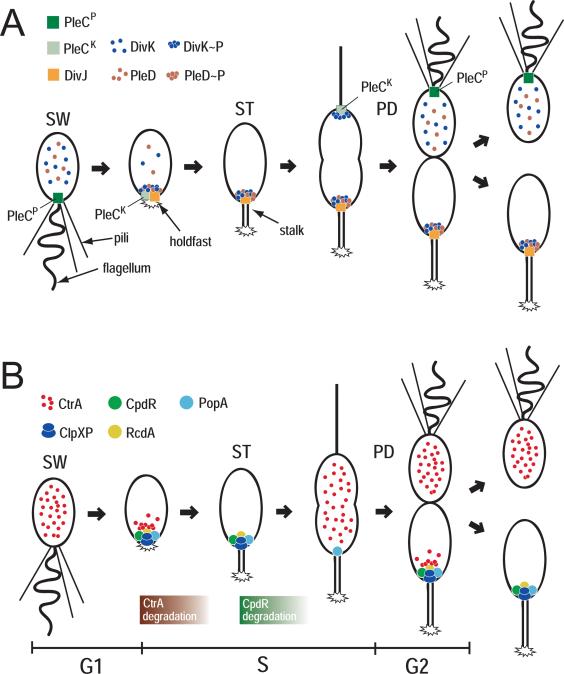
Figure 4. Role of SD-RRs in C. crescentus development and cell cycle.
(A) Schematic of the C. crescentus cell cycle with polar appendices and localization patterns for PleC, DivJ, DivK, and PleD. Kinase (PleCK) and phosphatase state (PleCP) of PleC are indicated. For details see text. (B) Schematic of the C. crescentus cell cycle with spatial distribution of CtrA, ClpXP CpdR, PopA, and RcdA. The succession of CtrA and CpdR degradation is indicated. For details see text.
Phosphorylation of PleD during stalked cell differentiation results in dimerization-based activation of its catalytic output domain and the concomitant sequestration of the regulator to the differentiating cell pole where it directs flagellar ejection, hold-fast biogenesis, and stalk formation [43,51]. Although it is not clear yet how synthesis of cyclic di-GMP orchestrates pole morphogenesis, temporal control of PleD activity during the cell cycle is critical for Caulobacter cell fate determination. But how could PleC and DivJ together mediate activation of PleD during cell differentiation? This is where the essential SD-RR DivK comes into play. DivK localizes to both poles of the predivisional cell in a phosphorylation-dependent manner, but is released from the flagellated pole after completion of cytokinesis (Fig. 4A) [52]. While the DivJ kinase is the main phosphodonor for DivK and responsible for its sequestration to the cell poles [45••,53], PleC acts as phosphatase that displaces DivK from the flagellated pole by maintaining DivK~P levels low in the swarmer cell [53,54••].
Recent in vitro and in vivo studies provided convincing evidence that DivK acts as allosteric regulator of both PleC and DivJ kinase activities [45••]. DivK stimulates PleC autophosphorylation but not phosphatase activity, arguing that it can switch PleC from the default phosphatase into the kinase state. Through allosteric activation of the DivJ autokinase, DivK also stimulates its own phosphorylation and polar localization [45••]. Together these data suggested that DivK is the central effector of an integrated circuit that operates via spatially organized feedback loops to determine C. crescentus cell fate. In swamer cells DivJ levels are low and PleC is in its phosphatase mode. As a result, DivK and PleD are inactive and delocalized. As DivJ levels rise during G1-to-S DivJ-mediated phosphorylation localizes DivK~P to the cell pole, where it forces PleC into the autokinase mode (Fig. 4A). At the same time DivK mediated activation of DivJ creates a positive feedback loop that may promote a sharp G1-to-S transition and, through the activation of PleD, rapidly and robustly commit cells to the stalked program. Conversely, the proposed feedback loops could explain how the system is reset as cells enter G1. During cytokinesis DivK localized at the flagellated pole is separated from its main kinase DivJ. This results in a rapid reduction of DivK~P levels in this compartment and a decrease of the DivK concentration at the cell pole below a threshold level required for PleC kinase activation. As a consequence, PleC switches back into its default phosphatase mode, thereby installing the swarmer cell program (Fig. 4A). According to this model the role of DivK is to facilitate long-range communication between the asymmetric DivJ and PleC antagonists and by that coordinate their activities. Ultimately, this mechanism is responsible for C. crescentus cell fate determination through PleD and possibly additional response regulators.
