Abstract
Pituitary GH-secretory profiles are sex dependent and regulate the sexually dimorphic expression of a large number of genes in the liver. The slow response of many sex-specific liver genes to changes in plasma GH status suggests that GH acts in the liver via both direct and indirect mechanisms organized in a hierarchical regulatory network. Presently, genome-wide liver transcription profiling was conducted to elucidate the global impact of pituitary hormone ablation on the sex specificity of rat liver gene expression and to identify sex-specific genes that respond rapidly to GH as candidates for direct targets of GH action. Hypophysectomy abolished the sex specificity of approximately 90% of 1032 sex-dependent genes, consistent with the dominant role of pituitary GH in regulating liver sexual dimorphism. Two major classes of sex-specific genes were identified: genes that were down-regulated after hypophysectomy and may be subject to positive GH regulation (461 class I genes), and genes that were up-regulated after hypophysectomy and may be subject to negative GH regulation (224 class II genes). Fifty class I sex-specific genes were induced, and 38 class II sex-specific genes were suppressed within 90 min of a physiological GH pulse, suggesting they are primary GH response genes. A further 71 sex-specific genes responded after a second GH treatment and may correspond to secondary response genes. Twenty four DNA-binding proteins were identified as early GH response genes, of which 15 were induced and nine were suppressed by GH. Five of these 24 genes displayed sex-specific expression, consistent with a hierarchical transcriptional network controlling sex-specific liver gene expression. Class II male-specific genes, such as Cyp2a2 and Cyp2c13, were down-regulated within 30 min of GH pulse treatment, as determined by heterogeneous nuclear RNA analysis, suggesting that transcription of these genes is restricted to the GH-free interpulse period in adult male rat liver. We conclude that GH acts via both positive and negative regulatory mechanisms to establish and maintain the sex specificity of liver gene expression.
GH REGULATES THE sexually dimorphic patterns of a large number of liver-expressed genes including various plasma and urinary proteins, cytochromes P450 (CYPs), and other enzymes of steroid and foreign compound metabolism, and various receptors and signaling molecules (1,2). Sex differences in liver gene expression are dictated by the temporal patterns of circulating GH, which are sex dependent and under gonadal control. In the adult male rat, GH is secreted in a highly regular, pulsatile manner, with high peaks of hormone (200–300 ng GH/ml plasma) occurring approximately every 3.5 h, separated by periods during which GH levels are undetectable (≤2 ng/ml). By contrast, pituitary GH release in the adult female rat is more frequent and results in a near continuous presence of GH in circulation (3,4). A key difference between male and female GH profiles is the length of the interpulse interval that is characteristic of adult males. This GH-free recovery period is required for the expression of male-specific genes such as Cyp2c11 (5), whereas a more continuous plasma GH profile, characteristic of female rats, activates the transcription of female-specific genes such as Cyp2c12 (6,7). Continuous infusion of GH in male rats and mice overrides the pulsatile, male plasma GH pattern and feminizes the expression of many liver genes (8,9).
GH binds to its plasma membrane receptor, which leads to the activation/tyrosine phosphorylation of signal transducer and activator of transcription (STAT) transcription factors. Upon activation, STAT proteins form homo- and heterodimers by reciprocal and complementary phosphotyrosine-SH2 domain interactions and, within minutes, translocate into the nucleus where they bind target DNA response elements and stimulate gene transcription (10). STAT5b responds directly to the male pulsatile GH pattern and is proposed to be a key regulator of the GH-responsive male-specific and female-specific liver Cyp genes (11). In adult male rats, high levels of active, tyrosine-phosphorylated STAT5b are induced by each successive plasma GH pulse, with little or no tyrosine-phosphorylated STAT5b detected during the GH-free interpulse period (12,13). By contrast, in livers of female rats, which are stimulated by GH in a more continuous manner, nuclear STAT5b levels are generally much lower than the peak levels seen in male liver (14). The importance of STAT5b for sex-specific Cyp expression is highlighted by the loss of sexually dimorphic Cyp gene expression in STAT5b-deficient male mice (15,16) and by the resistance of STAT5b-deficient hypophysectomized (Hypox) mice to GH pulse induction of sex-dependent liver Cyps normally expressed in intact mouse liver (8,17). However, STAT5b alone is not sufficient to induce the adult male pattern of liver gene expression when activated precociously in prepubertal rats given periodic (pulsatile) injections of GH (12) or when activated by very low GH pulses given to Hypox rats (18), suggesting a requirement for additional factors. Furthermore, although STAT5b binding sites have been identified in the promoters of several male-specific genes, including rat Cyp2c11, Cyp2a2, and Cyp4a2 and hamster Cyp3a10, these STAT5b-binding sites confer weak transcriptional responses in GH-stimulated cells (19,20). Moreover, the down-regulation of male-specific genes after continuous GH treatment of male mice requires several days to take effect, suggesting an indirect regulatory mechanism (8).
Taken together, the above studies suggest that GH-activated STAT5b regulates sex-specific genes directly, in cooperation with other transcription factors, and also indirectly, via early response genes that may encode transcriptional activators and repressors or other factors that mediate the effects of STAT5b. Hepatocyte-enriched nuclear factor (HNF)4α is an example of a liver-enriched transcription factor that can cooperate with STAT5b to positively regulate certain male-specific genes while inhibiting the expression of a subset of female-specific genes in male liver (8,21). HNF6, encoded by the Onecut gene, is a female-predominant liver transcription factor that is itself subject to GH regulation and may also contribute to the female-specific expression of genes such as Cyp2c12 (22,23). Two other continuous GH-dependent, female-specific nuclear factors, Cutl2/Cux2 and Trim24, reportedly act as transcriptional repressors and have been proposed to contribute to the loss of GH-regulated, male-specific liver genes expression seen in male mice deficient in either STAT5b or HNF4α (24).
The global regulatory role of GH with respect to sex-specific liver gene expression has been established by DNA microarray analysis (25). DNA microarrays have also been used to investigate the impact of hypophysectomy on liver gene expression in male rats, with changes in gene expression of up to approximately 2-fold reported for 29% of 720 detectable liver transcripts after hypophysectomy (26). However, the impact of pituitary hormone ablation in females, and any sex-differences in the effects of hypophysectomy, in particular on genes expressed in a sex-dependent manner, was not investigated. Microarray technology has been also applied to identify direct targets of GH/STAT5b-dependent gene induction (27) and gene suppression (28); however, whether sex-specific genes respond directly to GH was not investigated in these studies.
The present study uses microarray technology to probe approximately 41,000 feature microarrays to characterize the effect of hypophysectomy on the sex specificity of rat liver gene expression on a global scale, and to identify sex-specific genes that respond to GH rapidly, and thus may be direct targets of GH action. Hypophysectomy is shown to result in a near global loss of liver sex specificity, and short-term GH pulse replacement studies enabled us to identify genes likely to be primary targets of GH action. These include several male-specific Cyps the expression of which was unexpectedly found to be rapidly down-regulated by a GH pulse, as well as several sex-specific transcription factors with the potential to mediate the effects of GH on downstream sex-specific targets.
RESULTS
Experimental Design
A large-scale expression study was conducted to characterize the effect of Hypox on the sex-specificity of liver gene expression, and to identify sex-specific genes that respond to GH rapidly and are thus likely to be direct targets of GH action. RNA was isolated from livers of male (M) and female (F) rats that were: 1) intact and untreated (UT); 2) hypophysectomized at adulthood (Hypox); 3) Hypox adult male rats that were treated with a single GH pulse and killed either 30, 60, or 90 min later (M-Hypox + GH); or 4) were treated with two GH injections, spaced 3–4 h apart, and killed 60 min after the second GH injection (M-Hypox + 2GH). RNA samples representing each of these four conditions were analyzed in six sets of competitive hybridizations to 41,012-feature oligonucleotide microarrays: 1) M-UT vs. F-UT; 2) M-Hypox vs. M-UT; 3) F-Hypox vs. F-UT; 4) M-Hypox vs. F-Hypox; 5) M-Hypox + GH vs. M-Hypox; 6) M-Hypox + 2GH vs. M-Hypox. Normalized ratios and P values were calculated for all six data sets using Rosetta Resolver software. Of the 41,012 probes, 4,150 met the threshold criteria [average expression ratio >2 (or <0.5) and a significance of P < 0.005] for at least one of the six data sets after elimination of redundant probes. A complete listing of these genes, along with their expression ratios, measured signal intensities in all six microarray experiments, and gene annotations is provided in Table S2 (published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org).
