Heavy-atom derivatization is routinely used in protein structure determination and is thus of critical importance in structural biology. In order to replace the current trial-and-error heavy-atom derivative screening with a knowledge-based rational derivative-selection method, the reactivity of more than 40 heavy-atom compounds over a wide range of buffer and pH values was systematically examined using peptides which contained a single reactive amino-acid residue.
Keywords: heavy-atom derivatization, heavy-atom screening
Abstract
Heavy-atom derivatization is routinely used in protein structure determination and is thus of critical importance in structural biology. In order to replace the current trial-and-error heavy-atom derivative screening with a knowledge-based rational derivative-selection method, the reactivity of more than 40 heavy-atom compounds over a wide range of buffer and pH values was systematically examined using peptides which contained a single reactive amino-acid residue. Met-, Cys- and His-containing peptides were derivatized against Hg, Au and Pt compounds, while Tyr-, Glu-, Asp-, Asn- and Gln-containing peptides were assessed against Pb compounds. A total of 1668 reactive conditions were examined using mass spectrometry and were compiled into heavy-atom reactivity tables (http://sis.niaid.nih.gov/cgi-bin/heavyatom_reactivity.cgi). The results showed that heavy-atom derivatization reactions are highly linked to buffer and pH, with the most accommodating buffer being MES at pH 6. A group of 21 compounds were identified as most successful irrespective of ligand or buffer/pH conditions. To assess the applicability of the peptide heavy-atom reactivity to proteins, lysozyme crystals were derivatized with a list of peptide-reactive compounds that included both known and new compounds for lysozyme derivatization. The results showed highly consistent heavy-atom reactivities between the peptides and lysozyme.
1. Introduction
Determination of crystallographic phases is vital in the solution of a protein crystal structure. In principle, the phase problem can be solved by four existing techniques: direct methods, molecular replacement, isomorphous replacement and anomalous diffraction. The direct-methods option is still largely in a stage of development for macromolecular crystallography and structure solution via molecular replacement requires the pre-existence of a homologous structure. Hence, for most novel structure solutions phases must be determined experimentally by the attachment of heavy atoms to the protein molecule or by the presence of strong anomalous scattering atoms within the protein structure itself. One of the difficulties associated with heavy-atom-based phasing is the process of obtaining a heavy-atom derivative. The search for heavy-atom derivatives is still an empirical method and is very often a lengthy process that may require trials with dozens of compounds and the subsequent acquisition of multiple diffraction data sets which ultimately can still be unsuccessful. A number of techniques have been used over the years to overcome the inherent difficulties, such as the introduction of accessible cysteine residues by mutagenesis specifically for heavy-atom derivatization and subsequent structure solution (Sun et al., 1987 ▶; Hatfull et al., 1989 ▶). Known heavy-metal cofactors within a protein can be used to bypass the tedious heavy-metal screening process (Zhang et al., 1995 ▶). In recent years, selenomethionine (SeMet) or solvent halide (Br− and I−) based MAD/SAD phasing have also become important methods for protein structure determination (Hendrickson et al., 1990 ▶; Dauter et al., 2000 ▶). However, both SeMet and halide phasing have their limitations (Smith & Thompson, 1998 ▶). In particular, SeMet phasing is only applicable to recombinant proteins derived from bacterial or insect-cell expression systems. Proteins with too few SeMet sites often yield inadequate phasing, while those with too many SeMet sites may complicate the identification of the sites. More recently, attempts to use native S atoms for phasing have also been quite successful, although the method is most suitable for crystals diffracting to high resolution owing to the intrinsic low sulfur anomalous signal (∼1.5%) at the in-house copper wavelength (Dauter et al., 1999 ▶; Debreczeni et al., 2003 ▶; Wang et al., 2006 ▶). Thus, phasing by conventional heavy-atom derivatization still remains an indispensable method.
Heavy-metal compounds are commonly classified as class A or class B (Blundell & Johnson, 1976 ▶; Blundell & Jenkins, 1977 ▶). Class A compounds such as lanthanides and actinides tend to be electronegative and bind to their protein targets without covalent modification, e.g. the uranyl ion binds to the negatively charged carboxylate groups of glutamate and aspartate, as observed with insulin (Blundell et al., 1971 ▶) and prealbumin (Blake et al., 1974 ▶). Class B metals such as Pt, Au and Hg atoms are polarizable and in contrast bind covalently to sulfhydryl, imidazole and thiol groups and are the most commonly used derivatives in protein crystallography (Islam et al., 1998 ▶; Rould, 1997 ▶). Thallium and lead differ from the other B metals by favoring ligands such as carboxylate rather than imidazole and sulfur groups.
Despite our understanding of heavy-atom chemistry, the search for heavy-metal derivatives is still carried out in a trial-and-error manner. The lack of predictability often makes derivative screening a lengthy process, although the recent utilization of mass spectrometry (Sun & Hammer, 2000 ▶; Krishna et al., 1994 ▶) and native gel electrophoresis (Boggon & Shapiro, 2000 ▶) should accelerate the identification of potential derivatives for structure determination. The success of heavy-atom derivatization depends not only on the availability of specific amino-acid ligands in a given protein but also to a great extent on the crystallization conditions. It is often the case that components in the crystallization mother liquor mask potential protein ligands or even chelate heavy-atom compounds, preventing successful derivatization. Similarly, pH influences both the protonation state of functional groups and the solubility of heavy-atom reagents. Although examples of buffer effects have been provided by Blundell & Johnson (1976 ▶), to date there has been no large-scale systematic investigation of buffer influence on protein-derivatization reactions.
Mass spectrometry has increasingly found many roles within protein crystallography (Cohen & Chait, 2001 ▶) and on occasion it has been used to validate protein heavy-atom derivatives (Krishna et al., 1994 ▶). A systematic study utilizing three mercury-containing compounds and five diverse proteins further indicated that electrospray ionization mass spectrometry (ESI-MS) is the method of choice (Cohen et al., 2000 ▶). The great potential of this technique was shown by Sun & Hammer (2000 ▶) and it can be routinely used to identify protein heavy-atom derivatives.
Here, we report a systematic study using eight model peptides and mass spectrometry to analyze the effect of six commonly used buffers in the pH range 4–8.5 on the reactivity of over 40 heavy-atom compounds. The resulting pH- and buffer-dependent heavy-atom reactivity profiles can be used to facilitate the selection of heavy-atom compounds for derivatization experiments in specific crystallization conditions and should act as a reference for protein-derivatization experiments.
2. Materials and methods
2.1. Preparation of heavy-atom-reactive monoligand peptides
Peptides of 12 amino acids in length with the sequence GEAGXASAGGAG, where X represents a histidine, methionine or cysteine residue, were synthesized by the NIAID Research Technology Branch as lone functional ligands for the screening of gold-, platinum- and mercury-containing heavy-atom compounds. In the case of the lead-compound derivatization experiments, individual peptides with the sequence GAAGXASAGGAG were synthesized with the variable residue (X) being either aspartate, glutamate, asparagine, glutamine or tyrosine. The peptides for use with the gold-, platinum- and mercury-containing compounds have a glutamate substitution at position 2 when compared with the peptides used in lead-derivatization experiments in order to increase their water solubility. All peptide sequences and masses were confirmed by sequencing and ESI-MS. Peptide heavy-atom derivative experiments were carried out by mixing 2 µl of a heavy-atom compound solution at either 10 mM or saturated concentration (Table 1 ▶) in H2O with 3 µl of a peptide solution in H2O at a 1:2 peptide:heavy-atom molar ratio. 2 µl of a 34 mM specific buffer stock was added to this solution to give a final 10 mM buffer solution. After 10 min at room temperature, the derivatization reactions were quenched by the addition of 2 µl of a 10 mM imidazole solution before infusion of the sample into the mass spectrometer. Similar results were obtained for a higher peptide:heavy metal molar ratio (1:8) and also for 24 h derivatization reactions.