SD-RR as spatial regulators
Recent studies of the C. crescentus cell cycle indicate that SD-RR can actively contribute to the spatial organization of bacterial cells. The C. crescentus master cell cycle regulator CtrA impacts both cell cycle and development by regulating the transcription of a large set of genes and by blocking the initiation of chromosome replication (for recent reviews see: [55,56]). Upon entry into S-phase, the CtrA response regulator is inactivated via a dual level mechanism involving dephosphorylation and degradation [57]. Cell cycle dependent degradation of CtrA requires a specific spatial arrangement of the proteolytic machinery. During the G1-to-S transition, both CtrA and its cognate protease ClpXP [58,59] sequester to the future stalked cell pole where CtrA is degraded [60,61••]. CtrA is stabilized in mutants that fail to localize either the protease or the substrate to this subcellular site, suggesting that the timing of CtrA degradation is intimately linked to the dynamic spatial arrangement of these proteins during the cell cycle [61••, 62••,63•]. Polar targeting of ClpXP is controlled by the SD-RR CpdR, which itself sequesters to the stalked cell pole subject to its phosphorylation state [62••]. CpdR is phosphorylated and dispersed in swarmer cells. During the G1-to-S transition CpdR is dephosphorylated and, as a result, sequesters to the stalked pole to recruit the ClpXP protease (Fig. 4B). Later, in the predivisional cell, CpdR is re-synthesized but kept in the inactive, phosphorylated form by the CckA phosphorylation cascade that also activates CtrA. Through this elegant mechanism CtrA activity and stability are directly coordinated at this stage of the cell cycle [64••]. The observation that immediately after CtrA proteolysis, ClpXP also degrades CpdR at the cell pole implies that at this stage the protease detaches from the pole to further protect newly synthesized CtrA [65] (Fig. 4B). Interestingly, cell cycle dependent phosphorylation and dephosphorylation of CpdR appear to be antagonistically controlled by the essential SD-RR DivK [64••,65].
CtrA localizes to the ST cell pole coincident with CpdR and ClpXP. CtrA localization is regulated by PopA, a paralog of the PleD response regulator harboring two receiver-like domains fused to a GGDEF output domain. In contrast to PleD, the PopA output domain lacks diguanylate cyclase activity but has a conserved high-affinity binding site for c-di-GMP, the I-site. Like CpdR, PopA itself localizes to the ST pole and directs CtrA to this subcellular site via the direct interaction with an additional degradation factor, RcdA [61••,63•]. But in contrast to CpdR, PopA localization does not depend on its phosphorylation state, but requires binding of c-di-GMP to its I-site. Although epistasis experiments suggested that PopA and CpdR define two distinct polar recruitment avenues for the CtrA substrate and the ClpXP protease, respectively [63•], how exactly these pathways are interlinked is not clear. Mutants lacking CpdR, in addition to being unable to direct ClpXP to the cell pole, also fail to localize CtrA and show a partial defect in localizing RcdA to the cell pole [62••]. Conversely, CpdR and ClpX frequently mislocalize in popA mutants. Based on this and on the finding that CpdR weakly interacts with RcdA it has been proposed that all of these proteins might be part of a macromolecular complex that transiently assembles at the cell pole to activate protein degradation in response to the appropriate signals during the cell cycle [63•].