Sex Specificity in Intact and Hypox Rat Liver
Comparison of M-UT and F-UT liver gene expression profiles revealed sex specificity for 1032 of the 4150 genes of interest, similar to the number of sex-specific genes seen in mouse liver (15,29,30). Of the 1032 genes, 589 showed male-specific expression (M-UT:F-UT > 2.0), and 443 genes showed female specificity (M-UT:F-UT < 0.5). Pituitary hormone ablation led to a loss of sex specificity for approximately 90% of the sex-specific genes. Thus, 517 of the 589 male-specific genes (88%) and 418 of the 443 female-specific genes (94%) lost sex specificity after hypophysectomy (Table 1). Furthermore, many of the 71 male-specific genes and 22 female-specific genes that retained sex specificity after hypophysectomy showed a decrease in the sex specificity ratio. Of the sex-specific genes, less than 1% showed a reversal of sex specificity after hypophysectomy, and only 4% of the 3118 sex-independent genes of interest acquired sex specificity after hypophysectomy (Table 1).
Table 1.
Sex Specificity of 4150 Genes of Interest in Intact and in Hypox Rat Liver
| Sex Specificity in Intact, Untreated Rats | Sex Specificity in Hypox Rats | Gene Count | % |
|---|---|---|---|
| Male-specific | M > F | 71 | 12 |
| F > M | 1 | 0 | |
| Non-sex-specific | 517 | 88 | |
| Total | 589 | ||
| Female-specific | M > F | 3 | 1 |
| F > M | 22 | 5 | |
| Non-sex-specific | 418 | 94 | |
| Total | 443 | ||
| Sex-independent | M > F | 66 | 2 |
| F > M | 48 | 2 | |
| Non-sex-specific | 3004 | 96 | |
| Total | 3118 |
Liver-expressed genes (4150) meeting the criteria of male-female ratio greater than 2.0 (or < 0.5) and P< 0.005 (genes of interest) are grouped according to the sex specificity of their expression in intact (UT) rats and secondarily according to the sex specificity of their expression in Hypox rats.
Sex-Specific Response to Hypophysectomy
Many of the sex-specific genes responded to hypophysectomy in a highly sex-dependent manner (Table 2). Thus, of those sex-specific genes that responded to hypophysectomy in male liver only, 164 of 166 male-specific genes were decreased in expression and all 58 female-specific genes were increased in expression. In contrast, of those sex-specific genes that responded to hypophysectomy in female liver only, 120 of 121 male-specific genes were increased in expression whereas 137 of 138 female-specific genes were decreased. These patterns differ markedly from those of the sex-independent genes, where the distribution between genes increasing and genes decreasing in expression after hypophysectomy was independent of whether the change in expression occurred in males only, females only, or both sexes (Table 2). Moreover, for all 697 sex-independent genes that responded to hypophysectomy in both males and females, the response to hypophysectomy was in the same direction, i.e. either an increase or a decrease, in both sexes.
Table 2.
Impact of Hypophysectomy on Gene Expression
| Impact of Hypox | Change in Expression in
|
||
|---|---|---|---|
| M-Hypox Onlya | F-Hypox Onlya | M-Hypox and F-Hypoxa | |
| Male-specific genes | |||
| Decrease (class I) | 164 | 1 | 70 |
| Increase (class II) | 2 | 120 | 26 |
| Other | 0 | 0 | 17b |
| Female-specific genes | |||
| Decrease (class I) | 0 | 137 | 59 |
| Increase (class II) | 58 | 1 | 20 |
| Other | 0 | 0 | 14c |
| Sex-independent genes | |||
| Increase | 346 | 240 | 302 |
| Decrease | 289 | 233 | 395 |
Liver-expressed genes of interest (Table 1) were grouped based on their response to hypophysectomy in both males and females, and secondarily, based on whether gene expression was increased or decreased. The designation of class I and class II genes is as described in the text and in Table 3. Not tabulated are data for 1763 other genes of interest, namely, 189 male-specific genes, 154 female-specific genes, and 1313 sex-independent genes, the expression of which was not significantly changed after hypophysectomy (also see Table 3). A majority of these unchanged sex-specific genes actually lost sex specificity after hypophysectomy (i.e. male-female difference <2-fold), but the changes in gene expression did not reach the specified criteria of more than 2.0-fold and P < 0.005.
All numbers shown are gene counts.
Male-specific genes down-regulated in Hypox males and up-regulated in Hypox females.
Female-specific genes down-regulated in Hypox females and up-regulated in Hypox males.
Clustering by Significance and Differential Expression
The general trends in gene expression, summarized above, and the impact of short-term GH treatment were further investigated by classification of the 4150 genes of interest using a binary flagging system (15), whereby each gene is assigned to a specific category, termed TFS (total flagging sum), based upon its expression ratio and P value in each of the six microarray experiments. Genes were thus classified into groups and subgroups based on their sex specificity in intact rats, their response to hypophysectomy, their sex specificity in Hypox rat liver, and their response to either one or two GH injections. Major gene groups are shown in supplemental Table S3, and a complete listing of gene groups is presented in supplemental Table S4. Analysis of the major sex-specific gene groupings (supplemental Table S3A) revealed two classes of sex-specific genes: 1) class I sex-specific genes, which are down-regulated in one, or both, sexes after hypophysectomy and thus require pituitary hormones for full expression; and 2) class II genes, which are up-regulated in either one or both sexes after hypophysectomy and are thus suppressed by pituitary hormones (Tables 2 and 3). An example of a well-characterized class I male-specific gene is Cyp2c11, the expression of which is dependent on the repeated stimulation by male plasma GH pulses and is down-regulated in male liver after hypophysectomy (5). Cyp2a2 is a prototypic class II male gene; its expression does not require stimulation by plasma GH pulses but, rather, is repressed by the female plasma GH pattern (31,32,33). Cyp2c12, Onecut1/Hnf6, and A1bg are continuous GH-stimulated genes that serve as examples of class I female genes (6,7,23,34). Class II female-specific genes have not been previously described in the rat; however, mouse Cyp2b9 is representative of an analogous class of genes in mouse liver: it is highly female specific and is strongly up-regulated in Hypox male liver (8,35). Class II female genes presently identified in rat liver include Adh3, Adh4, and Igfbp1.
Table 3.
Classification of Sex-Specific Genes Based on Response to Hypophysectomy in Males and in Females
| Class | Response to M-Hypox | Response to F-Hypox | Gene Count | % | Examples |
|---|---|---|---|---|---|
| Male-specific genes | |||||
| IA | Down | − | 164 | 41 | Ncam2, Gluld1, Fgf21 |
| IB | Down | Down | 70 | 18 | Mup4 |
| IC | Down | Up | 17 | 4 | Cyp2c11 |
| IIA | − | Up | 120 | 30 | Cyp2a2, Cyp2C13, Ca3 |
| IIB | Up | Up | 26 | 6 | Cyp17a1 |
| Other | 3a | 1 | |||
| Total responsive to Hypox | 400 | ||||
| Unresponsive to Hypox | 189 | ||||
| Total male-specific | 589 | ||||
| Female-specific genes | |||||
| IA | − | Down | 137 | 48 | Cutl2/Cux2 |
| IB | Down | Down | 59 | 21 | Cyp2c12, Onecut1/Hnf6, A1bg |
| IC | Up | Down | 14 | 4 | Sult2a2 |
| IIA | Up | − | 58 | 19 | Adh3 |
| IIB | Up | Up | 20 | 7 | Adh4, Igfbp1 |
| Other | 1b | 0 | |||
| Total responsive to Hypox | 289 | ||||
| Unresponsive to Hypox | 154 | ||||
| Total female-specific | 443 |
Male-specific (400 genes) and 289 female-specific genes showing pituitary dependence are grouped into each of two classes (I, II) and subclasses (A, B, C) based on their response to hypophysectomy [up-regulation, down-regulation, or no significant change (−)] in males and females, as indicated. Class I sex-specific genes are positively regulated by pituitary factors, as shown by their down-regulation after hypophysectomy, whereas class II sex-specific genes are negatively regulated (i.e. up- regulated after hypophysectomy). The subclass designates whether the impact of hypophysectomy is seen in only one sex (A) or in both sexes (B). Genes that respond to hypophysectomy in both sexes but in the opposite direction in each sex are placed in subclass C.
