Abstract
The two-component systems (TCS), or histidine-to-aspartate phosphorelays, are evolutionarily conserved common signal transduction mechanisms that are implicated in a wide variety of cellular responses to environmental stimuli in both prokaryotes and eukaryotes including plants. Among higher plants, legumes including Lotus japonicus have a unique ability to engage in beneficial symbiosis with nitrogen-fixing bacteria. We previously presented a genome-wide compiled list of TCS-associated components of Mesorhizobium loti, which is a symbiont specific to L. japonicus (Hagiwara et al. 2004, DNA Res., 11, 57–65). To gain both general and specific insights into TCS of this currently attractive model legume, here we compiled TCS-associated components as many as possible from a genome-wide viewpoint by taking advantage that the efforts of whole genome sequencing of L. japonicus are almost at final stage. In the current database (http://www.kazusa.or.jp/lotus/index.html), it was found that L. japonicus has, at least, 14 genes each encoding a histidine kinase, 7 histidine-containing phosphotransmitter-related genes, 7 type-A response regulator (RR)-related genes, 11 type-B RR-related genes, and also 5 circadian clock-associated pseudo-RR genes. These results suggested that most of the L. japonicus TCS-associated genes have already been uncovered in this genome-wide analysis, if not all. Here, characteristics of these TCS-associated components of L. japonicus were inspected, one by one, in comparison with those of Arabidopsis thaliana. In addition, some critical experiments were also done to gain further insights into the functions of L. japonicus TCS-associated genes with special reference to cytokinin-mediated signal transduction and circadian clock.
Keywords: circadian clock, cytokinin signal transduction, L. japonicus, two-component signal transduction
The so-called two-component systems (TCS), or histidine-to-aspartate (His-Asp) phosphorelays, are evolutionarily conserved common signal transduction mechanisms that are implicated in a wide variety of cellular responses to environmental stimuli.1,2 To date, numerous instances of such TCS have been uncovered and characterized not only in many prokaryotic species but also in certain eukaryotic species, including higher plants.3,4 In particular, recent studies on TCS of the model plant Arabidopsis thaliana provided us with unprecedented progress toward a better understanding of the mechanisms underlying plant hormone (i.e. ethylene and cytokinin) responses, and also circadian clock.5–11
Since we discovered the first instance of histidine kinases (HKs) from Escherichia coli (i.e. the osmotic sensor EnvZ) in 1989, we have been characterizing TCS-associated components extensively by employing many prokaryotic and eukaryotic species, including E. coli, Synechocystis (a cyanobacterium), Rhizobia including Mesorhizobium loti that is a symbiont specific to Lotus japonicus, Schizosaccharomyces pombe (a fission yeast), Aspergillus nidulans (a fungus), A. thaliana, and Oryza sativa.12–22 On the basis of many years of our experience, we have conducted a series of genome-wide analyses to compile systematically TCS-associated components to gain both general and specific insights into the biological roles of TCS in given prokaryotic and eukaryotic spices, as cited above. To extend this line of approach, it is timely to compile and analyze TCS-associated components of L. japonicus from a genome-wide viewpoint. The special reasons are 2-fold; (i) L. japonicus, together with Medicago truncatula, is a model legume, which has a unique ability to engage in beneficial symbiosis with nitrogen-fixing bacteria,23 and (ii) L. japonicus and M. truncatula are the legumes of choice to conduct comparative genome-wide researches, because the efforts of whole genome sequencing are almost at final stage.24–26 On the basis of these rationales, here we compiled L. japonicus TCS-associated coding-sequences by adopting the currently available database (http://www.kazusa.or.jp/lotus/index.html/).
First, it is necessary to introduce general ideas about plant TCS-associated components, briefly. As schematically shown in Fig. 1, the plant TCS-associated components mainly consist of three phosphorelay signal transducers, such as hybrid HK, histidine-containing phosphotransmitter (HPt or HP), and response regulator (RR). The members of RRs are classified into two groups, at least; one is designated type-A RR, and the other type-B RR. The type-A RR consists of only a receiver domain containing an invariant phospho-accepting Asp (D) residue, whereas the type-B RR contains an additional common domain within a C-terminal extension. The motif is generally designated GARP (GOLDEN2, ARR, PSR1), which serves as a sequence-specific DNA-binding domain.27 Therefore, the former would function as a modulator of phosphorelay signal transduction,28,29 whereas the latter would serve as a gene-specific transcription factor.30,31 In addition to these typical TCS-associated components, A. thaliana has a small family of atypical TCS-associated components, which were named pseudo-RRs (PRRs),6,32 because the phospho-accepting aspartate (D) residue had been converted to glutamates (E). These PRRs do not appear to be involved in the phosphorelay signal transduction. However, they coordinately play essential roles within the circadian clock, which are implicated in many characteristic light signal transduction mechanisms of plants growing in natural habitats on the earth exhibiting the 24 h photoperiod.33,34 These views as to the TCS-associated components have been induced mainly from recent intensive studies on the model plant A. thaliana, and many excellent reviews are currently available, in which comprehensive overviews were presented with regard to the respective subjects, including ethylene signal transduction, cytokinin signal transduction, and plant circadian clock.5,6,8–11 On the basis of these backgrounds, here we attempted to compile the genome-wide framework of L. japonicus TCS-associated components.
Figure 1.
A schematic representation of the structural features of plant TCS-associated signal transduction components. HKs (e.g. cytokinin receptors) consists of three domains; N-terminal cytokinin-binding domain, central HK domain containing an invariant phospho-accepting histidine (H) residue, and C-terminal receiver domain containing an invariant aspartate (D) residue, which is capable of accepting a phosphoric group from a phospho-histidine. The phosphoryl group on the receiver domain is transferred to a histidine containing phospho-transmitter (HPt or HP), which subsequently serves as a phospho-donor toward RR. Other details are given in the text.
To compile a list of L. japonicus TCS-associated genes (or coding-sequence), an extensive computer-aided search was conducted using the current databases (http://www.kazusa.or.jp/lotus/index.html/). The logic and strategy of bioinformatics with regard to TCS-associated genes were described previously.13,14 We identified a number of coding-sequences, each of which most likely encodes a component of TCS (or related one). As summarized in Table 1, L. japonicus has, at least, 14 genes each encoding an HK, 7 HPt-related genes, 7 type-A RR-related genes, 11 type-B RR-related genes, and 5 putative circadian clock-associated PRR genes.
Table 1.