Conclusions
One could consider SD-RRs and their downstream targets as split response regulators with receiver and output domains being physically separated. As illustrated by the DivK example, this allows for interactions with multiple, spatially separated targets. The possibility for diverging signal transduction might be one reason for the relative abundance of this class of proteins. Another might be the possibility to freely diffuse and communicate information in the three-dimensional space of a cell. The role of CheY proteins in relaying information between chemoreceptors and motors represents a well-studied paradigm for such a mechanism. But apparently, CheY represents only the beginning of our appreciation of this large and versatile protein family. Like CheY, many of these proteins might work as simple switch modules for protein-protein interaction networks that operate through the phosphorylation-dependent remodeling of their α4-β5-α5 surfaces. One of the most exciting recent findings is that SD-RRs can contribute to cellular dynamics and organization. They do so not only by acting as spatial regulators to recruit proteins to a specific subcellular site, but they can also function as connectors by facilitating crosstalk, feedback, and long-range communication within the two-component phosphorylation network. Undoubtedly, additional functions for SD-RR will emerge in the future. It will be interesting to elucidate the molecular, cellular and structural properties of some of these molecules in order to uncover how extensively nature has made use of this simple protein-protein interaction module.
Supplementary Material
Acknowledgments
We would like to thank Tilman Schirmer for his help with figure 2. UJ is supported by Swiss National Science Foundation Fellowship 3100A0-108186. MYG is supported by the Intramural Research Program of the National Library of Medicine at the National Institutes of Health.
References
- 1.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 2.Gao R, Mack TR, Stock AM. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem. Sci. 2007;32:225–234. doi: 10.1016/j.tibs.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kern D, Volkman BF, Luginbuhl P, Nohaile MJ, Kustu S, Wemmer DE. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature. 1999;402:894–898. doi: 10.1038/47273. [DOI] [PubMed] [Google Scholar]
- 4.Simonovic M, Volz K. A distinct meta-active conformation in the 1.1-A resolution structure of wild-type ApoCheY. J. Biol. Chem. 2001;276:28637–28640. doi: 10.1074/jbc.C100295200. [DOI] [PubMed] [Google Scholar]
- 5.Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 6.Bachhawat P, Stock AM. Crystal structures of the receiver domain of the response regulator PhoP from Escherichia coli in the absence and presence of the phosphoryl analog beryllofluoride. J. Bacteriol. 2007;189:5987–5995. doi: 10.1128/JB.00049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wassmann P, Chan C, Paul R, Beck A, Heerklotz H, Jenal U, Schirmer T. Structure of BeF3--modified response regulator PleD: implications for diguany-late cyclase activation, catalysis, and feedback inhibition. Structure. 2007;15:915–927. doi: 10.1016/j.str.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 8.Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus RP, Dickerson RE. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–110561. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 9.Djordjevic S, Goudreau PN, Xu Q, Stock AM, West AH. Structural basis for me-thylesterase CheB regulation by a phosphorylation-activated domain. Proc. Natl.Acad. Sci. USA. 1998;95:1381–1386. doi: 10.1073/pnas.95.