One gene down-regulated in F-Hypox only and two genes up-regulated in M-Hypox only.
Gene is up-regulated in F-Hypox only.
Class I and class II sex-specific genes were further subdivided as shown in Table 3. Class IA male-specific genes are down-regulated in Hypox males but not females (164 genes). Thus, these genes are positively regulated by the male but not by the female pituitary hormone profile. In contrast, class IB male genes are down-regulated by hypophysectomy in both males and females, indicating that they require pituitary hormone for full expression in both sexes (70 genes). Class IC male genes are down-regulated by hypophysectomy in male liver, but are up-regulated by hypophysectomy in females (17 genes). Class IIA male-specific genes are selectively up-regulated in Hypox female liver, indicating strong negative regulation by the female pituitary hormone profile (120 genes), whereas class IIB male genes are up-regulated by hypophysectomy in both males and females, indicating negative regulation by pituitary hormones in both sexes (26 genes) (Table 3).
Class IA female-specific genes are down-regulated in Hypox females but not males, indicating positive regulation by the female, but not by the male, pituitary hormone profile (137 genes), whereas class IB female genes are down-regulated by hypophysectomy in both sexes (59 genes). Class IC female genes are down-regulated by hypophysectomy in female liver, but are up-regulated by hypophysectomy in males (14 genes). Class IIA female genes are up-regulated in Hypox male liver only, indicating a requirement for the male but not female pituitary hormone pattern for full expression (58 genes). Finally, class IIB female genes are up-regulated by hypophysectomy in both male and female liver, indicating negative regulation by pituitary hormones in both sexes (20 genes) (Table 3).
Response of Sex-Specific Genes to Short-Term GH Treatment
Of the sex-specific genes that showed a change in expression after hypophysectomy, 25–31% responded to short-term GH treatment in Hypox male liver, similar to the percentage of sex-independent genes of interest that responded rapidly to GH (24%) (supplemental Table S5). Moreover, 18–19% of the sex-specific genes the expression of which decreased after hypophysectomy were up-regulated after either one or two GH injections (49 of 251 class I male genes and 37 of 210 class I female genes; Table 4A and supplemental Table S6A). The induction seen in the case of the class I female genes may be a manifestation of their responsiveness to continuous GH stimulation.
Table 4.
Impact of a Single or Two GH Injections Given to Hypox Male Rats on GH-Responsive Class I (panel A) and Class II (panel B) Sex-Specific Genes
| A. Class I Genes | Up-Regulation by GH | All Class I Genes
|
Class IA
|
Class IB
|
Class IC
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Gene Count | % | Gene Count | % | Gene Count | % | Gene Count | % | ||
| Class I Male genes (251) | After first injection | 26 | 53 | 13 | 41 | 12 | 92 | 1 | 25 |
| After second injection only | 23 | 47 | 19 | 59 | 1 | 8 | 3 | 75 | |
| Total: | 49 | 32 | 13 | 4 | |||||
| Class I Female genes (210) | After first injection | 24 | 65 | 8 | 44 | 14 | 88 | 2 | 67 |
| After second injection only | 13 | 35 | 10 | 56 | 2 | 13 | 1 | 33 | |
| Total: | 37 | 18 | 16 | 3 | |||||
| B. Class II Genes | Down-Regulation by GH | All Class II Genes
|
Class IIA
|
Class IIB
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Gene Count | % | Gene Count | % | Gene Count | % | ||||
| Class II male genes (146) | After fist injection | 33 | 61 | 24 | 65 | 9 | 53 | ||
| After second injection only | 21 | 39 | 13 | 35 | 8 | 47 | |||
| Total: | 54 | 37 | 17 | ||||||
| Class II female genes (78) | After first injection | 5 | 26 | 2 | 15 | 3 | 50 | ||
| After second injection only | 14 | 74 | 11 | 85 | 3 | 50 | |||
| Total: | 19 | 13 | 6 | ||||||
Classes and subclasses of sex-specific genes are as defined in Table 3. Shown are class I genes induced by either one or two GH injections and class II genes suppressed by either one or two GH injections. “After first injection” designates genes that are up-regulated or down-regulated after both the first and the second GH injections. “After second injection only” designates genes that responded to the second GH injection but not to the first GH injection. Total number of genes in each class is shown in parentheses at left.
Similarly, GH suppressed 24–36% of the sex-specific genes that were up-regulated after hypophysectomy (54 of 146 of class II male genes and 19 of 78 class II female genes; Table 4B and supplemental Table S6B). Class I and class II sex-specific genes that did not respond to short-term GH treatment may be secondary GH response genes or perhaps primary genes characterized by a delayed response; alternatively, these genes may require pituitary hormones other than GH for expression. In addition, class II genes not showing a GH response may include direct targets of GH, the mRNA half-life of which is too long to allow for a significant decrease within the 4–5 h time frame of the GH treatment.
A substantial fraction (41–44%) of the GH-responsive class IA genes were induced after the first GH injection; the other 56–59% were induced after the second GH injection. In contrast, 88–92% of the class IB sex-specific genes induced by short-term GH treatment were already increased after the first GH injection, indicating they are likely to be direct GH targets (Table 4A). Supplemental Tables S2E and S2F list the top 100 early GH response genes (both sex specific and sex independent), with a complete listing presented in supplemental Tables S7–S9. In the case of the class IIA sex-specific genes suppressed by short-term GH treatment, 65% of the male-specific genes were down-regulated after the first GH injection, as compared with only 15% of the female-specific genes (P < 0.005; Fisher’s exact test) (Table 4B). The high level of expression of these class IIA male genes in Hypox liver, and their rapid suppression by a physiological GH pulse, raises the possibility that transcription of these genes is restricted to the GH-free interpulse interval. Of note, six of the 12 early GH-responsive class IB male-specific genes belong to the α2u-globulin/Mup family; the expression of these genes is decreased by hypophysectomy in both sexes but retains partial intrinsic male specificity in Hypox rat liver. Moreover, 24 genes coding for DNA-binding proteins (a majority of them transcription factors) were early GH response genes, of which 15 were rapidly induced by GH treatment and nine were down-regulated (supplemental Table S10). Three of these early GH response genes were expressed in a female-specific manner (Cux2/Cutl2, Onecut1/HNF6, and Ncl), and three were expressed in a male-specific manner (Zfp37, Bcl6, Pparg). Seventy other genes coding for DNA-binding proteins were either induced or suppressed at the second GH time point only; of these, only two showed sex-specific expression (Bhlhb8, male-specific; and Rfxdc1, female-specific).
Gene Ontology (GO) Analysis
The sex-specific genes were found to be enriched for a variety of biological processes and metabolic functions, with class I male genes enriched for genes related to cell division/cell cycle, axonal fasciculation, and pheromone binding (e.g. α-2u globulin/Mup genes, in male class IB), whereas the class II female genes showed enrichment for alcohol metabolism (supplemental Table S11). Interestingly, the class I female genes, which are down regulated in Hypox liver, are enriched in several of the same processes and functions related to steroid and lipid metabolism as the class II male genes, which are up-regulated in Hypox liver. This suggests that the up-regulation of the latter genes may be, in part, a compensatory response to the loss of the class I female genes. Sex-specific genes were also enriched in several important hepatic KEGG metabolic pathways, including xenobiotic metabolism by cytochromes P450, fatty acid metabolism, bile acid metabolism, and androgen and estrogen metabolism. Early GH response genes identified in this study were enriched in several of the above-mentioned biological processes and metabolic functions, but in addition, were enriched for genes related to acute phase response, aldo-keto reductase activity, and Janus family of tyrosine kinases-STAT signaling pathway (genes rapidly up-regulated by GH) and genes involved in digestion and exopeptidase activity (genes rapidly down-regulated by GH) (supplemental Table S11).