Table 1. List of ligands, metals and buffers used in peptide mass-spectrometry experiments.
| Heavy-atom compounds | Chemical formula | Vendor/catalog No.† | Peptides | Buffers |
|---|---|---|---|---|
| Platinum | All platinum-containing compounds were from Hampton Research HR2-422 | GEAGMASAGGAG | HEPES pH 78 | |
| Potassium tetrachloroplatinate(II)‡ | K2PtCl4 | GEAGCASAGGAG | MES pH 6 | |
| Platinum potassium thiocyanate | K2Pt(SCN)6 | GEAGHASAGGAG | Sodium acetate pH 45 | |
| Platinum potassium iodide§ | K2PtI6 | Sodium cacodylate pH 5.56.5 | ||
| Potassium hexabromoplatinate(IV)‡ | K2PtBr6 | Sodium citrate pH 46 | ||
| Potassium tetrabromoplatinate(II)‡ § | K2PtBr4 | TrisHCl pH 7.58.5 | ||
| Diamino platinum dinitrate‡ § | Pt(NH3)2(NO2)2 | |||
| Potassium tetranitroplatinate(II) | K2Pt(NO2)4 | |||
| Dichloro(ethylenediamine)platinum(II)§ | PtCl2(H2NC3H6NH2) | |||
| Potassium tetracyanoplatinate(II) | K2Pt(CN)4 | |||
| Ammonium tetrachloroplatinate(II) | (NH4)PtCl4 | |||
| Potassium hexachloroplatinate(IV) | K2PtCl6 | |||
| Gold | ||||
| Potassium tetrachloroaurate(III) | KAuCl4 | HR2-444 | GEAGMASAGGAG | HEPES pH 78 |
| Sodium tetrachloroaurate(III) | NaAuCl4 4 | HR2-444 | GEAGCASAGGAG | MES pH 6 |
| Gold(III) chloride‡ § | AuCl3 | HR2-444 | GEAGHASAGGAG | Sodium acetate pH 45 |
| Gold chloride | HAuCl4 | HR2-444 | Sodium cacodylate pH 5.56.5 | |
| Gold sodium bromide | NaAuBr4 | HR2-444 | Sodium citrate pH 46 | |
| Potassium tetrabromoaurate(III) | KAuBr4 | Strem Chemicals 79-3250 | TrisHCl pH 7.58.5 | |
| Gold sodium thiosulfate§ | Na3Au(S2O3)2 | Alfa Aesar 39741 | ||
| Gold(I) potassium cyanide | KAu(CN)2 | HR2-444 | ||
| Gold potassium thiocyanide§ | KAu(SCN)4 | Alfa Aesar 39666 | ||
| Mercury | ||||
| Thiomersal‡ | C9H9HgNaO2S | HR2-446 | GEAGMASAGGAG | HEPES pH 78 |
| Mersalyl‡ § | HOHgCH2CH(OCH3)CH2NHCOC6H4OCH2CO2H | HR2-446 | GEAGCASAGGAG | MES pH 6 |
| Potassium mercury tetraiodide§ | K2(HgI4) | HR2-446 | GEAGHASAGGAG | Sodium acetate pH 45 |
| Methylmercury(II) acetate‡ § | CH3HgO2CCH3 | Pfaltz Bauer M21875 | Sodium cacodylate pH 5.56.5 | |
| Ethylmercury(II) phosphate‡ | (C2H5HgO)HPO2 | HR2-446 | Sodium citrate pH 46 | |
| p-Chloromercuribenzenesulfonic acid (PCMBS) | C6H5ClHgSO3 | Sigma C4503 | TrisHCl pH 7.58.5 | |
| Mercury(II) acetate‡ § | Hg(OOCCH3)2 | HR2-446 | ||
| Methylmercury(II) chloride‡ § | CH3HgCl | HR2-446 | ||
| Mercury(II) cyanide | Hg(CN)2 | HR2-446 | ||
| Mercury(II) bromide‡ § | HgBr2 | HR2-446 | ||
| Mercury(II) chloride | HgCl2 | HR2-446 | ||
| Methylmercury(II) bromide | Ch3HgBr | HR2-446 | ||
| p-Chloromercuribenzoic acid (PCMB) | C7H5ClHgO2 | HR2-446 | ||
| Ethylmercury(II) chloride | C2H5HgCl | HR2-446 | ||
| Mercury(II) iodide§ | HgI2 | HR2-446 | ||
| Mercury(II) oxide | HgO | HR2-446 | ||
| Other | ||||
| Potassium hexachloroosmate(IV)§ | K2OsCl6 | HR2-448 | GEAGMASAGGAG | HEPES pH 78 |
| Potassium hexachloroiridate(III)§ | K2IrCl6 | HR2-448 | GEAGCASAGGAG | MES pH 6 |
| GEAGHASAGGAG | Sodium acetate pH 45 | |||
| Sodium cacodylate pH 5.56.5 | ||||
| Sodium citrate pH 46 | ||||
| TrisHCl pH 7.58.5 | ||||
| Lead | ||||
| Lead acetate‡ | Pb(CH3COO)2 | Sigma L3396 | GAAGDASAGGAG | HEPES pH 78 |
| Trimethyllead acetate | CH3CO2Pb(CH3)3 | Sigma 116815 | GAAGEASAGGAG | MES pH 6 |
| Hexaphenyl dilead§ | [(C6H5)3Pb]2 | Alfa 57132 | GAAGNASAGGAG | Sodium acetate pH 45 |
| Lead nitrate‡ | Pn(NO3)2 | Sigma L6258 | GAAGQASAGGAG | Sodium cacodylate pH 5.56.5 |
| Triethyllead acetate‡ | Ch3CO2Pb(C2H5)3 | Sigma 116823 | GAAGYASAGGAG | Sodium citrate pH 46 |
| TrisHCl pH 7.58.5 |
HR2-422, HR2-444, HR2-446 and HR2-448 are from Hampton Research.
Compounds used to derivatize lysozyme.
Compounds that were not fully soluble at 10mM.
2.2. Mass-spectrometry measurements
Intact mass analyses were performed on purified peptide or peptide–heavy metal complex solutions by ESI-MS. Samples were analyzed by coupling the Nanomate (Advion BioSciences, Ithaca, New York, USA), an automated chip-based nano-electrospray unit, to a quadrupole–time of flight (TOF) mass spectrometer (QStarXL MS/MS System; Applied Biosystems/Sciex, Framingham, Massachusetts, USA). AnalystQS and BioAnalyst software (Applied Biosystems/Sciex) were used for data acquisition and processing, respectively. TOF mass calibration was performed at regular intervals to ensure mass accuracy. A solution of Glu-fibrinopeptide B at 1 pmol µl−1 (F3261, Sigma–Aldrich, St Louis, Missouri, USA) and m + 2H/2 = 785.8 was acquired in product-ion mode (ms/ms) to obtain the 175.1190 and 1285.5444 m/z ions used for two-point calibration. Peaks corresponding to both the native and heavy-atom-derivatized peptides were identified based on their match to predicted molecular masses to within two mass units. The extent of heavy-atom reactivity was evaluated based on the peak heights of observed derivatives and were assigned on a four-level scale as either −, +, ++ or +++ for no significant derivative adduct formation and derivative adducts with intensity less than 25%, between 25 and 50% and above 50% of the native peak intensity, respectively.
In order to assess the applicability of peptide–heavy atom results to proteins, hen egg-white lysozyme derivatization was assessed as follows. 1 µl of a lysozyme solution (10 mg ml−1) in 50 mM sodium acetate buffer pH 4.6 was incubated with 1 µl of a 10 mM heavy-atom compound solution for 30 min and the reaction was quenched with 1 µl of a 10 mM imidazole solution and analyzed by ESI-MS as described previously (Sun & Hammer, 2000 ▶).
2.3. Heavy-atom derivatization of lysozyme crystals
Hen egg-white lysozyme (50 mg ml−1) from Sigma was crystallized in the tetragonal form by the hanging-drop vapor-diffusion technique in 0.8 M NaCl, 50 mM sodium acetate buffer pH 4.7. Crystals were derivatized by adding lead acetate or potassium tetracyanoplatinate(II) to the mother liquor to a final concentration of 10 mM and allowing the crystals to soak for 10 min (Sun et al., 2002 ▶; Sun & Radaev, 2002 ▶). Crystals were then placed in cryoprotectant solution containing mother liquor and 25% glycerol and flash-cooled in a liquid-nitrogen cryostream. X-ray diffraction data for the two derivatives were collected in-house using Cu Kα radiation from a Rigaku RU-200 generator and an R-AXIS IV++ detector. The data were scaled and processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). Heavy-atom sites were determined by difference Fourier calculation (F PH − F P) using CNS (Brünger et al., 1998 ▶).
3. Results
3.1. Design of monofunctional peptide ligands
The reactivity of heavy-atom compounds with proteins in crystals depends on a number of factors such as the accessibility of the reactive groups and the crystallization conditions. To systematically examine the heavy-atom/protein reactivity, we synthesized eight model peptides of 12 residues in length, each of which contains a single reactive amino acid. The use of peptide ligands represents the optimal situation in accessibility for heavy-atom derivatization. Peptides with a single cysteine, methionine or histidine residue were assessed for reactivity with platinum, gold and mercury compounds, whereas peptides containing a single aspartate, glutamate, asparagine, glutamine or tyrosine residue were used in the lead-derivatization experiments. In all, a total of 43 most successful and often used heavy-atom compounds were tested for derivatization of the peptides (Table 1 ▶). Since the solubility of a heavy-atom compound in a particular buffer and pH affects its reactivity towards peptides, the buffer and pH-dependent heavy-atom solubility was examined for each of the 43 heavy-atom compounds by mixing either 10 mM or saturated heavy-atom solutions with a 50 mM concentration of the individual buffers at all pH ranges and evaluating for visual precipitation under a light microscope. The compounds that exhibited reduced solubility under particular buffer and pH conditions are indicated by • in Tables 2 and 5. The peptide reactivity under these limited solubility conditions, however, frequently resulted in high levels of peptide–heavy atom adduct formation, indicating that the reduction in solubility need not affect the heavy-atom reactivity.