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Carlo S, Chen B, Hoover TR, Kondrashkina E, Nogales E, Nixon BT. The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev. 2006;20:1485–1495. doi: 10.1101/gad.1418306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batchelor JD, Doucleff M, Lee CJ, Matsubara K, De Carlo S, Heideker J, Lamers MH, Pelton JG, Wemmer DE. Structure and regulatory mechanism of Aquifex aeolicus NtrC4: variability and evolution in bacterial transcriptional regulation. J. Mol. Biol. 2008;384:1058–1075. doi: 10.1016/j.jmb.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 12.Park S, Meyer M, Jones AD, Yennawar HP, Yennawar NH, Nixon BT. Two-component signaling in the AAA+ ATPase DctD: binding Mg2+ and BeF3 selects between alternate dimeric states of the receiver domain. FASEBJ. 2002;16:1964–1966. doi: 10.1096/fj.02-0395fje. [DOI] [PubMed] [Google Scholar]
- 13.Toro-Roman A, Mack TR, Stock AM. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: a symmetric dimer mediated by the α4-β5-α5 face. J. Mol. Biol. 2005;349:11–26. doi: 10.1016/j.jmb.2005.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toro-Roman A, Wu T, Stock AM. A common dimerization interface in bacterial response regulators KdpE and TorR. Protein Sci. 2005;14:3077–3088. doi: 10.1110/ps.051722805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyer CM, Dahlquist FW. Switched or not?: the structure of unphosphorylated CheY bound to the N terminus of FliM. J. Bacteriol. 2006;188:7354–7363. doi: 10.1128/JB.00637-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoch JA, Varughese KI. Keeping signals straight in phosphorelay signal transduction. J. Bacteriol. 2001;183:4941–4949. doi: 10.1128/JB.183.17.4941-4949.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varughese KI. Conformational changes of Spo0F along the phosphotransfer pathway. J Bacteriol. 2005;187:8221–8227. doi: 10.1128/JB.187.24.8221-8227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koshland DE., Jr. A response regulator model in a simple sensory system. Science. 1977;196:1055–1063. doi: 10.1126/science.870969. [DOI] [PubMed] [Google Scholar]
- 20.Park SY, Lowder B, Bilwes AM, Blair DF, Crane BR. Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc. Natl.Acad. Sci. USA. 2006;103:11886–11891. doi: 10.1073/pnas.0602811103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sourjik V, Berg HC. Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc. Natl.Acad. Sci. USA. 2002;99:12669–12674. doi: 10.1073/pnas.192463199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bren A, Eisenbach M. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J. Mol. Biol. 1998;278:507–514. doi: 10.1006/jmbi.1998.1730. [DOI] [PubMed] [Google Scholar]
- 23.Toker AS, Macnab RM. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J. Mol. Biol. 1997;273:623–634. doi: 10.1006/jmbi.1997.1335. [DOI] [PubMed] [Google Scholar]
- 24.Dyer CM, Quillin ML, Campos A, Lu J, McEvoy MM, Hausrath AC, Westbrook EM, Matsumura P, Matthews BW, Dahlquist FW. Structure of the constitutively active double mutant CheYD13K Y106W alone and in complex with a FliM peptide. J. Mol. Biol. 2004;342:1325–1335. doi: 10.1016/j.jmb.2004.07.084. [DOI] [PubMed] [Google Scholar]
- 25.Lee SY, Cho HS, Pelton JG, Yan D, Henderson RK, King DS, Huang L, Kustu S, Berry EA, Wemmer DE. Crystal structure of an activated response regulator bound to its target. Nat. Struct. Biol. 2001;8:52–56. doi: 10.1038/83053. [DOI] [PubMed] [Google Scholar]
- 26.Porter SL, Wadhams GH, Armitage JP. Rhodobacter sphaeroides: complexity in chemotactic signalling. Trends Microbiol. 2008;16:251–260. doi: 10.1016/j.tim.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 27•.Porter SL, Wadhams GH, Martin AC, Byles ED, Lancaster DE, Armitage JP. The CheYs of Rhodobacter sphaeroides. J. Biol. Chem. 2006;281:32694–32704. doi: 10.1074/jbc.M606016200. [DOI] [PubMed] [Google Scholar]; This study dissects the specific role of six CheY homologs in Rhodobacter chemotaxis. The data suggest distinct functions for different CheYs in motor control to integrate information from two chemotaxis signaling clusters.