Validation of Sex-Specific Early GH Response Genes
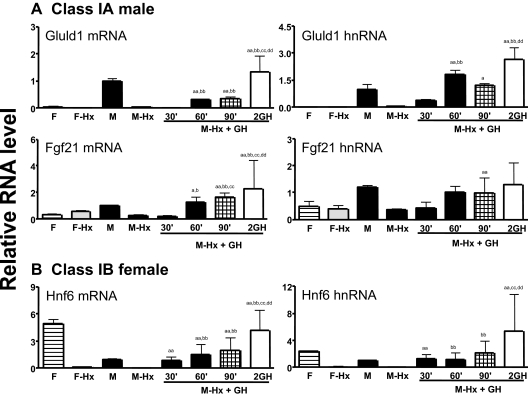
Microarray analysis identified 26 class I male genes and 24 class I female genes induced within 90 min of a single GH injection to Hypox rats (Table 4A), suggesting these are direct GH response genes. To confirm these findings, we performed quantitative real-time PCR (qPCR) analysis to measure mRNA levels, as well as heterogeneous nuclear RNA (hnRNA) levels, which correlate closely with results obtained in run-on transcription assays (36). In addition, the time course of induction was investigated by qPCR analysis carried out at individual time points, i.e. 30, 60, and 90 min after a single GH injection and 1 h after a second GH injection given 3–4 h after the first injection. Analysis of mRNA and hnRNA levels for two class IA genes showing male specificity verified their sex specificity and responsiveness to hypophysectomy and revealed GH-induced expression by 30 min (Gluld1 hnRNA) and by 60 min (Fgf21 mRNA and hnRNA) (Fig. 1A). Substantial increases in mRNA and hnRNA levels of Onecut1/Hnf6, a class IB female gene, were also seen 30 min after a single GH injection (Fig. 1B), in agreement with an earlier study of this gene (37) and validating the microarray data identifying it as an early GH response gene.
Figure 1.
qPCR Analysis of Class I Sex-Specific Genes Induced by Short-Term GH Treatment
Liver RNA samples were prepared from the liver of the following groups of rats: untreated males (M, n = 8); Hypox males (M-Hx, n = 6); Hypox males treated with a single GH injection and killed after 30 min (M-Hx+GH, 30′, n = 2), 60 min (M-Hx+GH, 60′, n = 3), and 90 min (M-Hx+GH, 90′, n = 3); Hypox males killed 1 h after a second GH injection given 3–4 h after the first injection (M-Hx+2GH, n = 3), untreated females (F, n = 7), and Hypox females (F-Hx, n = 7). RNAs were reverse transcribed to cDNA and assayed for the indicated mRNAs and hnRNAs by qPCR using primers shown in supplemental Table S1. Data shown are mean ± sd values (analysis of pooled RNA from UT male and female and Hypox male and female liver RNAs) or mean ± se values (analysis of individual liver RNAs prepared from Hypox rats treated with GH). Data were normalized to the 18S RNA content of each pooled cDNA sample, with the UT male group set at a value of 1. For M-Hx + GH (30′, 60′, 90′, and 2GH) vs. M-Hx: a, P < 0.05; and aa, P < 0.01; for M-Hx + GH (60′, 90′ and 2GH) vs. M-Hx + GH (30′): b, P < 0.05; and bb, P < 0.01; for M-Hx + GH (90′ and 2GH) vs. M-Hx + GH (60′): c, P < 0.05; and cc, P < 0.01; for M-Hx + 2GH vs. M-Hx + GH (90′): d, P < 0.05; and dd, P < 0.01.
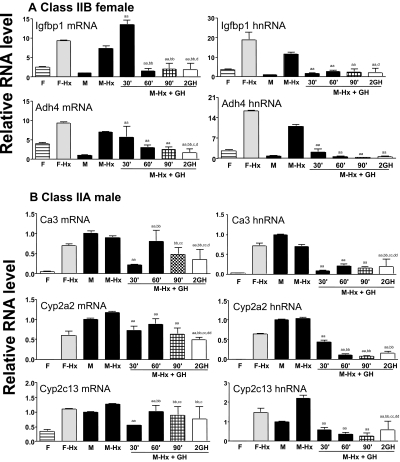
The suppressive action of GH was investigated by qPCR analysis of several class II sex-specific mRNAs and their cognate hnRNAs. Previous studies have shown that lgfbp1 expression rapidly decreases in response to supraphysiological GH treatment (28,38). Based on our microarray data, Igfbp1 is a class IIB female gene that is up-regulated in both male and female liver after hypophysectomy, by 8.2-fold and 2.9-fold, respectively. Igfbp1 did not meet the 2-fold microarray threshold for a response to short-term GH treatment, although a trend of a decrease was observed (M-Hypox + GH/M-Hypox ratio = 0.6; M-Hypox + 2GH/M-Hypox ratio = 0.7). Consistent with these array results, Igfbp1 mRNA was not decreased in Hypox rat liver until 60 min after GH treatment; however, Igfbp1 hnRNA was strongly down-regulated within 30 min of a single, physiological GH injection (Fig. 2A). Similar observations were made for Adh4, another class IIB female gene. Based on the microarray data, Adh4 was up-regulated after hypophysectomy in both males and females and was down-regulated after two GH injections. A modest and progressive decrease of Adh4 mRNA was seen 60 min after a single GH injection in Hypox males (Fig. 2A). However, hnRNA measurements showed that Adh4 expression was substantially suppressed 30 min after a single GH injection (Fig. 2A), indicating that this gene is likely to be a direct target of GH suppression.
Figure 2.
qPCR Analysis of Class II Sex-Specific Genes Repressed by Short-Term GH Treatment
Liver RNA samples described in Fig. 1 were assayed for the indicated mRNAs and hnRNAs by qPCR. Data presentation and statistical analyses are as described in Fig. 1.
Three class IIA male-specific genes were also analyzed: Ca3, Cyp2a2, and Cyp2c13. Based on the microarray data, these three genes are male specific, up-regulated (derepressed) in Hypox females, unchanged in Hypox males, and either suppressed by a single GH pulse (Ca3), suppressed after two GH pulses (Cyp2a2), or largely unresponsive to short-term GH treatment (Cyp2c13). These overall trends were validated by mRNA qPCR (Fig. 2B, left). Moreover, hnRNA analyses show that all three male-specific genes were rapidly suppressed, within 30 min of a single GH pulse (Fig. 2B, right). Thus, these genes are likely to be direct targets of the negative regulatory action of GH. Results were validated using a second set of hnRNA primers for the three genes showing GH suppression of hnRNA, but not mRNA, at the 30-min time point (Igfbp1, Adh4, and Cyp2a2; supplemental Fig. S1 published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). Sex specificity and responsiveness to hypophysectomy and GH treatment were also verified for six other class I male genes, three class II male genes, and two class I female genes (supplemental Figs. S2–S4).
DISCUSSION
Microarray technology was used to characterize the impact of pituitary hormone ablation on sex-specific gene expression in rat liver and to identify early GH response genes (primary targets of GH) that are sex specific and may potentially mediate the regulatory effects of GH on liver sex specificity. Approximately 90% of more than 1000 sex-specific liver genes were shown to be under pituitary control, demonstrating a near-global dependence of sex-specific gene expression on the pituitary gland, and consistent with the widespread regulatory role of GH with respect to the sex specificity of liver-expressed genes reported earlier (25). Two distinct classes of sex-specific liver genes were identified: genes positively regulated by pituitary hormones, as evidenced by their down-regulation after hypophysectomy (class I genes), and genes negatively regulated by pituitary hormones, as shown by their up-regulation after hypophysectomy (class II genes). Individual class I and class II sex-specific genes described previously at both the RNA and protein level include Cyp2c11, a class I male gene that requires pulsatile plasma GH stimulation for expression, and Cyp2c12, a class I female gene that is induced by the more continuous female plasma GH pattern (1,2). Class II male genes, such as Cyp2a2, Cyp3a2, and Cyp4a2, do not require male plasma GH pulse stimulation, as evidenced by their full expression in Hypox male liver. Rather, these genes are strongly repressed by the female plasma GH profile, which explains their up-regulation to near male levels in Hypox female liver (31,32,33,39). The present studies also identified a second, novel class of female-specific genes in rat liver, termed class II female genes, which are repressed by the male pituitary hormone profile, as indicated by their up-regulation in Hypox male liver. This response is analogous to that of a corresponding set of female-specific genes described earlier in mouse liver and exemplified by mouse Cyp2b9 (8,35). These class I and class II sex-specific genes play distinct but overlapping roles in liver metabolism and physiology, as determined by GO analysis. The greatest functional similarity was seen between class I female genes and class II male genes, suggesting the latter group of male-specific genes, the expression of which is up-regulated in Hypox female liver, compensates, in part, for the associated loss of the class I female genes.