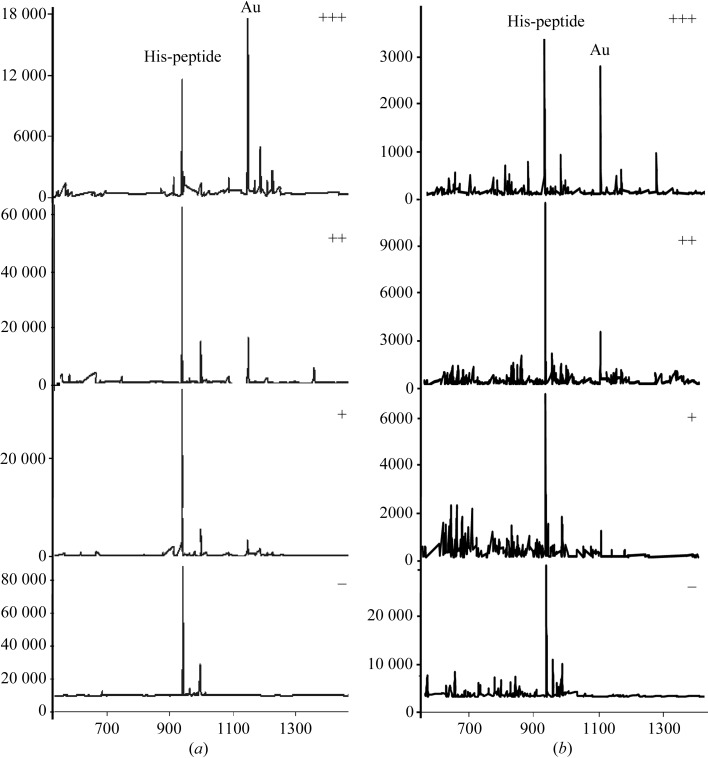
To analyze the effects of pH and buffer on derivatization, reactions were carried out in the presence of a 10 mM concentration of six commonly used crystallization buffers: HEPES (pH 7.0 and pH 8.0), MES (pH 6.0), sodium acetate (pH 4.0 and pH 5.0), sodium cacodylate (pH 5.5 and pH 6.5), sodium citrate (pH 4.0, pH 5.0 and pH 6.0) and Tris–HCl (pH 7.5 and pH 8.5). The extent of derivatization was scored based on the height of the expected derivative peak on a four-level scale: −, +, ++ and +++, representing no observable adduct (nonreactive) formation and adducts with a peak height less than 25% (minimally reactive), of between 25% and 50% (moderately reactive) and of greater than 50% (highly reactive) of the native peptides, respectively (Fig. 1 ▶). Although typical crystallization conditions contain a 50 mM or higher concentration of salts, the extent of heavy-atom derivatization was similar in 10 and 50 mM buffer concentrations, although the overall ionization efficiency was reduced by around fivefold in 50 mM buffer compared with 10 mM buffer (Fig. 1 ▶ b). Thus, to ensure the performance of ESI-MS, all buffer concentrations used in this study were limited to a 10 mM concentration of salts.
Figure 1.
Examples of mass spectra illustrating the four-level scale of heavy-atom reactivity based on the derivative peak height. (a) 10 mM buffer; (b) test case carried out in 50 mM buffer. The His-peptide derivatized by potassium tetrachloroaurate in highly reactive MES buffer at pH 6.0 (+++), moderately reactive sodium acetate buffer at pH 5.0 (++), minimal reactive sodium citrate buffer at pH 5.0 (+) and nonreactive HEPES buffer at pH 8.0 (−).
Unlike class B metals, which often form covalent adducts with one primary ligand on proteins, many class A heavy metals react noncovalently with proteins on binding sites that are formed by multiple ligands with specific stereochemistry, such as tetrahedral and octahedral coordinations (Blundell & Johnson, 1976 ▶). Owing to the difficulty of short peptides in assuming any defined stereo-geometry needed for this type of heavy-atom derivatization and the limitation of mass spectrometry in detecting ionic adducts, the reactivities of class A hard metal compounds are beyond the current approach.
3.2. Methionine derivatized by heavy-atom compounds
Heavy-atom derivatizations of the methionine-containing peptide (Met-peptide) are listed in Table 2 ▶. Amongst the compounds tested, K2PtCl4, K2PtBr6, Pt(NH3)2(NO3)2, K2Pt(NO3)4 and (NH4)2PtCl4 derivatized Met-peptide strongly in all buffers analyzed, indicating the strong reactivity of these platinum compounds with methionine. Potassium hexachloroosmate(IV) also formed derivatives with the Met-peptide in all the buffer solutions tested, but the level of derivatization was much lower than that observed for the platinum compounds.
Table 2. Heavy-atom reactivity of the methionine-containing peptide.
Compounds with reduced solubility in particular buffer and pH conditions are indicated by .
| Sodium acetate | Sodium citrate | MES | Sodium cacodylate | HEPES | TrisHCl | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | 4.0 | 5.0 | 4.0 | 5.0 | 6.0 | 6.0 | 5.5 | 6.5 | 7.0 | 8.0 | 7.5 | 8.5 |
| Platinum potassium thiocyanate | +++ | ++ | + | + | + | ++ | ++ | ++ | ++ | ++ | ++ | + |
| Platinum potassium iodide | + | |||||||||||
| Potassium hexabromoplatinate(IV) | +++ | +++ | +++ | ++ | ++ | +++ | +++ | ++ | ++ | ++ | ++ | ++ |
| Potassium tetrachloroplatinate(II) | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | ++ | + |
| Potassium tetrabromoplatinate(II) | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | ++ | ++ | +++ | +++ |
| Diamino platinum dinitrate | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Potassium tetranitroplatinate(II) | ++ | ++ | + | +++ | +++ | ++ | ++ | +++ | ++ | +++ | ++ | +++ |
| Dichloro(ehylenediamine)platinum(II) | + | + | + | + | + | + | + | + | + | ++ | + | |
| Potassium tetracyanoplatinate(II) | + | |||||||||||
| Ammonium tetrachloroplatinate(II) | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ |
| Potassium hexachloroplatinate(IV) | + | + | + | + | + | +++ | ++ | + | ++ | ++ | ||
| Potassium tetrachloroaurate(III) | + | ++ | ++ | +++ | ++ | + | + | +++ | +++ | ++ | ||
| Sodium tetrachloroaurate(III) | ++ | ++ | ++ | +++ | +++ | + | ++ | +++ | +++ | ++ | ||
| Gold(III) chloride | + | + | + | + | ++ | ++ | - | - | ||||
| Gold chloride | + | ++ | ++ | + | + | |||||||
| Gold sodium bromide | ++ | + | + | |||||||||
| Potassium tetrabromoaurate(III) | + | + | ++ | ++ | + | + | ||||||
| Gold sodium thiosulfate | + | + | + | ++ | + | + | + | + | ||||
| Gold(I) potassium cyanide | + | |||||||||||
| Gold potassium thiocyanide | + | + | + | |||||||||
| Thiomersal | + | + | + | + | ++ | + | + | ++ | + | |||
| Mersalyl | + | + | + | + | + | ++ | + | + | ++ | |||
| Mercury(II) acetate | + | +++ | + | + | +++ | + | + | + | + | |||
| PCMBS | + | + | + | ++ | + | + | ++ | + | + | |||
| Potassium mercury tetraiodide | + | + | + | + | ++ | + | ||||||
| Methylmercury(II) chloride | + | + | + | ++ | + | + | ||||||
| Methylmercury(II) acetate | ++ | ++ | + | + | ++ | ++ | ++ | +++ | + | |||
| Ethylmercury(II) phosphate | ++ | ++ | + | + | + | ++ | + | +++ | ++ | |||
| Mercury(II) cyanide | + | + | + | + | + | + | ||||||
| Mercury(II) bromide | + | + | + | + | + | + | ||||||
| Mercury(II) chloride | ++ | + | + | |||||||||
| Methylmercury(II) bromide | + | + | + | |||||||||
| p-Chloromercuribenzoic acid | + | + | + | + | + | + | ||||||
| Ethylmercury(II) chloride | + | + | + | + | + | + | ||||||
| Mercury(II) iodide | + | |||||||||||
| Mercury(II) oxide | + | + | + | + | ||||||||
| Potassium hexachloroiridate(III) | + | + | + | + | + | + | + | + | ++ | + | + | |
| Potassium hexachloroosmiate(IV) | + | + | + | + | + | + | + | + | + | + | + | + |
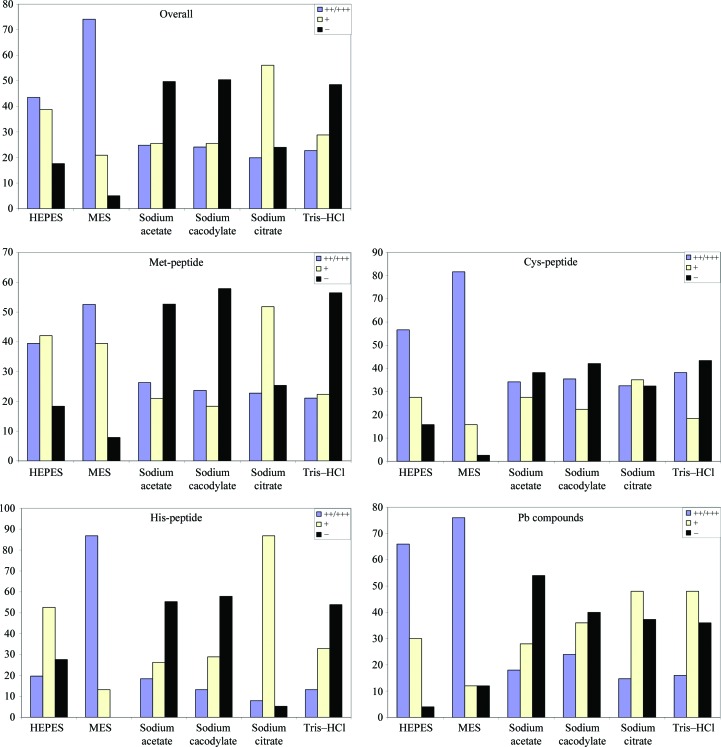
Blundell & Johnson (1976 ▶) noted that methionine rarely reacts with mercury, mentioning only the rare example of HgI3 − binding to rubredoxin (Herriott et al., 1970 ▶). In contrast, our experiments show reproducible derivatization with 13 out of 16 mercury-containing compounds in HEPES and MES buffers. In fact, greater than 40% success (>20% moderately to highly reactive) was observed even in poorly reactive buffer conditions such as sodium cacodylate and Tris–HCl (Fig. 2 ▶). Other mercury-containing compounds of note that show binding include methylmercury acetate and ethylmercury phosphate, which display high reactivity in sodium acetate and sodium citrate buffers. Gold-containing compounds reacted at a lower rate with the Met-peptide, with K2AuCl4 and NaAuCl4 being the most reactive examples with seven and nine out of 12 cases, respectively, shown to be highly reactive. This reactivity is likely to be a consequence of the ability of its lone pair of electrons to displace chloride from the gold chloride compounds, thus forming protein–gold adducts (Petsko, 1985 ▶).