A list of His-Asp phosphorelay-associated components found in the whole genome sequence of L. japonicus
| Signaling components | Clone and SGA-sequence | Nomenclature | Characteristic remarks |
|---|---|---|---|
| Histidine kinases | chr2.LjB11M03.80 | CK-1 | Cytokinin receptor |
| chr4.CM0042.1660 | CK-2 (LHK1) | Cytokinin receptor35,36 | |
| LjT24E07.10 | CK-3 | Cytokinin receptor (a part of N-terminus is missing) | |
| chr1.LjT14G19.40 | ET-1 | Ethylene receptor | |
| chr3.CM0634.490 | ET-2 | Ethylene receptor | |
| chr5.LjT09B23.140 and 150 | ET-3 | Ethylene receptor (possibly incorrect annotation) | |
| LjB14K02.50 | ET-4 | Ethylene receptor | |
| LjT24A04.50 | ET-5 | Ethylene receptor | |
| chr2.CM0695.410 | HK1a | Homologous to AtHK1 (a part of HK and RR domain is missing) | |
| LjSGA_031835.1 | HK1b | Homologous to AtHK1 (partial, highly similar to HK1a)) | |
| chr2.CM0545.200 | HK2a | Homologous to CKI1 (a part of RR is missing ) | |
| chr5.CM0239.360 | HK2b | Homologous to CKI1 | |
| chr1.CM0133.1270 | HK3a | Homologous to AHK5 | |
| chr5.CM0040.380 | HK3b | Homologous to AHK5 | |
| chr4.LjT06I07.90 | CSKp1 | Homologous to CSK (fragmental) | |
| chr4.LjT06I07.40 | CSKp2 | Homologous to CSK (fragmental) | |
| HPt factors | chr5.LjT17N18.30 | HP1 | |
| chr6.CM1829.110 | HP2 | ||
| LjSGA_017645.1 | HP3 | ||
| chr5.LjT17N18.30 | HP4 | ||
| chr2.CM0065.180 | HP5a | His residue is missing (180 and 190 are repetitive) | |
| chr2.CM0065.190 | HP5b | His residue is missing (180 and 190 are repetitive) | |
| chr3.CM0142.400 | HP6 | His residue is missing | |
| Type A RRs | chr1.CM0027.130 | RRa1 | Induced by cytokinin |
| chr1.CM0320.150 | RRa2 | Induced by cytokinin | |
| chr2.CM0028.90 | RRa3 | Induced by cytokinin | |
| chr3.LjT48P15.80 | RRa4 | Induced by cytokinin | |
| LjB09A03.90 | RRa5 | Induced by cytokinin | |
| LjT47H21.100 | RRa6 | Induced by cytokinin | |
| chr4.LjT30K03.10 | RRa7 | (A part of C-terminus is missing ) | |
| Type B RRs | chr5.CM0072.230 and 220 | RRb1 | (Possibly incorrect annotation) |
| chr5.CM0345.260 | RRb2 | ||
| chr5.LjT32I14.90 | RRb3 | ||
| chr6.CM0037.920 | RRb4 | ||
| CM1729.100 | RRb5 | ||
| LjSGA_022114.1 | RRb6 | ||
| LjSGA_066155.1 | RRb7 | Most likely identical to a part of RRb4 | |
| chr1.LjT03I11.150 | RRb8 | (GARP-motif is missing) | |
| chr4.CM0042.1440 | RRb9 | (Asp residue in RR is missing) | |
| chr4.CM0229.220 | RRb10 | (Asp residue in RR is missing) | |
| LjSGA_012431.2 | RRb11 | (Asp residue in RR is missing) may be identical to a part of RRb10 | |
| Type C RRs | chr1.LjT07G07.80 | RRc1 | (Similar to RR-A, but not induced by cytokinin) |
| Anomalous RRs | chr1.CM0133.1320 | (Fragmental) | |
| chr2.CM0695.400 | (Fragmental) | ||
| LjSGA_054723.1 | TCP34 | (Fragmental) | |
| Clock-associated PRRs | chr4.CM0087.600 | PRR1 | Circadian rhythm (Homologous to TOC1) |
| LjT08O17.180 + 130 + 120 | PRR3 | (A large insertion between 180 and 130) | |
| chr1.CM0105.560 | PRR5 | Circadian rhythm | |
| chr3.LjT05P05.60 | PRR7 | Circadian rhythm | |
| chr3.CM0208.230 | PRR9 | Circadian rhythm | |
| Other clock components | chr3.CM0208.430 | LHY | or CCA1 |
| chr3.CM0792.160 | LUX | Also known as PCL1 |
It is first worth mentioning that A. thaliana has 11 HK genes (including three genes encoding cytokinin receptors, five genes encoding ethylene receptors), 6 HPt-related genes, 10 type-A RR genes, 11 type-B RR genes, and 5 clock-associated genes.3,4 Considering the fact that the TCS-associated genes in higher plants are evolutionarily conserved, these results suggested that most of the L. japonicus TCS-associated genes have already been uncovered in this genome-wide analysis, if not all. These genes (or coding sequences) are listed in Table 1, about which it should be noted that each gene was named quite tentatively, because we believe that the final designation of their nomenclatures should be made more carefully and systematically in order to avoid any future confusion. In any case, by employing the readership friendly Supplementary Table S1, one may directly access each original database of L. japonicus TCS-associated cording-sequences through www hyperlinks (an Excel version of Supplementary Table S1 can be directly download through http://www.agr.nagoya-u.ac.jp/~microbio/phosphorelay/open data/Supplementary Table S1.xls ).