A significant number of the class II sex-specific genes were down-regulated by short-term GH treatment (supplemental Table S8). GH is thus the key pituitary factor the ablation of which leads to the up-regulation of these genes after hypophysectomy. Moreover, the rapid down-regulation of these genes by GH indicates they are direct targets of the suppressive action of GH and that their mRNAs have a short half-life. Presumably, there are other class II sex-specific genes that are rapidly down-regulated by GH but could not be detected over the 4–5 h time frame of the present study owing to their long mRNA half-lives. This conclusion is supported by our studies of Cyp2a2, Cyp2c13, Igfbp1, and Adh4 because the suppression of these genes by GH at 30 min was only apparent when examined at the level of hnRNA. Other sex-specific genes that did not respond to short-term GH treatment could be secondary response genes or perhaps may be primary response genes characterized by delays in transcription and/or subsequent stages of RNA elongation and processing (40). Alternatively, these genes may be dependent on other pituitary hormones for expression. Our finding that class II male-specific genes, such as Cyp2a2 and Cyp2c13, are directly repressed by a single physiological pulse of GH leads us to propose that gene expression is limited to the approximately 2-h GH-free internal between plasma GH pulses. Furthermore, the marked suppression of these and other male-specific genes in male rats given GH as a continuous infusion (9,31,41) supports the hypothesis that these genes are repressed at all times in female liver due to the near-continuous presence of GH in circulation. Further study is required to test this hypothesis, which predicts that transcription of these class II male-specific genes is intermittent in males and is restricted to the plasma GH-free interpulse interval.
Several of the primary GH target genes identified in the present study are induced by GH via either STAT5b or STAT5a. These genes include Igf1 (42), Cish (43), and Onecut1/Hnf6 (37). Rapid induction by GH (within 90 min) was presently observed for 50 class I sex-specific genes. These genes are candidates for direct regulation by STAT5b, which is rapidly activated by GH treatment in vivo (within 10–15 min) (44,45) and is required for sex-specific liver gene expression (15,46). Genes rapidly induced by GH (supplemental Table S7) include the male-specific genes Gluld1, Fgf21, and several α2u globulin/Mup genes, as well as female-specific genes such as Cux2/Cutl2 and Onecut1/Hnf6. The rapid suppressive effect of GH may also be regulated by STAT5b, as recently described for Igfbp1, where GH-activated STAT5b impairs the action of FoxO1, a transcription factor that positively regulates Igfbp1 (28). A STAT5b-dependent repression mechanism could also characterize the suppression of class II male genes, such as Cyp2a2, at the time of each plasma GH pulse, as discussed above. Indeed, in studies using adenoviral vectors delivering dominant-negative and constitutively active forms of STAT5b, STAT5b was shown to be a major mediator of the rapid suppressive effects of GH; however, STAT5b-independent mechanisms appeared to dominate the inductive effects of GH (27,28). STAT5-dependent repression may readily be identified through these types of studies, where a dominant-negative STAT5 only needs to partially inhibit endogenous STAT5 signaling to detectably reverse STAT5-dependent gene repression. However, to conclusively establish the STAT5 dependence of gene induction using this experimental approach, the dominant-negative STAT5 construct needs to be potent and fully effective in blocking endogenous STAT5 activity, which is difficult to achieve. Accordingly, STAT5b may mediate a larger fraction of the rapid gene induction responses that the 20% value reported earlier (27).
The rapid induction of several class I female genes by GH is probably a manifestation of their responsiveness to continuous GH stimulation, as seen here in the context of the GH-free background of a Hypox rat, rather than a response to GH pulse treatment per se. This response could involve STAT5a, which has been shown to play a role in sex-specific gene expression, particularly in female liver (29). Rapid GH induction characterized a significantly greater fraction (88–92%) of those class I genes that were dependent on pituitary hormone for expression in both males and females (designated class IB genes; Table 3), as compared with genes that showed a pituitary dependence in only one sex (class IA genes; 41–44%), indicating direct GH regulation of the class IB genes via a mechanism that is common to both sexes. Direct GH induction also characterizes the sex-independent GH-responsive genes Igf1 and Socs2, where STAT5b is a key transcriptional activator (27,47,48). Conceivably, class IB sex-specific genes may have strong binding sites for GH-activated STAT5b, which would enable them to be activated both in male liver, where the high but pulsatile plasma GH levels are associated with high nuclear concentrations of STAT5b, and in female liver, where the lower but more continuous plasma GH levels are associated with lower levels of active, nuclear STAT5b (12,14). Similarly, sex-independent genes that are STAT5b dependent, such as Igf1 and Socs2, may have stronger STAT5b binding sites than the STAT5b-regulated class IA male-specific genes, which could explain why the sex-independent genes are expressed in female liver at the same level as in male liver, despite the much lower peak STAT5b levels in the females. Further investigation is required to determine whether the sex-specific early GH response genes presently identified contain an overrepresentation of paired STAT5b-binding sites, which characterize many direct STAT5b target genes (43,49), or in the case of the class I female genes, if binding sites for STAT5a (50) are overrepresented.
At least some of the sex-specific genes that did not respond to short-term GH treatment may be regulated by GH via indirect mechanisms, as was previously suggested based on the rather long time (several days) required for certain sex-specific mouse liver mRNAs to respond to changes in plasma GH status (8). GH regulation of these genes may be mediated by sex-specific transcription factors that serve as direct targets of GH. Indeed, three class I female-specific DNA-binding proteins, Cutl2 (Cux2 gene product), HNF6 (Onecut1 gene product), and Nucleolin were found to be rapidly induced by GH, in agreement with an earlier study in the case of HNF6 (37). Cutl2 is a female-specific transcription factor that has the potential to repress male-specific genes in female liver (24), whereas HNF6 is a female-predominant factor that also has a Cut domain and may contribute to the female-specific expression of Cyp2c12 (22,23). Among the class IIA sex-specific genes suppressed by GH, two encode transcription factors showing male specificity (supplemental Table S10). Of these, the transcriptional repressor Bcl6 showed the highest sex-specificity (M/F = 15.8). Further studies are required to determine whether these or other sex-specific signaling molecules are involved in the regulation of downstream sex-specific targets by GH/STAT5b. Eighteen other DNA-binding protein genes that were rapidly induced (11 genes) or suppressed (seven genes) by GH did not show sex specificity in their expression (supplemental Table S10). Presumably, a subset of these genes contributes to the secondary effects of GH that are common to male and female liver.
In conclusion, GH confers sex specificity in rat liver through both positive and negative regulation, which may involve both direct and indirect mechanisms. Future studies will be required to determine the role of the early GH-responsive transcriptional regulators identified in this study in the global pattern of sex-specific liver gene expression. Finally, the new insights into the complex regulatory networks involved in the sex-dependent patterns of Cyps and other liver-expressed genes provided by the present studies may further our understanding of the role of GH in regulating the sex specificity in human liver of genes such as CYP3A4 (2), which plays a major role in human hepatic drug and steroid metabolism.
MATERIALS AND METHODS
Animal Treatments and Liver RNA Isolation
Male and female Fischer 344 rats, untreated or hypophysectomized at 8 wk of age (150–165 g), were purchased from Taconic Farms, Inc. (Germantown, NY). Rats were housed on Care Fresh bedding (catalog no. 2680; Scott’s Distributing, Inc., Hudson, NH) on a 12-h light, 12-h dark cycle with free access to food (Pro Lab Rat/Mouse/Hamster 3000; catalog no. 6600, Scott’s Distributing, Inc.) and drinking water. The completeness of hypophysectomy was confirmed by the absence of weight gain over a 4-wk period after surgery (weights monitored three times/wk, with final weights all ≤initial weight at time of surgery). Rats showing trends of increasing weight over the 4-wk monitoring period were excluded from the study. Hypox male rats were treated with a single ip injection of rat GH at 6 μg/100 g of body weight (rGH-B-14-SIAFP; National Hormone and Pituitary Program, NIDDK) and killed 30, 60, or 90 min later. A second group of Hypox male rats was given two ip injections of GH, spaced either 3 or 4 h apart, and then killed 60 min after the second GH injection. GH pulse treatments used in this study were given to Hypox male rats because a plasma GH pulse is the natural form of GH stimulation in males. GH pulse treatment of Hypox females, a nonphysiological form of GH replacement, was not tested in this study. Rats were euthanized by brief CO2 treatment followed by cervical dislocation. Livers were excised, flash frozen in liquid N2, and stored at −80 C. Total RNA was isolated from individual livers using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA). Livers from the following six groups were used for microarray analysis: intact, untreated male rats (M-UT; n = 7); intact, untreated female rats (F-UT; n = 6); Hypox male rats (M-Hypox; n = 6); Hypox female rats (F-Hypox; n = 7); Hypox male rats treated with a single GH injection and killed 30, 60, or 90 min later (M-Hypox + GH; n = 4), and Hypox male rats treated with two GH injections (M-Hypox + 2GH; n = 5). Liver RNA from M-Hypox rats given a single GH injection and killed 30, 60, and 90 min later were pooled for the microarray analysis because of the small numbers of individuals (n = 2 or 3) available in each group. These RNAs were assayed individually in the qPCR analysis, as described below.