Figure 2.
Percentage of conditions in each buffer that are moderately to highly reactive (++/+++), minimally reactive (+) and nonreactive (−) for each peptide group.
Analysis of buffer and pH preference shows that pH 6 is the optimum pH for derivatization reactions, with 95% success (74% moderately to highly reactive) in MES buffer and 76% success (20% moderately to highly reactive) in sodium citrate buffer at this pH. Noticeably, there is a specific buffer preference in the derivatization reactions, with HEPES preferred over Tris–HCl at higher pH values and sodium citrate preferred over sodium acetate at the lower values.
3.3. Cysteine derivatized by heavy-atom compounds
Analysis of the successful cysteine peptide derivatives showed a high degree of preference for mercury derivatization (Table 3 ▶). Ten of the 16 mercury compounds examined reacted strongly in all buffer conditions, producing near-complete derivatization (+++) in the majority of samples. Mercury(II) iodide and mercury(II) oxide were the least reactive compounds, reacting poorly in all buffers. The other compound types examined were quite comparable to each other, with only potassium tetrachloroplatinate(II), sodium tetrachloroaurate(III), gold(III) chloride and potassium hexachloroosmate(IV) showing high levels of adduct formation in almost all buffers examined.
Table 3. Heavy-atom reactivity of the cysteine-containing peptide.
| Sodium acetate | Sodium citrate | MES | Sodium cacodylate | HEPES | TrisHCl | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | 4.0 | 5.0 | 4.0 | 5.0 | 6.0 | 6.0 | 5.5 | 6.5 | 7.0 | 8.0 | 7.5 | 8.5 |
| Platinum potassium thiocyanate | + | + | +++ | ++ | +++ | + | ||||||
| Platinum potassium iodide | + | + | + | ++ | +++ | + | + | + | ||||
| Potassium hexabromoplatinate(IV) | + | ++ | + | ++ | ++ | + | ||||||
| Potassium tetrachloroplatinate(II) | +++ | + | ++ | +++ | +++ | +++ | ++ | +++ | + | +++ | ||
| Potassium tetrabromoplatinate | + | ++ | +++ | + | + | + | + | |||||
| Diamino platinum dinitrate | ++ | + | +++ | + | + | ++ | ++ | + | ||||
| Potassium tetranitroplatinate(II) | + | + | +++ | + | ++ | ++ | ||||||
| Dichloro(ethylenediamine)platinum(II) | + | + | + | + | ++ | |||||||
| Potassium tetracyanoplatinate(II) | + | + | + | ++ | + | + | ||||||
| Ammonium tetrachloroplatinate(II) | ++ | ++ | +++ | + | + | +++ | +++ | |||||
| Potassium hexachloroplatinate(IV) | + | + | +++ | + | + | ++ | ++ | ++ | ||||
| Potassium tetrachloroaurate(III) | + | + | + | ++ | +++ | +++ | ++ | ++ | ||||
| Sodium tetrachloroaurate(III) | + | ++ | + | + | + | +++ | ++ | ++ | +++ | ++ | +++ | +++ |
| Gold(III) chloride | ++ | ++ | ++ | + | + | ++ | +++ | +++ | + | + | ++ | ++ |
| Gold chloride | + | ++ | ++ | ++ | + | ++ | ||||||
| Gold sodium bromide | + | + | + | ++ | ++ | ++ | ||||||
| Potassium tetrabromoaurate(III) | + | ++ | ++ | + | + | + | ||||||
| Gold sodium thiosulfate | + | + | + | + | + | |||||||
| Gold(I) potassium cyanide | ++ | ++ | + | + | ||||||||
| Gold potassium thiocyanide | + | + | + | +++ | + | + | ||||||
| Thiomersal | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Mersalyl | +++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ | + | +++ | + | + |
| Mercury(II) acetate | +++ | +++ | +++ | ++ | ++ | +++ | +++ | + | +++ | ++ | ++ | ++ |
| PCMBS | ++ | ++ | ++ | ++ | ++ | +++ | ++ | ++ | +++ | +++ | ++ | ++ |
| Potassium mercury tetraiodide | + | +++ | ++ | ++ | + | +++ | +++ | + | ||||
| Methylmercury(II) chloride | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Methylmercury(II) acetate | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ |
| Ethylmercury(II) phosphate | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Mercury(II) cyanide | +++ | + | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Mercury(II) bromide | +++ | +++ | +++ | ++ | ++ | +++ | +++ | +++ | +++ | ++ | +++ | ++ |
| Mercury(II) chloride | + | ++ | + | |||||||||
| Methylmercury(II) bromide | + | + | + | + | + | + | + | |||||
| p-Chloromercuribenzoic acid | + | + | + | + | + | |||||||
| Ethylmercury(II) chloride | + | + | ++ | ++ | ++ | ++ | + | + | + | + | + | +++ |
| Mercury(II) iodide | + | |||||||||||
| Mercury oxide | + | + | + | |||||||||
| Potassium hexachloroiridate(III) | + | + | + | + | + | + | + | + | + | ++ | ++ | |
| Potassium hexachloroosmiate(IV) | + | ++ | ++ | + | + | +++ | ++ | +++ | ++ | + | ++ | ++ |
Previous studies had indicated the low reactivity of cysteine with PtCl4 2− ion (Petsko et al., 1978 ▶) at pH 6 and below. Further analysis of reactions over the pH range 4.2–9.1 (Petsko, 1985 ▶) showed that reactivity increased rapidly above pH 7. In our study, however, MES pH 6 was the most successful buffer, with heavy-atom adducts observed in 37 out of 38 cases. At high pH most metals clearly favored HEPES (84% success; 57% moderately to highly reactive) rather than Tris–HCl (56% success; 38% moderately to highly reactive) as the derivatization buffer (Fig. 2 ▶). At lower pH values (pH 4 and 5) sodium acetate and sodium citrate had very similar success rates, but differentiation was observed with specific compounds such as methylmercury(II) bromide and p-chloromercuribenzoic acid (PCMB), which reacted preferentially in sodium acetate and sodium cacodylate buffers, while other compunds only produced heavy-atom adducts in sodium citrate buffer.
3.4. Histidine binding by heavy-atom compounds
Histidine provides an imidazole ligand which is protonated at pH 6 or lower and is thus a poor nucleophile at low pH, limiting its ability to react with heavy-atom compounds. However, in this study histidine showed good reactivity with gold, platinum and mercury compounds in the range of pH values examined (Table 4 ▶). Interestingly, in contrast to the Met- and Cys-peptides there was not a single compound which reacted universally across the 12 buffer and pH ranges examined. However, there were a number of gold and mercury compounds in specific buffers which gave extremely high levels of derivatization e.g. gold(III) chloride in sodium acetate and sodium cacodylate buffers (all pH values). Platinum and mercury compounds reacted with the histidine peptide in quite a similar fashion. Remarkably, derivatization reactions with all compounds carried out in MES buffer pH 6 were successful.
