Abstract
The Drosophila Augmin complex localizes γ-tubulin to the microtubules of the mitotic spindle, regulating the density of spindle microtubules in tissue culture cells. Here, we identify the microtubule-associated protein Msd1 as a new component of the Augmin complex and demonstrate directly that it is required for nucleation of microtubules from within the mitotic spindle. Although Msd1 is necessary for embryonic syncytial mitoses, flies possessing a mutation in msd1 are viable. Importantly, however, in the absence of centrosomes, microtubule nucleation from within the spindle becomes essential. Thus, the Augmin complex has a crucial role in the development of the fly.
Keywords: Augmin, Drosophila, Msd1, γ-tubulin
Results and Discussion
Msd1 is required for localization of γ-tubulin to the mitotic spindle
In a previous functional proteomic study on the Drosophila “microtubule (MT) interactome,” we identified the gene CG13914/mitotic spindle density 1 (msd1) as encoding a MT-associated protein (MAP) present in early embryos (Hughes et al. 2008). We showed that treatment of S2 tissue culture cells with dsRNA against msd1 for 5 d led to the formation of monopolar spindles, and of long, bipolar mitotic spindles that showed a reduced density of MTs in their central region (Hughes et al. 2008). As msd1 was not identified in a genome-wide RNAi screen for genes involved in spindle formation in S2 cells (Goshima et al. 2007), we first sought to confirm and extend these original observations. As expected, cells fixed 2 d following treatment with msd1 dsRNA showed an increase in bipolar spindles possessing the weak spindle density phenotype, when compared with control cells (Fig. 1A–C). To assess the dynamics of spindle formation after msd1 depletion, we followed MTs in cells expressing GFP-α-tubulin. In control cells, a bipolar spindle formed, which progressed through mitosis within 30 min (Fig. 1D; Supplemental Movie S1). In contrast, cells treated with msd1 dsRNA failed to build a robust spindle, although astral MTs were clearly visible, arresting in a metaphase-like state for the duration of observation (Fig. 1E; Supplemental Movie S2).
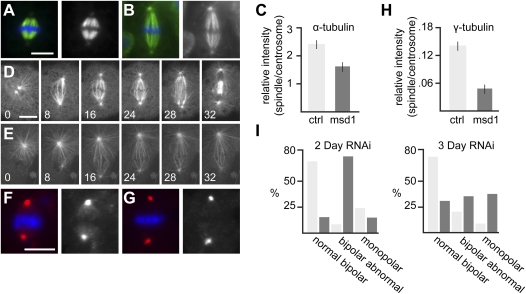
Figure 1.
Msd1 is a dim γ-tubulin (Dgt) protein required for regulating mitotic spindle density in S2 cells. (A,B) Metaphase in control S2 cell (A) and S2 cell treated with msd1 dsRNA (B), showing α-tubulin (green) and DNA (blue). Bar, 10 μm. (C) Bar chart showing ratio of intensity of α-tubulin on the spindle and centrosomes in control S2 cells and cells following msd1 knockdown. The ratio in msd1 dsRNA-treated cells (1.62 ± 0.17 SEM; n = 17) is significantly lower than in controls (2.40 ± 0.16 SEM; n = 13; P < 0.005). (D,E) Frames taken from time lapse of mitotic spindle formation in a control (D) and in an msd1 dsRNA-treated (E) S2 cell, expressing GFP-α-tubulin. Times are shown in minutes. Bar, 10 μm (Supplemental Movies S1, S2). (F,G) Metaphase in control S2 cell (F) and S2 cell treated with msd1 dsRNA (G), showing γ-tubulin (red) and DNA (blue). Bar, 10 μm. (H) Bar chart showing ratio of intensity of γ-tubulin on the spindle and centrosomes in control S2 cells and cells following msd1 knockdown. The ratio in msd1 dsRNA-treated cells (0.0469 ± 0.010 SEM; n = 28) is significantly lower than in controls (0.140 ± 0.01 SEM; n = 32; P < 0.0005). (I) Bar chart showing proportion of cells displaying normal metaphase spindles (as in A), bipolar weak density mitotic spindles (as in B), or monopolar spindles in control cells (light bars) or cells treated with msd1 dsRNA (dark bars), fixed 2 d (left graph) and 3 d (right graph) following treatment.
Our initial studies also showed an increase of monopolar spindles in msd1 dsRNA-treated cells, in comparison with control-treated cells (Hughes et al. 2008). We found that, after incubation of cells with dsRNA against msd1 for 2 d, the presence of monopolar spindles was similar to control-treated cells. However, an additional day of msd1 dsRNA treatment led to a significant increase in monopolar spindles and a decrease in cells with low density spindles (P > 0.001 for both cases) (Fig. 1I). This suggests that the weak spindle density phenotype reflects a reduction, but not absence, of Msd1 and that in S2 cells, loss of Msd1 leads to an inability to form a stable bipolar spindle.
We originally placed msd1 within a set of genes showing similar phenotypes, three of which were also identified as dim γ-tubulin (dgt) genes (Goshima et al. 2007; Hughes et al. 2008). Recently, the Dgt proteins have been shown to form a complex, termed Augmin, that is required to recruit the MT nucleator, γ-tubulin, to the mitotic spindle in S2 cells (Goshima et al. 2007, 2008). We therefore hypothesized that Msd1 might also be involved in this process. Indeed, when cells treated with msd1 dsRNA were fixed and stained with an antibody against γ-tubulin, a consistent reduced ratio of γ-tubulin signal was observed between mitotic spindle and centrosomes when compared with control cells (Fig. 1F–H). Thus, Msd1 has a role in γ-tubulin recruitment to the mitotic spindle in S2 cells.