Microarray Platform
The Agilent Whole Rat Genome Microarray platform (catalog no. G4131F, 4 x 44K slide format; Agilent Technology, Palo Alto, CA) was used for global gene expression analysis. This microarray contains 41,012 rat cDNA probes (features), each comprising a single 60-oligomer oligonucleotide sequence. In annotation assignments made by Aarathi Sugathan of this laboratory, accession numbers could be assigned to 39,308 of the 39,688 probes for which the manufacturer provided chromosomal location information: rat GenBank accession numbers were assigned for 36,383 of the probes; rat Ensembl transcript identifications (IDs) for 168 other probes; and nonrat accession numbers for 2,757 probes for which no rat annotations were available. Together, these probes encompass 23,642 unique rat accession numbers and 2,270 unique nonrat accession numbers and represent 16,947 rat Unigene IDs plus 5,941 nonrat Unigene IDs (Unigene build 166). Two or more probes mapping to a common accession number indicate redundancy in the microarray platform. Each of the probes assigned a unique accession number is herein referred to as representing a distinct gene (i.e. gene product). The true number of unique transcripts represented on the array is likely to be fewer than this number due to probes mapping to regions of the rat genome that are poorly (or incorrectly) annotated. Liver RNA samples having an RNA Integrity Analysis number greater than 8.0, determined using an Agilent Bioanalyzer 2100, were used for microarray analysis.
Sample Preparation, Hybridization, and Data Acquisition
Liver RNA pools were prepared for two independent sets of biological replicates for the M-UT, F-UT, M-Hypox, and F-Hypox groups (n = 3–4 livers per pool; two pools per group). A single pool of liver RNA (n = 4 livers from M-Hypox rats treated with a single GH injection and killed either 30, 60, or 90 min later) was used for the M-Hypox + GH group, and a single pool (n = 5 livers) for the M-Hypox + 2GH group. These RNA pools were used in six separate sets of competitive hybridization experiments in a loop design: 1) M-UT vs. F-UT; 2) M-Hypox vs. M-UT; 3) F-Hypox vs. F-UT; 4) M-Hypox vs. F-Hypox; 5) M-Hypox + GH vs. M-Hypox; 6) M-Hypox + 2GH vs. M-Hypox. Sample labeling, hybridization to microarrays, scanning, analysis of TIFF images with Agilent’s feature extraction software, calculation of linear and LOWESS normalized expression ratios, and initial data analysis using Rosetta Resolver (version 5.1; Rosetta Biosoftware) were carried out at the Wayne State University Institute of Environmental Health Sciences microarray facility (Detroit, MI) as detailed elsewhere (21). Dye-swapping experiments were carried out for each of the six hybridization experiments, as follows. The Alexa 555-labeled cDNA from one of the two M-UT pools was mixed with the Alexa 647-labeled cDNA from one of the two F-UT pools. Similarly, Alexa 647-labeled cDNA from the second M-UT pool was mixed with Alexa 555-labeled cDNA from the second F-UT pool. Together, these two mixed cDNA samples are considered a fluorescent reverse pair (dye swap). Dye swaps were similarly carried out for each of the five other competitive hybridization experiments, except that for experiments 5 and 6, a single pool of M-Hypox + GH liver cDNA, or a single pool of M-Hypox + 2GH liver cDNA, was used in each half of the fluorescent reverse pair. Two microarrays, one for each mixed cDNA sample, were hybridized for each of the six fluorescent reverse pairs, giving a total of 12 microarrays. Expression ratios obtained in this study are available for query or download from the Gene Expression Omnibus web site (http://www. ncbi.nlm.nih.gov/geo) as GEO series GSE-11529.
Statistical Analysis
A filter (P < 0.005) was applied to the P values obtained from Rosetta Resolver to determine the statistical significance of each gene’s differential expression for each of the six microarray experiments. A fold-change filter of 2.0-fold was combined with the above P value filter to reduce the false discovery rate to less than 1%. Thus, of the 41,012 features (probes) included on the array, 6,932 met the 2.0-fold expression filter for at least one of the six microarray comparisons. The number of probes expected to meet the combined threshold (P < 0.005 and >2.0-fold change in expression) by chance is 0.005 × 6,932, or 35 genes. The actual number of probes meeting the combined threshold was 4,621, corresponding to an apparent false discovery rate of 35 per 4,621 or 0.76%. No major change in the overall gene distribution patterns was observed when the threshold stringency was either relaxed (>1.5-fold change and P < 0.05) or increased (>2.25-fold change and P < 0.001).
A system of binary and decimal flags was used to cluster the microarray data based on the expression ratios and P values obtained for each gene in all six microarray experiments, as described previously (15). Average ratios meeting the threshold of 2.0-fold change and P < 0.005 contributed to the binary- and decimal-based flag. Thus, genes with a M-UT vs. F-UT microarray ratio meeting both criteria were assigned a binary flag value of 1, whereas genes meeting the criteria for the microarrays that analyzed M-Hypox vs. M-UT, F-Hypox vs. F-UT, M-Hypox vs. F-Hypox, M-Hypox + GH vs. M-Hypox, and M-Hypox + 2GH vs. M-Hypox were assigned, respectively, binary flag values of 2, 4, 8, 16, and 32. Genes not meeting these criteria were assigned flag values of 0. The sum of these binary-based flag values defines the whole number portion of the flag and was used as a simple method to identify which of the six microarrays met our criteria for inclusion for any given gene of interest, regardless of the direction (up or down) of the regulation. The flag value was then extended using decimal values of 0.1, 0.01, 0.001, 0.0001, 0.00001, and 0.000001 or 0.2, 0.02, 0.002, 0.0002, 0.00002, and 0.000002 for each of the six microarrays, to indicate the direction of regulation between the two conditions on the microarray. Thus, average ratios for M-UT vs. F-UT microarray greater than 2.0 were assigned a decimal value of 0.1, to indicate up-regulation, whereas average ratios less than 0.5 were assigned a value of 0.2, to indicate down-regulation. The five other microarray ratios were similarly flagged, by advancing to a new decimal position for each microarray (i.e. the M-Hypox: M-UT flag is in the hundredths position, and so on). For each gene, the resulting binary sum describes which microarray ratios met the selection criteria, and the six-digit decimal value describes the direction of regulation (Total Flagging Sum, TFS; see supplemental Tables S3 and S4). Of the 41,012 probes, 4,621 met the threshold criteria (average expression ratio >2-fold and a significance of P < 0.005) for at least one of the six data sets and were included in our analysis. In each case where two or more probes for the same transcript (as defined by a common accession number assignment) gave the same pattern of regulation, as indicated by mapping to the same TFS group (i.e. redundant probes), only the probes with the best P values were retained. The number of genes meeting the threshold criteria was thus reduced to from 4621 to 4150. These 4150 genes of interest, as well as the 471 redundant probes excluded from further analysis, are presented in supplemental Table S2.