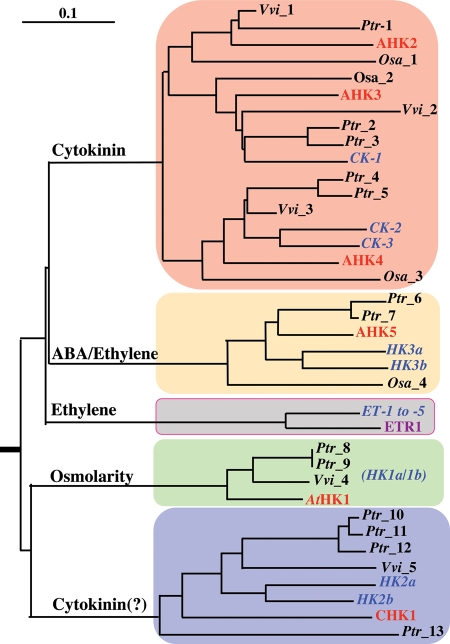
As judged by the knowledge about the cytokinin receptor HKs of A. thaliana (i.e. AHK2, AHK3, AHK4/CRE1/WOL), all of them are highly conserved in L. japonicus. One of cytokinin receptor HK has already been characterized experimentally with special reference to the root nodule organogenesis in response to a Rhizobium (named Lhk1, chr4.CM0042.1660 in Table 1).35,36 Likewise, five orthologs of ethylene receptor HKs (e.g. ETR1) are also found in L. japonicus. However, one of them seems to be incorrectly annotated in the current database. In this case, it is possible to deduce a full length of ethylene receptor HK by combining chr5.LjT09623.140 and 150 into a single coding-sequence. At present, no experimental report on these putative ethylene receptor HKs is available, although some reports on ethylene-mediated signaling in L. japonicus are present.37,38 In addition to these well-characterized hormone receptor HKs, A. thaliana has three more HKs, named AtHK1, CKI1, and AHK5/CKI2.39–41 In this regard, L. japonicus has also a set of HKs orthlogous to these A. thaliana HKs, respectively, although some of them might be incomplete (e.g. AtHK1 orthologs of L. japonicus), as noted specifically in Table 1. Interestingly, L. japonicus appears to have two copies for each of these orthologs. In summary, both the members and numbers of HK genes are highly conserved between A. thaliana and L. japonicus. This view is reasonably generalized for many other higher plants when a phylogenetic tree of HK amino acid sequences was constructed through integrating HKs from a grape tree (Vitis vinifera, Vvi; http://www.genoscope.cns.fr/spip/Vitis-vinifera-e.html), a popular (Populus truchocarpa, Ptr; http://genome.jgi-psf.org/Poptr1/Poptr1.home.html), as well as a rice (Oryza sativa, Osa; http://rapdb.dna.affrc.go.jp/) (Fig. 2).
Figure 2.
A phylogenetic tree of the HK family in some representative plants. Amino acid sequences of A. thaliana HKs were characterized previously, whereas those of L. japonicus were done in this study. In addition to these, a set of putative HK amino acid sequences was analyzed by adopting the databases of some other plants; a grape tree (Vitis vinifera, Vvi; http://www.genoscope.cns.fr/spip/Vitis-vinifera-e.html), a popular (Populus truchocarpa, Ptr; http://genome.jgi-psf.org/Poptr1/Poptr1.home.html), and a rice (Oryza sativa, Osa; http://rapdb.dna.affrc.go.jp/). Using these inferred amino acid sequences, a neighbor-joining phylogenetic tree was constructed by the program of ClustalW. The HKs of A. thaliana and L. japonicus were highlighted by red and blue colors, respectively. Ethylene receptor HKs were not integrated extensively for clarity of this tree. Positions of AtHK1 orthologs of L. japonicus were not certain because their inferred entire amino acid sequences are ambiguous (named HK1a and HK1b, Table 1).
Lotus japonicus has seven HPt-related genes, each encoding a typical phosphotransfer intermediate. Notably, two of them (chr2.CM0065.180 and 190) are highly homologous to each other, and they are located next to each other on the same chromosome, suggesting that they were the resultants of recent duplication. Note also that three out of seven inferred HPt factors, including the repetitive ones, are atypical in that they luck the invariant and phospho-accepting histidine residue in their amino acid sequences, as noted in Table 1. Arabidopsis thaliana has also an atypical HPt gene, which encodes an HPt factor without the phospho-accepting histidine residue. In the case of A. thaliana, this atypical one (named AHP6) plays an important role in the cytokining signal transduction, which is responsible for the developmental regulation of vasculature in roots.42 Hence, the atypical HPt factors of L. japonicus might also play some roles.
Arabidopsis thaliana has 10 genes encoding type-A RRs, expressions of which are induced very rapidly upon the onset of cytokinin treatment of plants.43–45 So far, seven type-A RR genes were found in L. japonicus, one of which is fragmental though. In any case, the most critical question is: whether these type-A RR genes of L. japonicus respond to cytokinin rapidly at the level of transcription, like those of A. thaliana. Here, this issue was addressed experimentally. In A. thaliana, one of the hallmarked cytokinin actions is inhibition of primary root elongation. This can be observed when young seedlings were grown on agar-plates containing an appropriated concentration of cytokinin (e.g. BA; 6-benzylaminopurine). We confirmed that this is also the case in L. japonicus (Fig. 3A, upper panel), and the effective BA concentrations for inhibition of root elongation is nearly the same in both A. thaliana and L. japonicus (Fig. 3A, lower panel). On the basis of this preliminary experiment, L. japonicus young seedlings were sprayed with cytokinin (t-zeatin), and then RNA samples were prepared at intervals, followed by semi-quantitative reverse-transcriptase–polymerase chain reaction (RT–PCR) with each specific pair of primer for type-A RR genes of L. japonicus (for primers used, see Supplementary Table S2). As shown in Fig. 3B, all identified type-A RR genes, except for RRa7, were confirmed to be cytokinin responsive. Note that we failed amplifying cDNA of RRa7, suggesting that this coding-sequence might be non-functional (Table 1). As positive and negative references, it was also shown that an example of L. japonicus type-B RR genes (named RRb4, Table 1) did not respond to cytokinin, whereas an example of L. japonicus cytokinin oxidase genes (named tentatively CKX3, LjT02N03.150) did so markedly under the experimental conditions used, as expected (Fig. 3B). These results were further confirmed by quantitative real-time PCR (qRT–PCR) (Fig. 4).
Figure 3.