- 28.Del Campo AM, Ballado T, de la Mora J, Poggio S, Camarena L, Dreyfus G. Chemotactic control of the two flagellar systems of Rhodobacter sphaeroides is mediated by different sets of CheY and FliM proteins. J. Bacteriol. 2007;189:8397–8401. doi: 10.1128/JB.00730-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sourjik V, Schmitt R. Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry. 1998;37:2327–2335. doi: 10.1021/bi972330a. [DOI] [PubMed] [Google Scholar]
- 30.Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 2008;6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 31.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus(Myxobacterales): genes controlling movement of single cells. Mol. Gen. Genet. 1979;171:177–191. [Google Scholar]
- 32.Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, Ronning CM, Barbazuk WB, Blanchard M, Field C, et al. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. USA. 2006;103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mignot T, Kirby JR. Genetic circuitry controlling motility behaviors of Myxococcusxanthus. BioEssays. 2008;30:733–743. doi: 10.1002/bies.20790. [DOI] [PubMed] [Google Scholar]
- 34.Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcusxanthus. Nat. Rev. Microbiol. 2007;5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 35.Mignot T, Merlie JP, Jr., Zusman DR. Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science. 2005;310:855–857. doi: 10.1126/science.1119052. [DOI] [PubMed] [Google Scholar]
- 36.Nudleman E, Wall D, Kaiser D. Polar assembly of the type IV pilus secretin inMyxococcus xanthus. Mol. Microbiol. 2006;60:16–29. doi: 10.1111/j.1365-2958.2006.05095.x. [DOI] [PubMed] [Google Scholar]
- 37•.Mignot T, Shaevitz JW, Hartzell PL, Zusman DR. Evidence that focaladhesion complexes power bacterial gliding motility. Science. 2007;315:853–856. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript proposes that internal motors power gliding motility in Myxococcus through force transmission by dynamic focal adhesions.
- 38•.Leonardy S, Freymark G, Hebener S, Ellehauge E, Sogaard-Andersen L. Coupling of protein localization and cell movements by a dynamically localized response regulator in Myxococcus xanthus. EMBOJ. 2007;26:4433–4444. doi: 10.1038/sj.emboj.7601877. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors identify RomR, a protein that in response to Frz pathway control dynamically localizes to the lagging pole during Myxococcus cell reversal where it orchestrates polar organization of A-motility.
- 39.Hartzell PL. Complementation of sporulation and motility defects in a prokaryote by a eukaryotic GTPase. Proc. Natl. Acad. Sci. USA. 1997;94:9881–9886. doi: 10.1073/pnas.94.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spormann AM. Gliding motility in bacteria: insights from studies of Myxococcus xanthus. Microbiol. Mol. Biol. Rev. 1999;63:621–641. doi: 10.1128/mmbr.63.3.621-641.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spormann AM, Kaiser D. Gliding mutants of Myxococcus xanthus with high reversal frequencies and small displacements. J. Bacteriol. 1999;181:2593–2601. doi: 10.1128/jb.181.8.2593-2601.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Inclan YF, Vlamakis HC, Zusman DR. FrzZ, a dual CheY-like response regulator, functions as an output for the Frz chemosensory pathway of Myxococcus xanthus. Mol. Microbiol. 2007;65:90–102. doi: 10.1111/j.1365-2958.2007.05774.x. [DOI] [PubMed] [Google Scholar]; This study demonstrates that the dual receiver domain protein FrzZ mediates the readout of the Frz signaling system to control Myxococcus cell reversal frequency. Both receiver domains of FrzZ are phosphorylated by FrzE and are required for Frz signaling.
- 43.Paul R, Abel S, Wassmann P, Beck A, Heerklotz H, Jenal U. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J. Biol. Chem. 2007;282:29170–29177. doi: 10.1074/jbc.M704702200. [DOI] [PubMed] [Google Scholar]
- 44•.Inclan YF, Laurent S, Zusman DR. The receiver domain of FrzE, a CheA- CheY fusion protein, regulates the CheA histidine kinase activity and downstream signalling to the A- and S-motility systems of Myxococcus xanthus. Mol. Microbiol. 2008;68:1328–1339. doi: 10.1111/j.1365-2958.2008.06238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that receiver domain of the FrzE hybrid kinase does not play a direct role in phospho-relay but rather modulates autophosphorylation activity of FrzE in dependence of its phosphorylation state.
- 45••.Paul R, Jaeger T, Abel S, Wiederkehr I, Folcher M, Biondi EG, Laub MT, Jenal U. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell. 2008;133:452–461. doi: 10.1016/j.cell.2008.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript provides in vitro and in vivo evidence for a direct role of the SD-RR DivK in allosteric control of PleC and DivJ kinase activities. The authors present a model where DivK-mediated kinase feedback loops control robust developmental transitions and cell fate determination.