GO Enrichment Analysis
Gene lists were analyzed for enrichment of GO categories and biological pathways using DAVID (http://david.abcc. ncifcrf.gov), a web-accessible bioinformatics database (51,52). Each gene was categorized per the GO Consortium designations using biological process and molecular function ontology levels (53). Categories enriched in each list as compared with the species background were identified using the DAVID P value, and those with a P value < 0.01 were selected for subsequent analysis. Enrichment in KEGG pathways was determined with DAVID, with results presented for pathways significant at P < 0.05.
qPCR Analysis
Liver RNA samples were prepared from the following eight groups of rats: untreated males (M, n = 8); Hypox males (n = 6); Hypox males treated with a single GH injection and killed after 30 min (n = 2), 60 min (n = 3) and 90 min (n = 3); Hypox males killed 1 h after a second GH injection given 3–4 h after the first injection (n = 3), untreated females (n = 7) and Hypox females (n = 7). Liver RNA samples were converted to cDNA and assayed for the expression of individual genes by qPCR (8). Amplification of a single, specific product during qPCR cycling was verified by examination of the dissociation curve of each amplicon. For untreated males, untreated females, Hypox males, and Hypox females, qPCR analysis was performed on pooled liver cDNA samples; for Hypox rats treated with GH, individual liver cDNA samples were used. Relative RNA levels were determined after normalization to the 18S RNA content of each sample. Statistical analysis was carried out by one-way ANOVA and Bonferoni posttest with the level of significance set at P < 0.05 using GraphPad Prism software version 4 (GraphPad Software, Inc., San Diego, CA). qPCR primer design was carried out using Primer Express software (Applied Biosystems, Foster City, CA), and all primers were verified with respect to their specificity for the target transcript by BLAST-like alignment Tool analysis (BLAT) of the rat genome (November 2004 assembly) at http://genome.ucsc.edu/ cgi-bin/hgBlat. Where indicated, primary nuclear transcripts (hnRNA) were assayed using qPCR primer sets that amplified across an exon-intron junction. Primer sequences are shown in supplemental Table S1.
Supplementary Material
Acknowledgments
We thank Aarathi Sugathan for probe annotation analysis and Dr. Rosana Meyer for assistance with qPCR analysis presented in the supplemental data.
Footnotes
This work was supported by National Institutes of Health Grant DK33765 (to D.J.W.) V.W. received fellowship support from the Belgian American Educational Foundation and from the Catholic University of Louvain.
Author Disclosure Summary: V.W. and D.J.W. have nothing to declare.
First Published Online May 15, 2008
Abbreviations: CYP, Cytochrome P450; F-Hypox, hypophysectomized female; F-UT, untreated female; GO, gene ontology; HNF, hepatocyte-enriched nuclear factor; hnRNA, heterogeneous nuclear RNA (primary RNA transcript); Hypox, hypophysectomized; ID, identification; M-Hypox, hypophysectomized male; M-UT, untreated male; qPCR, quantitative real-time PCR; STAT, signal transducer and activator of transcription; TFS, total flagging sum.
References
- Mode A, Gustafsson JA 2006 Sex and the liver—a journey through five decades. Drug Metab Rev 38:197–207 [DOI] [PubMed] [Google Scholar]
- Waxman DJ, O'Connor C 2006 Growth hormone regulation of sex-dependent liver gene expression. Mol Endocrinol 20:2613–2639 [DOI] [PubMed] [Google Scholar]
- Jansson JO, Eden S, Isaksson O 1985 Sexual dimorphism in the control of growth hormone secretion. Endocr Rev 6:128–150 [DOI] [PubMed] [Google Scholar]
- Shapiro BH, Agrawal AK, Pampori NA 1995 Gender differences in drug metabolism regulated by growth hormone. Int J Biochem Cell Biol 27:9–20 [DOI] [PubMed] [Google Scholar]
- Waxman DJ, Pampori NA, Ram PA, Agrawal AK, Shapiro BH 1991 Interpulse interval in circulating growth hormone patterns regulates sexually dimorphic expression of hepatic cytochrome P450. Proc Natl Acad Sci USA 88:6868–6872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundseth SS, Alberta JA, Waxman DJ 1992 Sex-specific, growth hormone-regulated transcription of the cytochrome P450 2C11 and 2C12 genes. J Biol Chem 267:3907–3914 [PubMed] [Google Scholar]
- Legraverend C, Mode A, Westin S, Strom A, Eguchi H, Zaphiropoulos PG, Gustafsson JA 1992 Transcriptional regulation of rat P-450 2C gene subfamily members by the sexually dimorphic pattern of growth hormone secretion. Mol Endocrinol 6:259–266 [DOI] [PubMed] [Google Scholar]
- Holloway MG, Laz EV, Waxman DJ 2006 Co-dependence of growth hormone-responsive, sexually dimorphic hepatic gene expression on signal transducer and activator of transcription 5b and hepatic nuclear factor 4-α. Mol Endocrinol 20:647–660 [DOI] [PubMed] [Google Scholar]
- Pampori NA, Shapiro BH 1996 Feminization of hepatic cytochrome P450s by nominal levels of growth hormone in the feminine plasma profile. Mol Pharmacol 50:1148–1156 [PubMed] [Google Scholar]
- O'Shea JJ, Gadina M, Schreiber RD 2002 Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell 109(Suppl):S121–S131 [DOI] [PubMed] [Google Scholar]
- Waxman DJ, Ram PA, Park SH, Choi HK 1995 Intermittent plasma growth hormone triggers tyrosine phosphorylation and nuclear translocation of a liver-expressed, Stat 5-related DNA binding protein. Proposed role as an intracellular regulator of male-specific liver gene transcription. J Biol Chem 270:13262–13270 [DOI] [PubMed] [Google Scholar]
- Choi HK, Waxman DJ 2000 Plasma growth hormone pulse activation of hepatic JAK-STAT5 signaling: developmental regulation and role in male-specific liver gene expression. Endocrinology 141:3245–3255 [DOI] [PubMed] [Google Scholar]
- Tannenbaum GS, Choi HK, Gurd W, Waxman DJ 2001 Temporal relationship between the sexually dimorphic spontaneous GH secretory profiles and hepatic STAT5 activity. Endocrinology 142:4599–4606 [DOI] [PubMed] [Google Scholar]
- Choi HK, Waxman DJ 1999 Growth hormone, but not prolactin, maintains, low-level activation of STAT5a and STAT5b in female rat liver. Endocrinology 140:5126–5135 [DOI] [PubMed] [Google Scholar]
- Clodfelter K, Holloway MG, Hodor P, Park S-H, Ray WJ, Waxman DJ 2006 Sex-dependent liver gene expression is extensive and largely dependent upon STAT5b: STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Mol Endocrinol 20:1333–1351 [DOI] [PubMed] [Google Scholar]
- Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, Waxman DJ, Davey HW 1997 Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci USA 94:7239–7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey HW, Park SH, Grattan DR, McLachlan MJ, Waxman DJ 1999 STAT5b-deficient mice are growth hormone pulse-resistant. Role of STAT5b in sex-specific liver p450 expression. J Biol Chem 274:35331–35336 [DOI] [PubMed] [Google Scholar]
- Verma AS, Dhir RN, Shapiro BH 2005 Inadequacy of the Janus kinase 2/signal transducer and activator of transcription signal transduction pathway to mediate episodic growth hormone-dependent regulation of hepatic CYP2C11. Mol Pharmacol 67:891–901 [DOI] [PubMed] [Google Scholar]
- Park SH, Waxman DJ 2001 Inhibitory cross-talk between STAT5b and liver nuclear factor HNF3β: impact on the regulation of growth hormone pulse-stimulated, male-specific liver cytochrome P-450 gene expression. J Biol Chem 276:43031–43039 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Teixeira J, Wang J, Gil G 1995 A STAT factor mediates the sexually dimorphic regulation of hepatic cytochrome P450 3A10/lithocholic acid 6 β-hydroxylase gene expression by growth hormone. Mol Cell Biol 15:4672–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway MG, Miles GD, Dombkowski AA, Waxman DJ 2008 Liver-specific HNF4-α deficiency: greater impact on gene expression in male than in female mouse liver. Mol Endocrinol 1274–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delesque-Touchard N, Park SH, Waxman DJ 2000 Synergistic action of hepatocyte nuclear factors 3 and 6 on CYP2C12 gene expression and suppression by growth hormone-activated STAT5b. Proposed model for female specific expression of CYP2C12 in adult rat liver. J Biol Chem 275:34173–34182 [DOI] [PubMed] [Google Scholar]