Table 4. Heavy-atom reactivity of the histidine-containing peptide.
| Sodium acetate | Sodium citrate | MES | Sodium cacodylate | HEPES | TrisHCl | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | 4.0 | 5.0 | 4.0 | 5.0 | 6.0 | 6.0 | 5.5 | 6.5 | 7.0 | 8.0 | 7.5 | 8.5 |
| Platinum potassium thiocyanate | + | + | + | +++ | + | + | ||||||
| Platinum potassium iodide | + | + | + | +++ | + | + | ||||||
| Potassium hexabromoplatinate(IV) | + | + | + | +++ | + | + | ||||||
| Potassium tetrachloroplatinate(II) | + | + | + | +++ | + | + | ||||||
| Potassium tetrabromoplatinate(II) | + | + | + | + | + | + | + | + | +++ | ++ | + | |
| Diamino platinum dinitrate | + | + | + | + | + | +++ | + | + | + | |||
| Potassium tetranitroplatinate(II) | + | + | ++ | +++ | + | + | ||||||
| Dichloro(ethylenediamine)platinate(II) | + | + | + | ++ | + | +++ | + | + | + | + | + | |
| Potassium tetracyanoplatinate(II) | + | + | + | +++ | + | + | + | |||||
| Ammonium tetrachloroplatinate(II) | + | + | + | +++ | + | + | ||||||
| Potassium hexachloroplatinate(IV) | + | + | + | +++ | + | + | ||||||
| Potassium tetrachloroaurate(III) | ++ | ++ | + | +++ | +++ | ++ | ++ | ++ | ||||
| Sodium tetrachloroaurate(III) | ++ | ++ | + | + | +++ | ++ | ++ | + | + | |||
| Gold(III) chloride | +++ | +++ | + | + | + | +++ | +++ | +++ | +++ | +++ | ||
| Gold chloride | + | +++ | + | + | + | +++ | +++ | +++ | +++ | ++ | ||
| Gold sodium bromide | + | + | + | + | + | + | + | + | ||||
| Potassium tetrabromoaurate(III) | + | + | + | + | + | +++ | + | + | ||||
| Gold sodium thiosulfate | + | + | + | ++ | + | + | + | + | ||||
| Gold(I) potassium cyanide | ++ | ++ | + | + | ++ | ++ | ++ | ++ | ++ | +++ | ||
| Gold potassium thiocyanide | + | + | + | + | ++ | + | + | ++ | ++ | |||
| Thiomersal | + | + | + | + | + | ++ | + | + | ++ | +++ | + | |
| Mersalyl | + | + | + | + | + | ++ | + | ++ | +++ | + | + | |
| Mercury(II) acetate | ++ | ++ | + | + | ++ | +++ | + | + | + | + | ||
| PCMBS | + | + | + | + | + | ++ | + | + | ++ | +++ | + | |
| Potassium mercury tetraiodide | + | + | + | +++ | + | +++ | + | + | ||||
| Methylmercury(II) chloride | + | + | + | +++ | + | + | + | |||||
| Methyl mercury(II) acetate | +++ | + | + | ++ | ++ | +++ | ++ | +++ | + | + | ||
| Ethylmercury(II) phosphate | +++ | +++ | ++ | ++ | ++ | +++ | +++ | +++ | + | + | ||
| Mercury(II) cyanide | + | + | + | +++ | + | + | ||||||
| Mercury(II) bromide | + | + | + | +++ | + | + | ||||||
| Mercury(II) chloride | + | + | + | +++ | + | + | + | + | ||||
| Methylmercury(II) bromide | + | + | + | +++ | + | + | ||||||
| p-Chloromercuribenzoic acid | + | + | + | +++ | + | + | + | + | ||||
| Ethylmercury(II) chloride | + | + | + | +++ | + | + | + | + | ||||
| Mercury(II) iodide | + | |||||||||||
| Mercury oxide | + | + | + | + | ||||||||
| Potassium hexachloroiridate(III) | + | + | + | + | + | + | ++ | + | + | |||
| Potassium hexachloroosmiate(IV) | + | + | + | + | ++ | + | ++ | + | ||||
Gold compounds exclusively prefer Tris–HCl as the high-pH buffer for histidine derivatization, failing to react in the presence of HEPES in all cases. In contrast, platinum and mercury compounds derivatize efficiently under HEPES buffering conditions, with additional high levels of derivatization also observed within a subsection of mercury-containing compounds.
Examination of the acidic pH sodium buffers indicates that sodium citrate allows a high percentage of minimally reactive (+) profiles for platinum, gold and mercury compounds. In contrast, sodium acetate and sodium cacodylate buffers tend to hinder mercury and platinum adduct formation, although gold derivatization was much more efficient in these conditions than in comparable sodium citrate conditions. In general, however, strongly derivatizing compounds such as PCMBS and thiomersal still display activity even at acidic pH, regardless of buffer type. The overall pH profile shows that buffer preference is minimal at pH 6 but evident at the higher pHs examined, where HEPES is much preferred over Tris–HCl (Fig. 2 ▶). At the lower pH values examined, sodium citrate gave a high level of derivatization (95% success; 8% moderately to highly reactive) in comparison to sodium acetate, which has 44% success overall but of these 26% are moderately to highly reactive.
3.5. Aspartate, glutamate, asparagine, glutamine and tyrosine derivatization by lead compounds
The mass-spectrometric analyses of Asp-, Glu-, Asn-, Gln- and Tyr-peptide derivatization with lead compounds are shown in Table 5 ▶. HEPES, MES and sodium citrate are the preferred buffers for Asp-peptide derivatization, with either no reaction or low levels observed for all compounds in sodium acetate, sodium cacodylate and Tris–HCl buffers (Fig. 2 ▶).
Table 5. Lead containing compound reactivity of the aspartate, glutamate, asparagine glutamine and tyrosine containing peptides.
| Sodium acetate | Sodium citrate | MES | Sodium cacodylate | HEPES | TrisHCl | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | 4.0 | 5.0 | 4.0 | 5.0 | 6.0 | 6.0 | 5.5 | 6.5 | 7.0 | 8.0 | 7.5 | 8.5 |
| Asp-peptide | ||||||||||||
| Lead acetate | + | + | + | + | + | +++ | + | + | + | + | + | + |
| TMLA | + | + | + | ++ | ++ | ++ | ||||||
| Hexaphenyl dilead | + | + | + | ++ | + | ++ | ||||||
| Lead nitrate | + | + | ++ | ++ | + | + | + | + | ++ | + | + | + |
| Triethyl lead acetate | + | + | + | + | + | ++ | + | |||||
| Glu-peptide | ||||||||||||
| Lead acetate | + | +++ | + | + | +++ | ++ | ++ | +++ | +++ | + | + | |
| TMLA | + | + | + | +++ | ++ | ++ | + | + | ||||
| Hexaphenyl dilead | + | + | + | ++ | ++ | ++ | + | + | ||||
| Lead nitrate | + | + | + | + | + | +++ | + | +++ | ++ | + | + | |
| Triethyl lead acetate | + | + | + | ++ | + | + | ++ | ++ | + | + | ||
| Asn-peptide | ||||||||||||
| Lead Acetate | + | + | ++ | + | + | +++ | +++ | + | ++ | |||
| TMLA | + | + | + | + | ++ | +++ | ||||||
| Hexaphenyl dilead | +++ | + | ++ | +++ | ++ | + | +++ | |||||
| Lead nitrate | + | + | ++ | + | + | ++ | ++ | +++ | + | |||
| Triethyl lead acetate | +++ | ++ | ++ | ++ | ++ | + | + | |||||
| Gln-peptide | ||||||||||||
| Lead Acetate | + | ++ | ++ | ++ | ||||||||
| TMLA | + | + | ++ | + | ||||||||
| Hexaphenyl dilead | + | + | + | |||||||||
| Lead nitrate | +++ | +++ | ++ | ++ | + | ++ | +++ | + | + | |||
| Triethyl lead acetate | + | + | + | + | + | |||||||
| Tyr-peptide | ||||||||||||
| Lead Acetate | +++ | +++ | ++ | ++ | ++ | +++ | +++ | +++ | ++ | ++ | ++ | +++ |
| TMLA | + | + | +++ | + | + | + | ||||||
| Hexaphenyl dilead | +++ | + | +++ | ++ | + | |||||||
| Lead nitrate | +++ | +++ | ++ | ++ | + | ++ | +++ | ++ | +++ | +++ | +++ | + |
| Triethyl lead acetate | ++ | +++ | ++ | + | + | +++ | +++ | +++ | +++ | + | + | + |
In the case of the glutamate-containing peptide, lead acetate reacted and was the only lead-containing compound to be highly reactive in sodium acetate and sodium cacodylate buffers. All other compounds were highly reactive in HEPES and MES buffers. At low pH, the Glu-peptide displayed similar activity to the Asp-peptide with some preference for sodium citrate, although the derivatizations were at the minimally reactive (+) level. With both the Asp- and Glu-peptides, trimethyllead acetate and hexaphenyl dilead failed to derivatize the ligand in sodium acetate and sodium cacodylate conditions.
This pattern was also observed with the asparagine peptide-derivatization reactions with both HEPES and MES conditions, generating substantial amounts of adduct when compared with the other buffers. All the lead compounds derivatized the Asn-peptide in sodium cacodylate conditions, while the reactivity of the compounds in sodium acetate and sodium citrate buffers was generally very poor. The reaction levels in HEPES and MES buffers were very good and some reasonable reaction levels were also seen in the presence of Tris–HCl buffer (with the exception of trimethyllead acetate). Glutamine was the least reactive group for lead-compound derivatization, with interestingly only lead nitrate showing significant levels at acidic pH in all buffer types.