- 46.Radhakrishnan SK, Thanbichler M, Viollier PH. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev. 2008;22:212–225. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc NatlAcadSci USA. 2002;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sommer JM, Newton A. Pseudoreversion analysis indicates a direct role of cell division genes in polar morphogenesis and differentiation in Caulobacter crescentus. Genetics. 1991;129:623–630. doi: 10.1093/genetics/129.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hecht GB, Newton A. Identification of a novel response regulator required for the swarmer- to-stalked-cell transition in Caulobacter crescentus. J. Bacteriol. 1995;177:6223–6229. doi: 10.1128/jb.177.21.6223-6229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hecht GB, Lane T, Ohta N, Sommer JM, Newton A. An essential single domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBOJ. 1995;14:3915–3924. doi: 10.1002/j.1460-2075.1995.tb00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jacobs C, Hung D, Shapiro L. Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc. Natl. Acad. Sci. USA. 2001;98:4095–4100. doi: 10.1073/pnas.051609998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lam H, Matroule JY, Jacobs-Wagner C. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev. Cell. 2003;5:149–159. doi: 10.1016/s1534-5807(03)00191-6. [DOI] [PubMed] [Google Scholar]
- 54••.Matroule JY, Lam H, Burnette DT, Jacobs-Wagner C. Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell. 2004;118:579–590. doi: 10.1016/j.cell.2004.08.019. [DOI] [PubMed] [Google Scholar]; This study demonstrates that the HPK PleC has a dominant role as a phosphatase and together with its antagonist kinase DivJ controls C. crescentus development via the dynamic cellular distribution of the SD-RR DivK.
- 55.Goley ED, Iniesta AA, Shapiro L. Cell cycle regulation in Caulobacter: location, location, location. J Cell Sci. 2007;120:3501–3507. doi: 10.1242/jcs.005967. [DOI] [PubMed] [Google Scholar]
- 56.Bowers LM, Shapland EB, Ryan KR. Who's in charge here? Regulating cell cycle regulators. Curr Opin Microbiol. 2008;11:547–552. doi: 10.1016/j.mib.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 57.Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 58.Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBOJ. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chien P, Perchuk BS, Laub MT, Sauer RT, Baker TA. Direct and adaptor-mediated substrate recognition by an essential AAA+ protease. Proc. Natl. Acad. Sci. USA. 2007;104:6590–6595. doi: 10.1073/pnas.0701776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ryan KR, Huntwork S, Shapiro L. Recruitment of a cytoplasmic response regulator to the cell pole is linked to its cell cycle-regulated proteolysis. Proc. Natl. Acad. Sci. USA. 2004;101:7415–7420. doi: 10.1073/pnas.0402153101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61••.McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell. 2006;124:535–547. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]; This study shows that the ClpXP protease complex dynamically sequesters to the C. crescentus cell pole during the G1-to-S cell cycle transition coincident with CtrA degradation and identifies a novel factor, RcdA specifically involved in CtrA turnover.
- 62••.Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho- signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc. Natl. Acad. Sci. USA. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]; This manuscript identifies the SD-RR CpdR as a key regulatory factor for the recruitment of the ClpXP protease to the C. crescentus cell pole and demonstrates that polar localization of CpdR is controlled by phosphorylation.
- 63•.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies a PopA as a novel player required for polar recruitment and cell cycle-dependent degradation of CtrA and provides evidence that polar localization of this spatial regulator depends on binding of the second messenger c-di-GMP.
- 64••.Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]; This study defines a phosphorylation cascade that concomitantly regulates activity and stability of CtrA during the C. crescentus cell cycle.
- 65.Iniesta AA, Shapiro L. A bacterial control circuit integrates polar localization and proteolysis of key regulatory proteins with a phospho-signaling cascade. Proc. Natl. Acad. Sci. USA. 2008;105:16602–16607. doi: 10.1073/pnas.0808807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. USA. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.