- Lahuna O, Fernandez L, Karlsson H, Maiter D, Lemaigre FP, Rousseau GG, Gustafsson J, Mode A 1997 Expression of hepatocyte nuclear factor 6 in rat liver is sex-dependent and regulated by growth hormone. Proc Natl Acad Sci USA 94:12309–12313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laz EV, Holloway MG, Chen CS, Waxman DJ 2007 Characterization of three growth hormone-responsive transcription factors preferentially expressed in adult female liver. Endocrinology 148:3327–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahluwalia A, Clodfelter KH, Waxman DJ 2004 Sexual dimorphism of rat liver gene expression: regulatory role of growth hormone revealed by deoxyribonucleic acid microarray analysis. Mol Endocrinol 18:747–760 [DOI] [PubMed] [Google Scholar]
- Flores-Morales A, Stahlberg N, Tollet-Egnell P, Lundeberg J, Malek RL, Quackenbush J, Lee NH, Norstedt G 2001 Microarray analysis of the in vivo effects of hypophysectomy and growth hormone treatment on gene expression in the rat. Endocrinology 142:3163–3176 [DOI] [PubMed] [Google Scholar]
- Vidal OM, Merino R, Rico-Bautista E, Fernandez-Perez L, Chia DJ, Woelfle J, Ono M, Lenhard B, Norstedt G, Rotwein P, Flores-Morales A 2007 In vivo transcript profiling and phylogenetic analysis identifies suppressor of cytokine signaling 2 as a direct signal transducer and activator of transcription 5b target in liver. Mol Endocrinol 21:293–311 [DOI] [PubMed] [Google Scholar]
- Ono M, Chia DJ, Merino-Martinez R, Flores-Morales A, Unterman TG, Rotwein P 2007 Signal transducer and activator of transcription (Stat) 5b-mediated inhibition of insulin-like growth factor binding protein-1 gene transcription: a mechanism for repression of gene expression by growth hormone. Mol Endocrinol 21:1443–1457 [DOI] [PubMed] [Google Scholar]
- Clodfelter KH, Miles GD, Wauthier V, Holloway MG, Zhang X, Hodor P, Ray WJ, Waxman DJ 2007 Role of STAT5a in regulation of sex-specific gene expression in female but not male mouse liver revealed by microarray analysis. Physiol Genomics 31:63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ 2006 Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res 16:995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman DJ, LeBlanc GA, Morrissey JJ, Staunton J, Lapenson DP 1988 Adult male-specific and neonatally programmed rat hepatic P-450 forms RLM2 and 2a are not dependent on pulsatile plasma growth hormone for expression. J Biol Chem 263:11396–11406 [PubMed] [Google Scholar]
- Agrawal AK, Shapiro BH 2000 Differential expression of gender-dependent hepatic isoforms of cytochrome P-450 by pulse signals in the circulating masculine episodic growth hormone profile of the rat. J Pharmacol Exp Ther 292:228–237 [PubMed] [Google Scholar]
- Agrawal AK, Shapiro BH 2001 Intrinsic signals in the sexually dimorphic circulating growth hormone profiles of the rat. Mol Cell Endocrinol 173:167–181 [DOI] [PubMed] [Google Scholar]
- Gardmo C, Persson B, Mode A 2001 Cloning of a novel growth hormone-regulated rat complementary deoxyribonucleic acid with homology to the human α1B-glycoprotein, characterizing a new protein family. Endocrinology 142:2695–2701 [DOI] [PubMed] [Google Scholar]
- Sakuma T, Kitajima K, Nishiyama M, Mashino M, Hashita T, Nemoto N 2004 Suppression of female-specific murine Cyp2b9 gene expression by growth or glucocorticoid hormones. Biochem Biophys Res Commun 323:776–781 [DOI] [PubMed] [Google Scholar]
- Bichell DP, Kikuchi K, Rotwein P 1992 Growth hormone rapidly activates insulin-like growth factor I gene transcription in vivo. Mol Endocrinol 6:1899–1908 [DOI] [PubMed] [Google Scholar]
- Lahuna O, Rastegar M, Maiter D, Thissen JP, Lemaigre FP, Rousseau GG 2000 Involvement of STAT5 (signal transducer and activator of transcription 5) and HNF-4 (hepatocyte nuclear factor 4) in the transcriptional control of the hnf6 gene by growth hormone. Mol Endocrinol 14:285–294 [DOI] [PubMed] [Google Scholar]
- Gronowski AM, Rotwein P 1995 Rapid changes in gene expression after in vivo growth hormone treatment. Endocrinology 136:4741–4748 [DOI] [PubMed] [Google Scholar]
- Waxman DJ, Ram PA, Pampori NA, Shapiro BH 1995 Growth hormone regulation of male-specific rat liver P450s 2A2 and 3A2: induction by intermittent growth hormone pulses in male but not female rats rendered growth hormone deficient by neonatal monosodium glutamate. Mol Pharmacol 48:790–797 [PubMed] [Google Scholar]
- Tullai JW, Schaffer ME, Mullenbrock S, Sholder G, Kasif S, Cooper GM 2007 Immediate-early and delayed primary response genes are distinct in function and genomic architecture. J Biol Chem 282:23981–23995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampori NA, Shapiro BH 1999 Gender differences in the responsiveness of the sex-dependent isoforms of hepatic P450 to the feminine plasma growth hormone profile. Endocrinology 140:1245–1254 [DOI] [PubMed] [Google Scholar]
- Woelfle J, Billiard J, Rotwein P 2003 Acute control of insulin-like growth factor-I gene transcription by growth hormone through Stat5b. J Biol Chem 278:22696–22702 [DOI] [PubMed] [Google Scholar]
- Verdier F, Rabionet R, Gouilleux F, Beisenherz-Huss C, Varlet P, Muller O, Mayeux P, Lacombe C, Gisselbrecht S, Chretien S 1998 A sequence of the CIS gene promoter interacts preferentially with two associated STAT5A dimers: a distinct biochemical difference between STAT5A and STAT5B. Mol Cell Biol 18:5852–5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram PA, Park SH, Choi HK, Waxman DJ 1996 Growth hormone activation of Stat 1, Stat 3, and Stat 5 in rat liver. Differential kinetics of hormone desensitization and growth hormone stimulation of both tyrosine phosphorylation and serine/threonine phosphorylation. J Biol Chem 271:5929–5940 [DOI] [PubMed] [Google Scholar]
- Miquet JG, Sotelo AI, Dominici FP, Bonkowski MS, Bartke A, Turyn D 2005 Increased sensitivity to GH in liver of Ames dwarf (Prop1df/Prop1df) mice related to diminished CIS abundance. J Endocrinol 187:387–397 [DOI] [PubMed] [Google Scholar]
- Holloway MG, Cui Y, Laz EV, Hosui A, Hennighausen L, Waxman DJ 2007 Loss of sexually dimorphic liver gene expression upon hepatocyte-specific deletion of Stat5a-Stat5b locus. Endocrinology 148:1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia DJ, Ono M, Woelfle J, Schlesinger-Massart M, Jiang H, Rotwein P 2006 Characterization of distinct Stat5b binding sites that mediate growth hormone-stimulated IGF-I gene transcription. J Biol Chem 281:3190–3197 [DOI] [PubMed] [Google Scholar]
- Wang Y, Jiang H 2005 Identification of a distal STAT5-binding DNA region that may mediate growth hormone regulation of insulin-like growth factor-I gene expression. J Biol Chem 280:10955–10963 [DOI] [PubMed] [Google Scholar]
- Meyer WK, Reichenbach P, Schindler U, Soldaini E, Nabholz M 1997 Interaction of STAT5 dimers on two low affinity binding sites mediates interleukin 2 (IL-2) stimulation of IL-2 receptor α gene transcription. J Biol Chem 272:31821–31828 [DOI] [PubMed] [Google Scholar]
- Soldaini E, John S, Moro S, Bollenbacher J, Schindler U, Leonard WJ 2000 DNA binding site selection of dimeric and tetrameric Stat5 proteins reveals a large repertoire of divergent tetrameric Stat5a binding sites. Mol Cell Biol 20:389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis Jr G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA 2003 DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4:P3 [PubMed] [Google Scholar]
- Hosack DA, Dennis Jr G, Sherman BT, Lane HC, Lempicki RA 2003 Identifying biological themes within lists of genes with EASE. Genome Biol 4:R70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G 2000 Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.