Tyrosine turned out to be the most reactive group for lead compounds, with lead acetate, lead nitrate and triethyllead acetate reacting to high levels in all buffers, while hexaphenyl dilead and trimethyllead acetate show comparably poor reactivity at low pH in cacodylate, acetate and citrate buffers.
3.6. Correlation of the peptide–heavy atom reactivities with lysozyme derivatizations
Undoubtedly, the presence of protein tertiary structure will modify the reactivity of a heavy-atom compound, primarily owing to the restricted access to the interior of the protein. To a certain extent, the peptide reactivities are most relevant to surface-exposed sites on proteins. Thus, proteins will in general be less reactive than peptides, suggesting that the peptide-derived heavy-atom reactivity represents the maximum derivatization for a given protein. To assess the validity of the peptide-derivatization data to proteins, we carried out a series of heavy-atom soaks using hen egg-white lysozyme crystals guided by the peptide-heavy atom reactivity table. The structure of lysozyme was originally solved using o-mercurihydroxytoluene p-sulfonate (MHTS), K2HgI4 and K2PdCl4 as phasing derivatives (Blake et al., 1962 ▶). Seven additional compounds, including K2AuCl4, K2HgBr4, PCMB, PCMBS, K2PtBr4, K2PtCl4 and K2PtCl6, were also known to react with lysozyme (Blake, 1968 ▶). Among the ten published lysozyme-reactive heavy-atom compounds, three (K2HgBr4, MHTS and K2PdCl4) are not present in the peptide–heavy atom reactivity table, while two compounds (K2HgI4 and PCMB) failed to derivatize the peptides. Among the remaining five compounds, their peptide reactivity follows the order K2PtCl4, K2PtBr4 (highly reactive) > K2AuCl4 (moderately reactive) > K2PtCl6 and PCMBS (minimally reactive) in sodium acetate buffers pH 4 and 5 (Table 6 ▶).
Table 6. Mass spectrometry, data collection and X-ray crystallographic statistics of lysozyme derivatives.
(a).
Mass spectrometry.
| Compound | Peptide reactivity | Lysozyme reactivity |
|---|---|---|
| MHTS | ND | Blake et al. (1962 ▶) |
| Potassium tetrachloropalladate(II) | ND | Blake et al. (1962 ▶) |
| Mercury(II) potassium bromide | ND | Blake (1968 ▶) |
| Mercury(II) potassium iodide | Blake et al. (1962 ▶) | |
| PCMB | Blake (1968 ▶) | |
| PCMBS | + | Blake (1968 ▶) |
| Potassium hexachloroplatinate(IV) | + | Blake (1968 ▶) |
| Potassium tetrachloroaurate(III) | ++ | Blake (1968 ▶) |
| Potassium tetrabromoplatinate(II) | +++ | +; Blake (1968 ▶) |
| Potassium tetrachloroplatinate(II) | +++ | +++; Blake (1968 ▶) |
| Potassium hexabromoplatinate(IV) | +++ | +++ |
| Methylmercury(II) acetate | +++ | +++ |
| Ethylmercury phosphate | +++ | +++ |
| Mercury(II) acetate | +++ | +++ |
| Triethyllead acetate | +++ | + |
| Lead nitrate | +++ | +++ |
| Lead acetate | +++ | +++ |
| Diamino platinum dinitrate | +++ | + |
| Gold(III) chloride | +++ | + |
| Thiomersal | +++ | |
| Mersalyl | +++ | |
| Mercury(II) bromide | +++ | |
| Methylmercury(II) chloride | +++ | |
| Mercury(II) iodide | ||
| Methylmercury(II) bromide | ||
| Potassium tetracyanoplatinate(IV) | +++ | |
| Platinum potassium iodide | ||
| Gold sodium thiosulfate | ||
| Hexaphenyl lead |
(b).
Data-collection and X-ray crystallographic statistics. Values in parentheses are for the high-resolution shell.
| Lead acetate | K2Pt(CN)4 | |
|---|---|---|
| Unit-cell parameters | ||
| a () | 78.967 | 77.977 |
| b () | 78.967 | 77.977 |
| c () | 37.104 | 36.983 |
| Resolution | 501.84 (1.911.84) | 502.5 (2.592.5) |
| Completeness (%) | 97.4 (94.3) | 87.5 (91.6) |
| R merge | 0.051 (0.165) | 0.11 (0.362) |
| I/(I) | 29.46 (9.77) | 11.41 (3.24) |
| R iso | 0.109 | 0.319 |
| FOM | 0.235 | 0.144 |
| Heavy-atom peak height (in ) | ||
| Site 1 | 14.6 | 4.91 |
| Site 2 | 10.92 | N/A |
| Site 3 | 5.16 | N/A |
The sequence of lysozyme contains two methionine, eight cysteine, one histidine and three tyrosine residues that are potential ligands for heavy-atom derivatization. Based on the lysozyme crystallization conditions and the presence of potential heavy-atom ligands in its sequence, a total of 15 heavy-atom compounds (Table 6 ▶) are found to be highly reactive against the Met-, Cys-, His- and Tyr-peptides (Tables 2–5 ▶ ▶ ▶ ▶). Only two of these 15 compounds, K2PtCl4 and K2PtBr4, overlap with the phasing derivatives described by Blake (1968 ▶), demonstrating the incompleteness of the number of derivatives found by the traditional trial-and-error method and the potential of finding new phasing compounds using the peptide–heavy atom reactivity table. All 15 compounds were assessed for their ability to derivatize lysozyme using mass spectrometry. Except for four mercury compounds selected based on their reactivities with the cysteine peptide, the remaining 11 compounds all reacted with lysozyme (Table 6 ▶, Fig. 3 ▶). The failure of the four mercury compounds, methylmercury(II) chloride, thiomersal, mersalyl and mercury(II) bromide, to derivatize lysozyme is probably a consequence of the lack of free cysteines in the protein as supported by its structure, in which all eight cysteines are present in their disulfide-bonded form. Mercury(II) acetate was reactive to both the Cys- and Met-peptides in sodium acetate buffer and indeed derivatized lysozyme as judged by mass spectrometry.
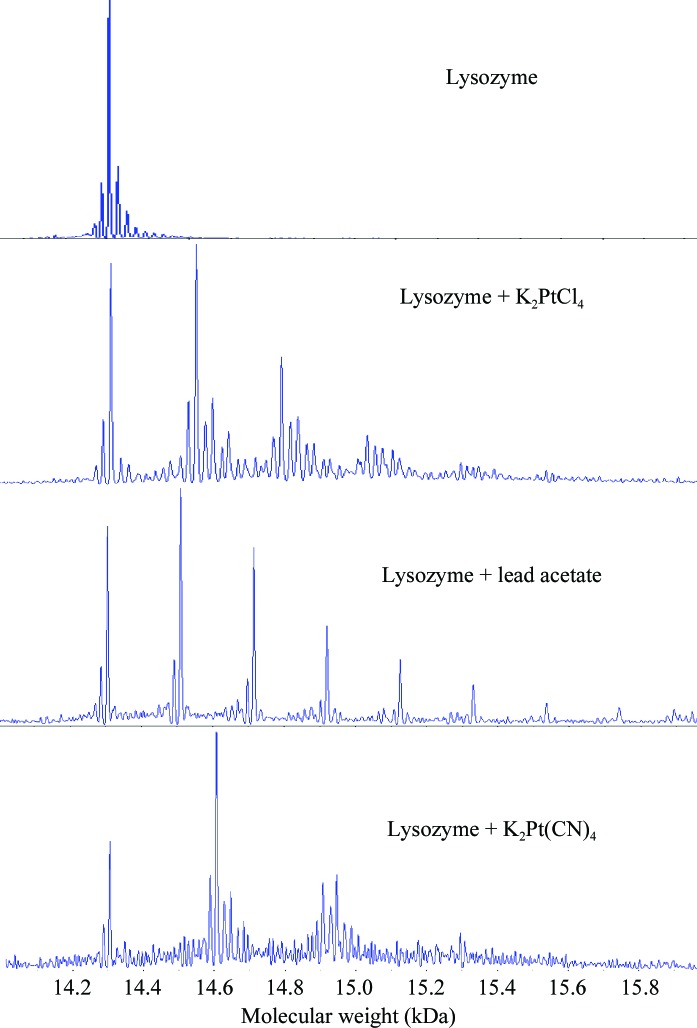
Figure 3.
Mass spectra for (a) native lysozyme, (b) potassium tetrachloroplatinate(II)-derivatized lysozyme, (c) lead acetate-derivatized lysozyme and (d) potassium tetracyanoplatinate(II)-derivatized lysozyme.