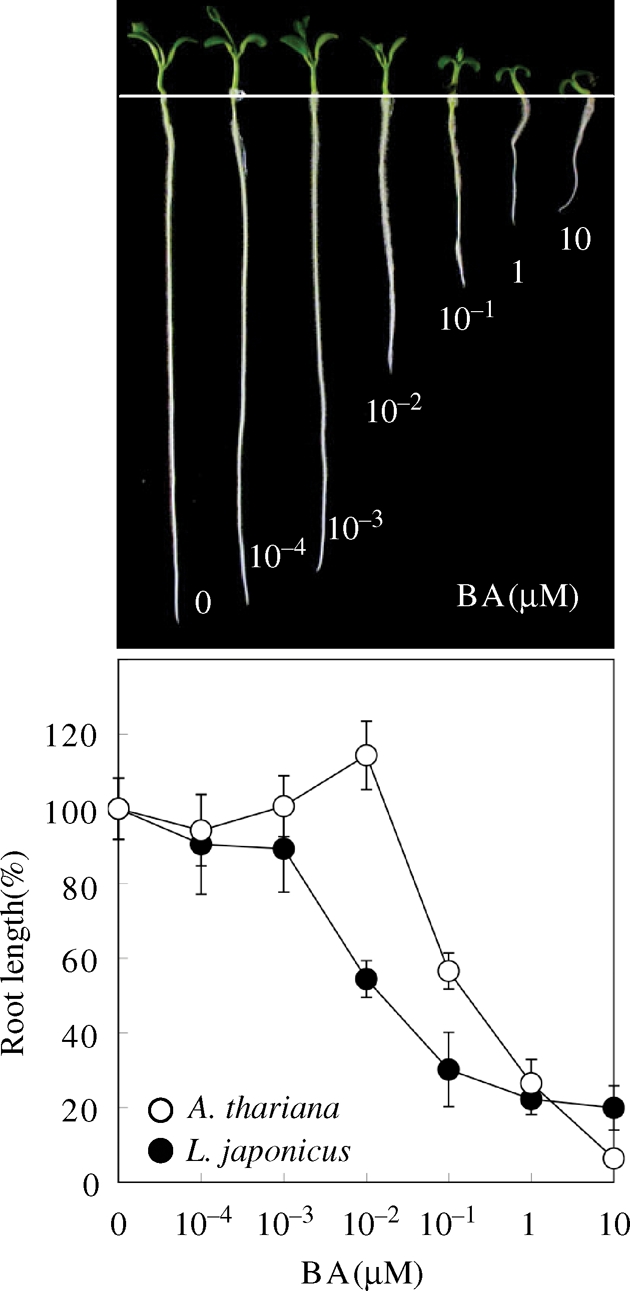
Cytokinin responses of L. japonicus. Cytokinin-induced inhibition of root elongation of L. japonicus seedlings. Seedlings were grown on vertically oriented MS agar-plates containing cytokinin (BA, 6-benzylaminopurine) at varied concentrations, as indicated. After being incubated for 9 days, the average lengths of their primary roots (n ≥ 20) were measured. Photographs were taken for each representative (upper panel), and the resulting lengths of root were plotted against the cytokinin concentrations tested. As a reference, essentially the same experiments were carried out with A. thaliana seedlings.
Figure 4.
Quantitative confirmation of cytokinin-induced expression of type-A RR genes in L. japonicus seedlings. (A) Cytokinin-induced expression of type-A RR genes in L. japonicus seedlings. Seedlings were grown MS gellan gum-plates under constant light conditions for 17 days, and then these seedlings were sprayed with cytokinin (20 µM t-zeatin in 0.02% DMSO). The seedlings were harvested immediately before and 0.5, 1.0, 3.0 h after the treatment and subjected to RNA preparation. A set of indicated RR transcripts were analyzed by means of semi-quantitative RT–PCR to measure the amounts of each transcript. The UBC transcript encoding ubiquitin-conjugating enzyme (chr1.LjT04O06.40, a homologue of Arabidopsis At5g25760) was used as an internal control, the type-B RRb4 transcript was a negative control, and the CKX3 encoding cytokinin oxidase enzyme (LjT02N03.150, a homologue of Arabidopsis CKX4) was a positive control. The results of reference samples treated with 0.02% DMSO for 1.0 h were also presented at the most right-hand side. The primer set, used for these PCR analyses, was listed in Supplementary Table S2. (B) The essentially the same experiments were repeated in a more quantitative manner by means of real-time quantitative RT–PCR. The experiments were replicated three times to obtain mean values with SD.
We identified 11 coding-sequences, each of which was inferred to specify a type-B RR protein, as judged by the fact that their deduced amino acid sequences (except for RRb8) contain the signature GARP DNA-binding motif, which is preceded by a receiver domain (Fig. 1). The current annotation for chr5.CM0072.220 and 230 must be reconsidered, because if these adjacent coding-sequences were combined together, one can deduce the existence of full-length type-B RR gene (here named RRb1). RRb8 (LiSGA contig) appears to be identical to a part of RRb4. It may be also noted that the invariant phospho-accepting aspartate residue is missing in the receiver domains of some RRb sequences (RRb9, RRb10, and RRb11). In any case, it was revealed that L. japonicus has a certain number of type-B RR genes, each of which is assumed to encode most likely a cytokinin-regulated transcription factor.
We also found some miscellaneous coding-sequences, which may or may not specify RR-related peptides (Table 1). Among them, chr.LjT07G07.80 seems to encode an intact receiver domain, whose amino acid sequence is related to the so-called type-C ARR24 of A. thaliana.46 We confirmed experimentally that this L. japonicus RR gene is transcribed in a manner independent of cytokinin (data not shown). Other two (chr.CM0133.120 and chr2.CM0695.44) might be derived from the C-terminal receiver domains of the hybrid sensors AtHK1-like and AHK5-like coding-sequences, respectively, as judged by the fact that the inferred amino acid sequences of these truncated forms of RRs are highly similar to those of AtHK1-like and AHK-like HKs, respectively. In any case, it is reasonable to expect that more RR-related genes will be uncovered when the whole genome sequence of L. japonicus has been entirely completed. Nevertheless, our results suggest that it is also certain that most of TCS-associated genes have already been uncovered in the current database of L. japonicus, as inspected above.
It may be worth mentioning that Puthiyaveeti and Allen47 claimed recently that A. thaliana has certain nuclear genes, which appear to encode chloroplast-localized HK (designated CSK, At1G67840) and RR (named TCP34, At3G26580), respectively. In this connection, there found L. japonicus coding sequences that encode amino acid sequences homologous to truncated portions of CSK. However, these coding sequences (chr4.LjT06I07.90 and chr4.LjT06I07.40) with different orientation to each other are separated by a large intervening sequence. Hence, it seems likely that L. japonicus lacks an intact CSK homologous gene. The inferred amino acid sequence (76 a.a.) from LjSGA_054723.1 is homologous to that of a small portion of TCP34 (350 a.a). Hence, it is also not certain that L. japonicus has an intact TCP34 homologous gene. In any case, they were included in Table 1.