To further assess the validity of the peptide-reactivity table, six compounds which failed to react with the peptides in sodium acetate buffer were selected for test reactions with lysozyme (Table 6 ▶). They included mercury(II) iodide, methylmercury(II) bromide, gold sodium thiosulfate, potassium hexaiodoplatinate(IV), potassium tetracyanoplatinate(II) and hexaphenyl dilead. Except for potassium tetracyanoplatinate(II), no adduct formation was observed between lysozyme and the remaining five compounds. To evaluate whether potassium tetracyanoplatinate(II) reacted with amino acids other than Met, Cys and His, the Asp-, Glu-, Asn-, Gln- and Tyr-peptides were tested for potassium tetracyanoplatinate(II) reactivity. However, no adduct formation was observed with each tested peptide.
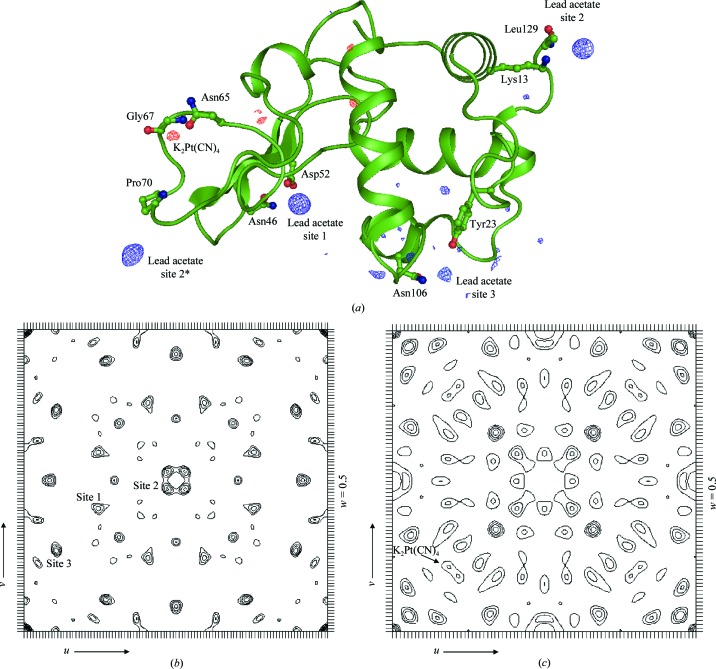
To further analyze and compare both the novel lead acetate derivative and the interesting potassium tetracyanoplatinate(II) example, lysozyme crystals were soaked with these compounds using the quick-soak method of Sun et al. (2002 ▶). Upon soaking, both derivatized crystals appeared visually to be undamaged; however, the diffraction limit of the crystal soaked in potassium tetracyanoplatinate(II) was reduced to 2.5 Å while the lead acetate-derivatized crystal diffracted to 1.84 Å. Difference Fourier maps were generated from individually collected diffraction data using CNS (Brünger et al., 1998 ▶) (Table 6 ▶, Fig. 4 ▶ a). The lead acetate derivative produced three binding sites, coordinated by Asn46 and Asp52 at site 1, by Lys13 and Leu129 at site 2, and by Tyr23 and Asn106 at site 3 (Fig. 4 ▶ a). In comparison, only a minor site, coordinated by Asn65 and Gly67, was observed in the potassium tetracyanoplatinate(II)-derivatized crystal. All three lead-binding sites appear to be more occupied than the platinum site and the lead derivative resulted in a better figure of merit for phasing. The superior phasing quality of the lead acetate is also apparent from a comparison between the Harker section w = 0.5 of the two difference Patterson maps (Figs. 4 ▶ b and 4 ▶ c). The results show that while compounds that failed to react with the model peptides may still derivatize a protein, they are likely to produce only minor binding sites for phasing. Although lysozyme represents a single specific protein and the general predictive ability of the peptide–heavy atom reactivity table on other proteins remains to be examined, the correlation with the peptide studies and the predicted derivatives based on the peptide-reactivity table are quite impressive.
Figure 4.
(a) Difference Fourier (F o − F c) maps calculated for lysozyme derivatized with lead acetate (in blue density) and with potassium tetracyanoplatinate(II) (in red density) and contoured at the 3σ level. The structure of lysozyme is shown in ribbon representation, with the residues coordinating heavy atoms shown as ball-and-stick models. PyMOL was used to generate the figure. Difference Patterson maps of the lead acetate (b) and potassium tetracyanoplatinate(II) (c) derivatives of lysozyme calculated for the Harker section w = 0.5 with diffraction data between 50 and 3.8 Å resolution. The sections are contoured with 1σ increments starting at 2σ. The respective self Patterson vectors from the individual heavy-atom-binding sites are indicated in the Harker sections.
4. Discussion
In the process of phasing of novel protein structures, the ability to efficiently prepare protein crystals containing heavy or anomalous scattering atoms is of fundamental importance. Although recent advances such as ‘native gel shift assays’ and mass-spectrometric analysis have provided crystal-free methods for determining heavy-atom derivatives and thus providing a basis for a more systematic search, the screening process still remains arduous. Crystallization parameters such as buffer, pH and precipitants play an important role in altering the chemical environment of heavy-atom ligands, thereby influencing the derivatization process. The results presented here will assist in the selection of heavy-metal compounds for derivatization trials given a specific set of crystallization buffer and pH conditions. To date, this is the first large-scale study that has experimentally assessed the effect of the buffer on derivatization experiments. This study builds on the crystallographic data provided by Blundell & Johnson (1976 ▶) and the recent compilation of this and other additional information in the Heavy Atom Databank (Islam et al., 1998 ▶) in a more complete and systematic manner.
Using model peptides, we examined the heavy-atom reactivities for a number of amino acids, including Met, Cys, His, Tyr, Glu, Asp, Asn and Gln. Thus, for proteins rich in methionine platinum compounds should be the first choice of screening, while mercury and gold compounds become the obvious candidates for those rich in free cysteine and histidine residues, respectively. Importantly, this study rules out a large number of compounds that are poorly reacting or nonreacting in specific buffer–pH combinations. Equally valuable, this study supported certain heavy-atom compound–pH combinations for derivatization that would not have been tested based on their conventional predicted chemical properties, a serious limitation raised by Petsko et al. (1978 ▶) who stated ‘experimental observations are in poor agreement with the expected observations’ in heavy-atom adduct reactions.
To correlate the peptide-reactivity results with protein derivatization, lysozyme was derivatized using compounds selected from the peptide-reactivity table. The results showed that 11 of these highly reactive compounds also reacted with lysozyme; however, four cysteine-reactive mercury compounds failed to derivatize the protein owing to the lack of free cysteines. In addition, the selection of heavy-atom compounds based on the peptide-reactivity table appeared to be much broader than the previously used compounds. For example, lead acetate was predicted as a new potent compound for adduct formation with lysozyme and this was confirmed by both mass spectrometry and a difference Fourier density map.
The peptide technique we have used allows direct analysis of a potential ligand and heavy-atom compound, but does not take into account the influence of a protein structure or of other residues that may stabilize heavy-atom binding and reaction, as has been noted by Sugahara et al. (2005 ▶). Although heavy-atom derivatizations do not generally require tight sequence motifs, a preference for certain amino acids was reflected by the motif-based prediction server HATODAS. However, the limitation of the HATODAS server is that it is biased towards frequently published compounds and thus is less accurate for compounds described in only a few publications. Protein steric hindrance can also interfere with and thus modify the peptide reactivity, in particular to the binding of larger heavy-atom compounds, as observed by Blundell & Johnson (1976 ▶) in the case of lactate dehydrogenase.
An obvious caveat of this study is that it does not account for the role of other crystallization components such as high salt or polyethylene glycol which are common in crystallization conditions; the effects of these compounds are not readily examined by ESI-MS and furthermore the immense number of additional parameters is beyond the scope of this study. Expanding on the study of Sun & Hammer (2000 ▶), it would be useful to carry out derivatization experiments with soluble protein in the presence of low concentrations of crystallization components as opposed to solely H2O in a case-by-case manner to find suitable derivatives. Furthermore, at the higher buffer concentrations commonly used in crystallization, the reactivity of the heavy-metal compounds may be lower but ultimately the general trend of reactivities should remain.
Among the metal compounds tested, Pt(NH3)2(NO2)2, K2PtCl4, Thiomersal, Mersalyl, Hg(OOCCH3)2, CH3Hg(OOCCH3), PCMBS and ethylmercury(II) phosphate produce almost near-complete derivatization under all buffer conditions. The compounds NH4PtCl4, K2PtBr6, K2PtBr4, K2Pt(NO2)4, K2AuCl4, NaAuCl4, gold(III) chloride, KAu(CN)2, Pb(CH3COO)2, Pb(NO3)2, CH3HgCl, Hg(CN)2 and HgBr2 also produce substantial amounts of covalent metal adducts when their preferred ligand is available (Table 7 ▶). These 21 heavy-metal compounds form an excellent starting point to increase the probability of obtaining useful derivatives. The ‘magic seven’ of heavy-atom derivatization has been mentioned in a number of publications (Garman & Murray, 2003 ▶; Boggon & Shapiro, 2000 ▶) and these were tested in our assays with success, further validating the ability of this study to correctly identify derivatizing compounds. This study expands the initial seven to 21, not including the uranium-containing compounds that are also highly successful. The compounds potassium hexachloroiridate(III) and potassium hexachloroosmate(IV) also react consistently with the Met-, Cys- and His-peptides, albeit at lower levels. This property may be desirable in cases where isomorphism or reduced diffraction quality is evident after derivatization with other compounds, as the most reactive compound may not invariably be the best phasing derivative.
Table 7. Success rate for compounds with moderate and high peptide reactivity.
| Compound | Success rate (%) |
|---|---|
| Platinum potassium thiocyanate | 33.3 |
| Platinum potassium iodide | 8.3 |
| Potassium hexabromoplatinate(IV) | 41.7 |
| Potassium tetrabromoplatinate(II) | 55.5 |
| Potassium tetrachloroplatinate(II) | 44.4 |
| Diamino platinum dinitrate | 47.2 |
| Potassium tetranitroplatinate(II) | 44.4 |
| Dichloro(ethylenediamine)platinum(II) | 11.1 |
| Potassium tetracyanoplatinate(II) | 5.5 |
| Ammonium tetrachloroplatinate(II) | 50 |
| Potassium hexachloroplatinate(IV) | 22.2 |
| Potassium tetrachloroaurate(III) | 52.7 |
| Sodium tetrachloroaurate(III) | 61.1 |
| Gold(III) chloride | 47.2 |
| Gold chloride | 33.3 |
| Gold sodium bromide | 11.1 |
| Potassium tetrabromoaurate(III) | 13.9 |
| Gold sodium thiosulfate | 5.5 |
| Gold(I) potassium cyanide | 27.8 |
| Gold potassium thiocyanide | 11.1 |
| Thiomersal | 47.2 |
| Mersalyl | 38.8 |
| Mercury(II) acetate | 47.2 |
| PCMBS | 47.2 |
| Potassium mercury tetraiodide | 22.2 |
| Methylmercury(II) chloride | 38.8 |
| Mercury(II) cyanide | 33.3 |
| Methylmercury(II) acetate | 66.6 |
| Ethylmercury(II) phosphate | 69.4 |
| Mercury(II) bromide | 36.1 |
| Mercury(II) chloride | 8.3 |
| Methylmercury(II) bromide | 2.7 |
| p-Chloromercuribenzoic acid | 2.7 |
| Ethylmercury(II) chloride | 16.6 |
| Mercury(II) iodide | 0 |
| Mercury(II) oxide | 0 |
| Potassium hexachloroiridate(III) | 11.1 |
| Potassium hexachloroosmate(IV) | 27.7 |
| Lead acetate | 43.3 |
| Trimethyllead acetate | 16.6 |
| Hexaphenyl dilead | 21.6 |
| Lead nitrate | 33.3 |
| Triethyllead acetate | 26.6 |
Among the buffers tested, MES had the highest success rate and stands out as the most complementary buffer for derivatization reactions. This indicates that proteins crystallized under MES conditions have a higher probability of successful derivatization. For proteins crystallized in different buffers, exchange of the buffer to MES, if possible, should increase the likelihood of derivatization. Rould (1997 ▶) suggested that all crystals destined for derivatization experiments should be transferred to a storage solution to improve their isomorphism. Utilization of our observations will allow the best choice of storage solution for success in derivatization. Among the basic pH buffers, HEPES shows a much better derivatization profile than Tris–HCl. However, depending on the peptide ligands available, heavy atoms may react preferentially in either HEPES or Tris–HCl. For example, cysteine–platinum derivatives are more readily formed under HEPES buffer conditions rather than Tris–HCl buffer conditions, while conversely histidine–gold derivatives are observed in Tris–HCl buffer but not in HEPES buffer. The current study also reveals the pH preference of some of the heavy metals. Gold sodium bromide, potassium tetrabromoaurate(III), gold potassium thiocyanide and trimethyllead acetate perform well at slightly acidic to basic pH, while potassium tetracyanoplatinate(II), gold sodium thiosulfate, mercury(II) chloride, methylmercury(II) bromide, p-chloromercuribenzoic acid, dichloro(ethylenediamine)platinum(II) and potassium hexachloroplatinate(IV) react strongly in acidic conditions.
Selecting appropriate heavy metals based on the pH of the crystallization condition in addition to trials with the most reactive 21 compounds will further enhance the probability of successful derivatization. The results of our study showed that potential heavy-atom derivatives can be selected simply based on the primary sequence of a protein and its crystallization conditions. Although successful derivatization is only certain after finding the phasing atoms using Patterson or other methods, the current study provides extremely useful leads which may be used to obtain a suitable phasing derivative and will certainly accelerate the process of protein structure determination.
Acknowledgments
A searchable version of the peptide–heavy atom reactivity tables can be accessed at http://sis.niaid.nih.gov/cgi-bin/heavyatom_reactivity.cgi. This work is supported by NIAID intramural research funding.
References
- Blake, C. C. (1968). Adv. Protein Chem. 23, 59–120. [DOI] [PubMed]
- Blake, C. C., Fenn, R. H., North, A. C., Phillips, D. C. & Poljak, R. J. (1962). Nature (London), 196, 1173–1176. [DOI] [PubMed]
- Blake, C. C., Geisow, M. J., Swan, I. D., Rerat, C. & Rerat, B. (1974). J. Mol. Biol. 88, 1–12. [DOI] [PubMed]
- Blundell, T. L., Dodson, G. G., Dodson, E., Hodgkin, D. C. & Vijayan, M. (1971). Recent Prog. Horm. Res. 27, 1–40. [DOI] [PubMed]
- Blundell, T. L. & Jenkins, J. A. (1977). Chem. Soc. Rev. 6, 139–171.
- Blundell, T. L. & Johnson, L. N. (1976). Protein Crystallography, pp. 183–239. London: Academic Press.
- Boggon, T. J. & Shapiro, L. (2000). Structure, 8, R143–R149. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Cohen, S. L. & Chait, B. T. (2001). Annu. Rev. Biophys. Biomol. Struct. 30, 67–85. [DOI] [PubMed]
- Cohen, S. L., Padovan, J. C. & Chait, B. T. (2000). Anal Chem. 72, 574–579. [DOI] [PubMed]
- Dauter, Z., Dauter, M., de La Fortelle, E., Bricogne, G. & Sheldrick, G. M. (1999). J. Mol Biol. 289, 83–92. [DOI] [PubMed]
- Dauter, Z., Dauter, M. & Rajashankar, K. R. (2000). Acta Cryst. D56, 232–237. [DOI] [PubMed]
- Debreczeni, J. É., Girmann, B., Zeeck, A., Krätzner, R. & Sheldrick, G. M. (2003). Acta Cryst. D59, 2125–2132. [DOI] [PubMed]
- Garman, E. & Murray, J. W. (2003). Acta Cryst. D59, 1903–1913. [DOI] [PubMed]
- Hatfull, G. F., Sanderson, M. R., Freemont, P. S., Raccuia, P. R., Grindley, N. D. & Steitz, T. A. (1989). J. Mol. Biol. 208, 661–667. [DOI] [PubMed]
- Hendrickson, W. A., Horton, J. R. & LeMaster, D. M. (1990). EMBO J. 9, 1665–1672. [DOI] [PMC free article] [PubMed]
- Herriott, J. R., Sieker, L. C., Jensen, L. H. & Lovenberg, W. (1970). J. Mol. Biol. 50, 391–406. [DOI] [PubMed]
- Islam, S. A., Carvin, D., Sternberg, M. J. E. & Blundell, T. L. (1998). Acta Cryst. D54, 1199–1206. [DOI] [PubMed]
- Krishna, T. S., Fenyo, D., Kong, X. P., Gary, S., Chait, B. T., Burgers, P. & Kuriyan, J. (1994). J. Mol. Biol. 241, 265–268. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Petsko, G. A. (1985). Methods Enzymol. 114, 147–156. [DOI] [PubMed]
- Petsko, G. A., Phillips, D. C., Williams, R. J. & Wilson, I. A. (1978). J. Mol. Biol. 120, 345–598. [DOI] [PubMed]
- Rould, M. A. (1997). Methods Enzymol. 276, 461–472. [DOI] [PubMed]
- Smith, J. L. & Thompson, A. (1998). Structure, 6, 815–819. [DOI] [PubMed]
- Sugahara, M., Asada, Y., Ayama, H., Ukawa, H., Taka, H. & Kunishima, N. (2005). Acta Cryst. D61, 1302–1305. [DOI] [PubMed]
- Sun, D. P., Alber, T., Bell, J. A., Weaver, L. H. & Matthews, B. W. (1987). Protein Eng. 1,115–123. [DOI] [PubMed]
- Sun, P. D. & Hammer, C. H. (2000). Acta Cryst. D56, 161–168. [DOI] [PubMed]
- Sun, P. D. & Radaev, S. (2002). Acta Cryst. D58, 1099–1103. [DOI] [PubMed]
- Sun, P. D., Radaev, S. & Kattah, M. (2002). Acta Cryst. D58, 1092–1098. [DOI] [PubMed]
- Wang, J., Dauter, M. & Dauter, Z. (2006). Acta Cryst. D62, 1475–1483. [DOI] [PubMed]
- Zhang, G., Kazanietz, M. G., Blumberg, P. M. & Hurley, J. H. (1995). Cell, 81, 917–924. [DOI] [PubMed]