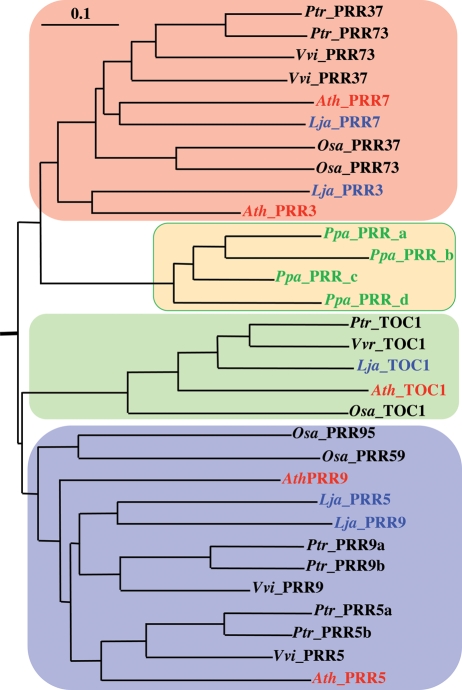
It is also of interest to search for the circadian clock-associated PRR genes in the current L. japonicus database (Fig. 1). Arabidopsis thaliana has five PRR genes (namely, TOC1/PRR1, PRR3, PRR5, PRR7, and PRR9), all of which play crucial roles within the central oscillator that generates fundamental circadian rhythms.6 Here, all of their counterparts (or orthologs) were found in the current database of L. japonicus, as listed in Table 1. Furthermore, one-to-one pairwise assignment was possible between PRRs of A. thaliana and L. japonicus, when phylogenetic tree was constructed for these PRR amino acid sequences (Fig. 5). However, it should be noted that the L. japonicus PRR3 gene might have been disrupted (or it is non-functional), because our inspection revealed that its coding-sequence is separated into three currently annotated ones (LjTo8o17.180, 130, and 120). These sequences are closely located on the same chromosome, but there are a few intervening unrelated coding-sequences between LjT08017.180 and130. Hence, it was assumed that L. japonicus had lost the functional PRR3 gene.
Figure 5.
A phylogenetic tree of the PRR family in some representative plants. Amino acid sequences of A. thaliana PRRs were characterized previously, whereas those of L. japonicus were done in this study. In addition to these, a set of putative PRR amino acid sequences were analyzed by employing the databases for some other plants, as noted in Fig. 2. In addition, we included a set of putative PRR sequences from Physcomitrella patens (Ppa, a moss) (http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html). Other details were essentially the same as those in Fig. 2.
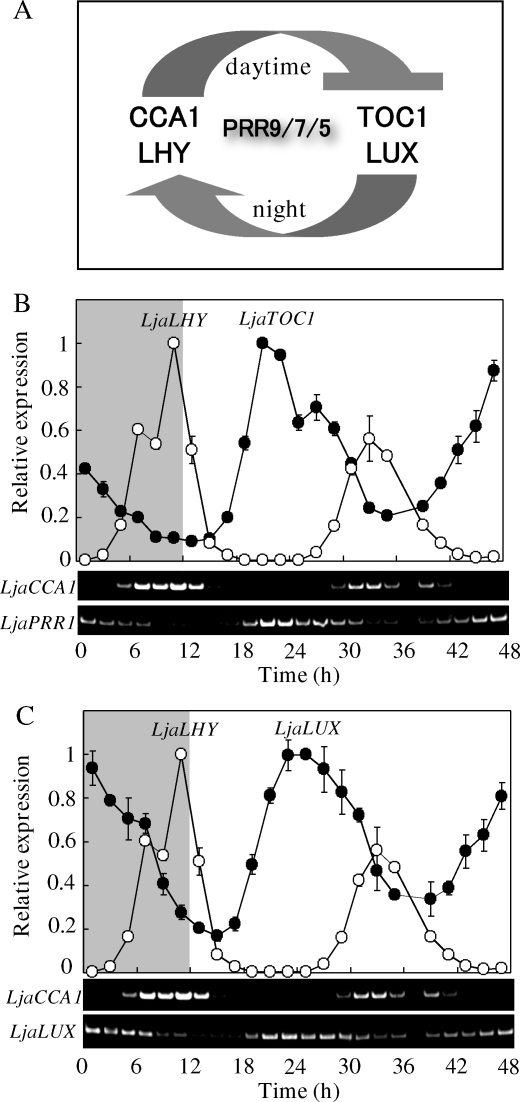
According to the phylogenetic tree (Fig. 5), it is clear that the PRR gene family is conserved among many higher plants, suggesting that the molecular mechanism underlying circadian clock is also highly conserved. This view is consistent with the fact that some highly related sequences are found even in Physcomitrella patens (Ppa, a moss), which is a basal lineage of land plant, and which had branched ∼450 millions years ago (Fig. 5).48 These inferred Ppa PRRs also have both the typical protein structure, in which a receiver domain is followed by a signature CCT [constans, constans-like, timing of cab expression 1 (TOC1)] motif (Fig. 1) (http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html). In any case, it is assumed that the molecular mechanism underlying the circadian clock appears to be conserved at least among higher plants, as has previously been demonstrated by comparing PRRs from A. thaliana and O. sativa.21 In this regard, it was shown that both A. thaliana and rice have additional clock-associated components, such as late elongated hypocotyl (LHY) (or circadian clock-associated 1) and LUX arrhythmo (LUX) (also known as PCL1).33,34 These clock-components, together with TOC1, constitute a fundamental core transcriptional feedback loop at the level of transcription, which generates a free-running and intrinsic rhythm, as schematically shown in Fig. 6A.49 Therefore, we searched for LHY-like and LUX-like genes in the L. japonicus database. Indeed, L. japonicus has a set of putative LHY and LUX orthologous genes, as also included in Table 1 (other putative clock-associated components in L. japonicus, such as GI, ELF3, ELF4, and ZTL, will be analyzed elsewhere, because they are not directly relevant to this subject).
Figure 6.
Characterization of free-running circadian rhythms of putative clock-associated genes of L. japonicus. (A) A schematic representation of the currently consistent model of central oscillator. This proposed model for A. thaliana means that the morning clock genes CCA1 and LHY act repressors for the evening clock genes TOC1 and LUX, in turn, these evening genes somehow activate the expression of CCA1/LHY. In addition, PRR9, PRR7, and PRR5 are the daytime clock genes, which are essential for the clock function. On the basis of this well-established clock model for A. thaliana, a set of putative clock-associated genes of L. japonicus was analyzed with regard to their free-running expression profiles. For this purpose, L. japonicus plants were grown in the 12 h light/12 h dark cycles for 17 days, and then they were released into the continuous light. RNA samples were prepared at intervals (every 3 h), as schematically shown in the panels (the shaded duration corresponds to the final dark period). The resulting expression profiles were analyzed with regard to the putative LHY and TOC1 genes (B) and the putative LHY and LUX genes (C), respectively, by means of both real-time and semi-quantitative RT–PCR (upper and lower parts, respectively). The primer set, used for these PCR analyses, was presented in Supplementary Table S2.
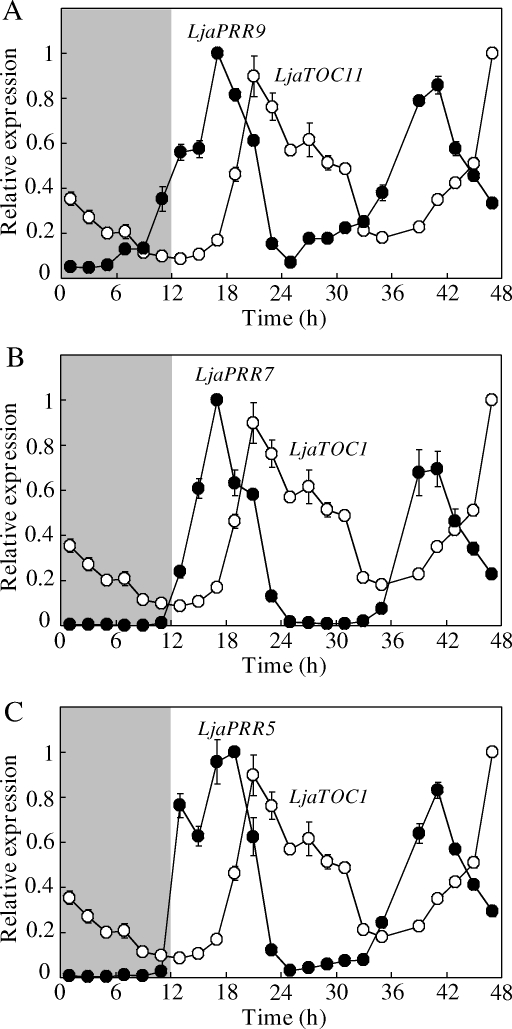
Then, we conducted a critical experiment, through which the diurnal expression profiles of these L. japonicus genes were examined. For this purpose, L. japonicus seedlings were grown under the 12 h light/12 h dark cycles for 17 days, and they were released into continuous light to see clock-controlled free-running rhythms. As shown in Fig. 6B and C, a robust oscillation at the level of transcription was observed for LHY of L. japonicus with a peak at the subjective dawn, whereas robust oscillations were observed for both TOC1 and LUX with a peak at the subjective dusk, respectively. These reciprocal expression profiles of LHY and TOC1 are quite similar to those observed for A. thaliana, suggesting the existence of common core oscillator in both L. japonicus and A. thaliana (Fig. 6A). This view was further confirmed by examining the circadian rhythms of putative PRR9, PRR7, and PRR5. These PRR genes showed each robust peak at daytime (e.g. after LHY and before TOC1) (Fig. 7). In this regard, a few specific comments should be made. (i) In contrast to the case of PRR9 of A. thaliana, PRR9 of L. japonicus did not markedly respond to a light exposure at morning.50 (ii) As mentioned above, we could not detect any transcript of putative PRR3 in L. japonicus (data not shown). In A. thaliana, it was recently shown that PRR3 plays a tissue-specific role in modulating the protein degradation of TOC1, and the clock-associated phenotype of a prr3 loss-of-function mutant is subtle.51,52 Hence, PRR3 might be dispensable for the fundamental clock function, and L. japonicus had lost such a PRR3 function, as mentioned above. Taken together, we concluded that the clock mechanism is well conserved between A. thaliana and L. japonicus, if not completely. In other words, the data compiled is informative for further studies on L. japonicus with special reference to the physiological functions of circadian clock, such as regulation of flowering time, as further discussed later.
Figure 7.
Further characterization of free-running circadian rhythms of putative PRR clock-associated genes of L. japonicus. To extend the results of Fig. 6, other putative clock genes (PRR9, PRR7, and PRR5) were also analyzed with regard to their circadian rhythms. Other details were the same as those given in Fig. 6.
It has been emphasized repeatedly that legumes are important plant species of choice for future genome-wide studies. One of many obvious reasons is that they have a unique ability to engage in beneficial symbiosis with nitrogen-fixing bacteria, which allows the host plant to utilize atmospheric nitrogen. Arabidopsis thaliana lacks this ability. This is one of the reasons why the genomes of not one but three leguminous species, namely L. japonicus (http://www.kazusa.or.jp/lotus/), M. truncatula (barrel medic; http://www.medicago.org/genome/), and Glycine max (soybean; http://www.phytozome.net/soybean), are currently the subjects of independent large-scale sequencing projects. Furthermore, the entire genome sequences of some symbiotic genera of Rhizobia (e.g. M. loti, Sinorhizobium meliloti, Bradyrhizobium japonicum) are also currently available (http://genome.kazusa.or.jp/rhizobase/). In this regard, we previously presented the compiled lists of TCS-associated components for these symbiotic genera. In this respect, notably, two independent groups recently demonstrated in L. japonicus that the cytokinin receptor HK is directly involved in root nodule organogenesis.35,36 Hence, the data compiled will provide us with a basis for further studies on L. japonicus with special reference to the symbiosis with nitrogen-fixing bacteria.
Not only these metabolic natures but also the phyllotaxy (or morphological nature) are quite different between A. thaliana and L. japonicus. It is currently general idea that the cytokinin-mediated signal transduction plays crucial roles in the maintenance of shoot apical meristem (SAM) and root meristematic niche, and also the pattern formation of leaf primordia (or phyllotaxy).53 Hence, L. japonicus would be one of the plant species of choice to study on mechanisms underlying phyllotaxy in connection with the cytokinin signal transduction.
Finally, it should be emphasized that A. thaliana is an annual herb, whereas L. japonicus is a perennial temperate pasture, suggesting that the mechanisms underlying circadian clock-controlled photoperiodic responses might also be different between these species.54,55 Such circadian clock-controlled photoperiodic responses should include the control of flowering time and the formation of freeze-tolerant buds (or SAM) of perennial pasture.56 These are just a few examples that fairly rationalize future studies on TCS-related components in L. japonicus. In short, a comprehensive overview was presented with regard to the TCS-associated components (HKs, HPs, RRs, together with PRRs), in comparison with those of A. thaliana. The database compiled here will provide us with a useful platform, on which one can ask many interesting questions with special reference to the general and optional biological roles of TCS-associated genes in L. japonicus.
Supplementary Data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This study was supported by a Grant-in-Aid for Scientific Research on Priority Areas (19039013 to T.M. and 20061016 to T.Y.) and a Grant-in-Aid for the GCOE Programs (Systems Biology) of the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Acknowledgments
We thank National BioResource Project (Legume Base) (http://www.legumebase.agr.miyazaki-u.ac.jp/index.jsp) for providing us with seeds of L. japonicus, and useful information for laboratory conditions to cultivate this legume. Thanks are also due to Dr M. Kawaguchi (National Institute of Basic Biology, Okazaki, Japan), Dr S. Sato (Kazusa DNA Research Institute, Kisarazu, Japan), and Dr S. Tabata (Kazusa DNA Research Institute, Kisarazu, Japan) for helpful advices.
Footnotes
Edited by Katsumi Isono
References
- 1.Mizuno T. His-Asp phosphotrasnsfer signal transduction. J. Biochem. (Tokyo) 1998;123:555–63. doi: 10.1093/oxfordjournals.jbchem.a021972. [DOI] [PubMed] [Google Scholar]
- 2.Stock A.M., Robinson V.L., Goudreau P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 3.Hwang I., Chen H.-I., Sheen J. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 2002;129:500–15. doi: 10.1104/pp.005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizuno T. Plant response regulators implicated in signal transduction and circadian rhythm. Curr. Opin. Plant Biol. 2004;7:499–505. doi: 10.1016/j.pbi.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 5.Mizuno T. Two-component phosphorelay signal transduction systems in plants: from hormone response to circadian rhythms. Biosci. Biotechnol, Biochem. 2005;69:2263–76. doi: 10.1271/bbb.69.2263. [DOI] [PubMed] [Google Scholar]
- 6.Mizuno T., Nakamichi N. Pseudo response regulators (PRR) or true oscillator components (TOC) Plant Cell Physiol. 2005;46:677–85. doi: 10.1093/pcp/pci087. [DOI] [PubMed] [Google Scholar]
- 7.Nakamichi N., Kita M., Niinuma K., et al. Arabidopsis clock-associated pseudo-response regulatorsPRR9, PRR7, and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 2007;48:822–32. doi: 10.1093/pcp/pcm056. [DOI] [PubMed] [Google Scholar]
- 8.Müller B., Sheen J. Arabidopsis cytokinin signalling pathway. Sci. STKE. 2007;9:407–8. doi: 10.1126/stke.4072007cm5. [DOI] [PubMed] [Google Scholar]
- 9.Kendrick M.D., Chang C. Ethylene signalling: new levels of complexity and regulation. Curr. Opin. Plant Biol. 2008;11:479–85. doi: 10.1016/j.pbi.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.To J.P., Kieber J.J. Cytokinin signalling: two-components and more. Trends Plant Sci. 2008;13:85–92. doi: 10.1016/j.tplants.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Z., Guo H. Genetic basis of ethylene perception and signal transduction in Arabidopsis. J. Integr. Plant Biol. 2008;50:808–15. doi: 10.1111/j.1744-7909.2008.00710.x. [DOI] [PubMed] [Google Scholar]
- 12.Mizuno T., Mizushima S. Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of porin genes. Mol. Microbiol. 1990;4:1077–82. doi: 10.1111/j.1365-2958.1990.tb00681.x. [DOI] [PubMed] [Google Scholar]
- 13.Mizuno T., Kaneko T., Tabata S. Compilation of all genes encoding bacterial two-component signal transducers in the genome of the cyanobacterium, Synechocystis sp. strain PCC 6803. DNA Res. 1996;3:407–14. doi: 10.1093/dnares/3.6.407. [DOI] [PubMed] [Google Scholar]
- 14.Mizuno T. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 1997;4:161–8. doi: 10.1093/dnares/4.2.161. [DOI] [PubMed] [Google Scholar]
- 15.Ohmiya R., Yamada H., Kato C., et al. The Prr1 response regulator is essential for transcription of ste11+ and sexual development in fission yeast. Mol. Gen. Genet. 2000;264:441–51. doi: 10.1007/s004380000305. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki T., Sakurai K., Imamura A., et al. Compilation and characterization of histidine-containing phosphotransmitters implication in His-to-Asp phosphorelay in plants: AHP signal transducers of Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2000;64:2486–9. doi: 10.1271/bbb.64.2486. [DOI] [PubMed] [Google Scholar]
- 17.Ueguchi C., Koizumi H., Suzuki T., et al. Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 2001;42:231–5. doi: 10.1093/pcp/pce015. [DOI] [PubMed] [Google Scholar]
- 18.Nakamichi N., Yamada H., Aoyama K., et al. His-to-Asp phosphorelay circuitry regulates sexual development in Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 2002;66:2663–72. doi: 10.1271/bbb.66.2663. [DOI] [PubMed] [Google Scholar]
- 19.Hagiwara D., Yamashino T., Mizuno T. Genome-wide comparison of the His-to-Asp phosphorelay signaling components of three symbiotic genera of Rhizobia. DNA Res. 2004;11:57–65. doi: 10.1093/dnares/11.1.57. [DOI] [PubMed] [Google Scholar]
- 20.Hagiwara D., Asano Y., Mari J., et al. The SskA and SrrA response regulators are implicated in oxidative stress response of hyphae and asexual spores in the phosphorelay signaling network of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2007;71:1003–14. doi: 10.1271/bbb.60665. [DOI] [PubMed] [Google Scholar]
- 21.Murakami M., Tago Y., Yamashino T., et al. Comparative overviews as to clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007;48:110–121. doi: 10.1093/pcp/pcl043. [DOI] [PubMed] [Google Scholar]
- 22.Schaller G.E., Doi K., Hwang I., et al. Nomenclature of two-component signaling elements of rice. Plant Physiol. 2007;143:555–7. doi: 10.1104/pp.106.093666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szczyglowski K., Stougaard J. Lotus genome: pod of gold for legume research. Trends Plant Sci. 2008;13:515–7. doi: 10.1016/j.tplants.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Sato S., Tabata S. Lotus japonicus as a platform for legume research. Curr. Opin. Plant Biol. 2005;9:128–32. doi: 10.1016/j.pbi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Sato S., Nakamura Y., Kaneko T., et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15:227–39. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Town C.D. Annotating the genome of Medicago truncatula. Curr. Opin. Plant Biol. 2006;9:122–7. doi: 10.1016/j.pbi.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Hosoda K., Imamura A., Katoh E., et al. Molecular structure of the GARP family of plant Myb-related DNA-binding motifs of the Arabidopsis response regulators. Plant Cell. 2002;14:2015–29. doi: 10.1105/tpc.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiba T., Yamada H., Sato S., et al. The type-A response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2003;44:868–74. doi: 10.1093/pcp/pcg108. [DOI] [PubMed] [Google Scholar]
- 29.To J.P., Haberer G., Ferreira F.J., et al. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16:658–71. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mason M.G., Mathews D.E., Argyros D.A., et al. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell. 2005;17:3007–18. doi: 10.1105/tpc.105.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishida K., Yamashino T., Yokoyama A., et al. Three hype-B response regulators, ARR1, ARR10, and ARR12 play essential but redundant roles in cytokinin signal transduction through the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 2008;49:47–57. doi: 10.1093/pcp/pcm165. [DOI] [PubMed] [Google Scholar]
- 32.Matsushika A., Makino S., Kojima M., et al. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol. 2000;41:1002–12. doi: 10.1093/pcp/pcd043. [DOI] [PubMed] [Google Scholar]
- 33.Gardner M.J., Hubbard K.E., Hotta C.T., et al. How plants tell the time. Biochem. J. 2006;397:15–24. doi: 10.1042/BJ20060484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McClung C.R. Comes a time. Curr. Opin. Plant Biol. 2008;11:514–20. doi: 10.1016/j.pbi.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 35.Murray J.D., Karas B.J., Sato S., et al. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science. 2007;315:101–4. doi: 10.1126/science.1132514. [DOI] [PubMed] [Google Scholar]
- 36.Tirichine L., Sandal N., Madsen L.H., et al. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science. 2007;315:104–7. doi: 10.1126/science.1132397. [DOI] [PubMed] [Google Scholar]
- 37.Asamizu E., Shimoda Y., Kouchi H., et al. A positive regulatory role for LjERF1 in the nodulation process is revealed by systematic analysis of nodule-associated transcription factors of Lotus japonicus. Plant Physiol. 2008;147:2030–40. doi: 10.1104/pp.108.118141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nukui N., Ezura H., Minamisawa K. Transgenic Lotus japonicus with an ethylene receptor gene Cm-ERS1/H70A enhances formation of infection threads and nodule primordia. Plant Cell Physiol. 2004;45:427–35. doi: 10.1093/pcp/pch046. [DOI] [PubMed] [Google Scholar]
- 39.Kakimoto T. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science. 1996;274:982–5. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- 40.Urao T., Yakubov B., Satoh R., et al. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell. 1999;11:1743–54. doi: 10.1105/tpc.11.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwama A., Yamashino T., Tanaka Y., et al. AHK5 His-kinase regulates root elongation through an ETR1-dependent integrated abscisic acid and ethylene signaling pathway in Arabidopsis thaliana. Plant Cell Physiol. 2007;48:375–80. doi: 10.1093/pcp/pcl065. [DOI] [PubMed] [Google Scholar]
- 42.Mahonen A.P., Bishopp A., Higuchi M., et al. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311:94–8. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 43.Imamura A., Hanaki N., Nakamura A., et al. Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 1999;40:733–42. doi: 10.1093/oxfordjournals.pcp.a029600. [DOI] [PubMed] [Google Scholar]
- 44.Bradstatter I., Kieber J.J. Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell. 1998;10:1009–19. doi: 10.1105/tpc.10.6.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taniguchi M., Kiba T., Sakakibara H., et al. Expression of Arabidopsis response regulator homologs is induced by cytokinins and nitrate. FEBS Lett. 1998;429:259–62. doi: 10.1016/s0014-5793(98)00611-5. [DOI] [PubMed] [Google Scholar]
- 46.Kiba T., Naitou T., Koizumi N., et al. Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His-Asp phosphorelay circuitry. Plant Cell Physiol. 2005;46:339–55. doi: 10.1093/pcp/pci033. [DOI] [PubMed] [Google Scholar]
- 47.Puthiyaveetil S., Allen J.F. Chloroplast two-component systems: evolution of the link between photosynthesis and gene expression. Proc. R. Soc. Lond. B. 2009;276:2113–47. doi: 10.1098/rspb.2008.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quatrano R.S., McDaniel S.F., Khandelwal A., et al. Physcomitrella patens: mosses enter the genomic age. Curr. Opin. Plant Biol. 2007;10:182–9. doi: 10.1016/j.pbi.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 49.Alabadi D., Oyama T., Yanovsky M.J., et al. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–3. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 50.Matsushika A., Imamura A., Yamashino T., et al. Aberrant expression of the light-inducible and circadian-regulated APRR9 gene belonging to the circadian-associated APRR1/TOC1 quintet results in the phenotype of early flowering in Arabidopsis thaliana. Plant Cell Physiol. 2002;43:833–43. doi: 10.1093/pcp/pcf118. [DOI] [PubMed] [Google Scholar]
- 51.Murakami M., Yamashino T., Mizuno T. Characterization of circadian-associated APRR3 pseudo-response regulator belonging to the APRR/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:645–50. doi: 10.1093/pcp/pch065. [DOI] [PubMed] [Google Scholar]
- 52.Para A., Farre E.M., Imaizumi T., et al. PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell. 2007;19:3462–73. doi: 10.1105/tpc.107.054775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fleming A.J. Formation of primordia and phyllotaxy. Curr. Opin. Plant Biol. 2005;8:53–8. doi: 10.1016/j.pbi.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 54.Hotta C.T., Gardner M.J., Hubbard K.E., et al. Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 2007;30:333–49. doi: 10.1111/j.1365-3040.2006.01627.x. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Q., Li H., Li R., et al. Association of the circadian rhythmic expression of GmCRY1a with a latitudinal cline in photoperiodic flowering of soybean. Proc. Natl Acad. Sci. USA. 2008;105:21028–33. doi: 10.1073/pnas.0810585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolters H., Jurgens G. Survival of the flexible: hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 2009;10:305–17. doi: 10.1038/nrg2558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.