Msd1 is a MAP required for correct localization of both Augmin and γ-TuRC components
msd1 encodes a 138-amino-acid MAP, the first 40 amino acids of which are predicted to encode a coiled-coil domain, using COILS (Lupas et al. 1991; data not shown). To confirm the interaction between Msd1 and MTs in vivo, we generated flies expressing an Msd1-GFP fusion protein in the early embryo and subjected extracts to MT cosedimentation. As expected, Msd1-GFP associated with MTs in the presence of taxol (Fig. 2A). In good agreement, we found Msd1-GFP to localize to MTs throughout mitosis (Fig. 2B; Supplemental Movie S3).
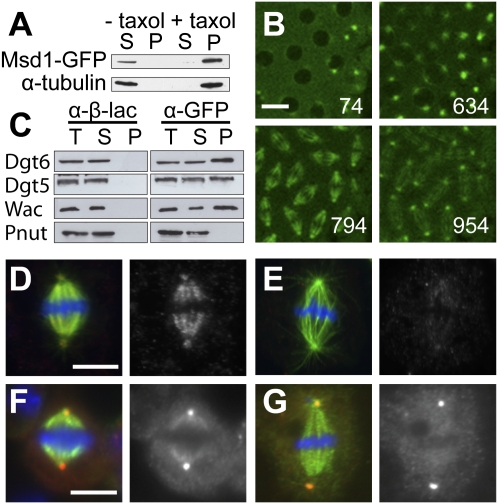
Figure 2.
Msd1 is a MAP and a component of the Augmin complex. (A) Msd1-GFP MT cosedimentation assay. Msd1-GFP and α-tubulin are found in the supernatant (S) in the absence of taxol, but are present in the pellet (P) following polymerization of MTs with taxol. (B) Subcellular localization of Msd1-GFP in a living embryo. The elapsed time (in seconds) after the beginning of the time-lapse recording is given in the bottom right corner of each image (Supplemental Movie S3). Bar, 10 μm. (C) Coimmunoprecipitation of members of the Augmin complex (Dgt5, Dgt6, and Wac), but not the unrelated MAP Pnut, from Msd1-GFP embryos, using anti-GFP. (T) Total extract; (S) supernatant; (P) pellet. Anti-β-galactosidase does not precipitate members of the Augmin complex. (D,E) Metaphase in control S2 cell (D) and S2 cell treated with msd1 dsRNA (E) showing α-tubulin (green), Dgt6 (red), and DNA (blue). Dgt6 localizes along the MTs of the mitotic spindle in control S2 cells (average intensity over half-spindles of 1200, n = 43, SEM ± 51). In msd1 dsRNA-treated cells, localization to the mitotic spindle is significantly reduced (average intensity over half spindles of 746, n = 39, SEM ± 27; P > 0.0005). Bar, 10 μm. (F,G) Metaphase in control S2 cell (F) and S2 cell treated with msd1 dsRNA (G) showing α-tubulin (green), Dgrip71 (red) and DNA (blue). The ratio of intensity of Dgrip71 on the spindle and centrosomes in msd1 dsRNA-treated cells (0.219 ± 0.019 SEM; n = 40) is significantly lower than in controls (0.388 ± 0.026 SEM; n = 32; P < 0.0005). Bar, 10 μm.
Next, we immunoprecipitated Msd1-GFP from embryos and investigated whether the fusion protein can interact physically with Augmin. While Augmin components did not coprecipitate with a control antibody (Fig. 2C, left panel), we found that Msd1-GFP coprecipitated all Augmin subunits tested: Dgt5, Dgt6, Wac (Fig. 2C, right panel; Goshima et al. 2007, 2008; Hughes et al. 2008; Meireles et al. 2009), and Dgt4 (data not shown). In addition, Msd1-GFP cofractionated with Augmin in embryo extracts subjected to sucrose gradient centrifugation (Supplemental Fig. S1). Moreover, we found that the localization of Augmin was dependent on Msd1; in control cells, the Augmin subunit Dgt6 is present along spindle MTs (Fig. 2D; Goshima et al. 2007), a localization that was markedly reduced in cells treated with msd1 dsRNA (Fig. 2E).
Cytosolic γ-tubulin exists in complexes (γ-TuSC and γ-TuRC) that regulate its capacity to nucleate MTs from specific cellular locations (Raynaud-Messina and Merdes 2007). Recently, the human homolog of Dgt6, FAM29A, has been shown to recruit γ-tubulin to the MTs of the mitotic spindle via the γ-TuRC subunit, NEDD1/GCP-WD (Zhu et al. 2008). We therefore investigated whether Msd1 is required for the spindle localization of Dgrip71, the Drosophila NEDD1/GCP-WD homolog. As reported previously, Dgrip71 localized to centrosomes and mitotic spindles (Fig. 2F; Verollet et al. 2006). However, in msd1 dsRNA-treated cells, the localization of Dgrip71 to the mitotic spindle was specifically reduced in comparison with that at centrosomes (Fig. 2G; Supplemental Fig. S2).
Together, the above studies show that Msd1 is an in vivo MAP that biochemically associates with, and regulates the localization of, Drosophila Augmin. Our analysis also demonstrates that Msd1 is required for localization of Dgrip71 and γ-tubulin to the mitotic spindle of S2 cells. These findings draw parallels with studies in human tissue culture cells and reinforce the suggestion that Augmin, and the mechanism by which it regulates a dense MT network in the mitotic spindle, may be evolutionarily conserved.
Msd1 is required for nucleation of MTs from within the mitotic spindle
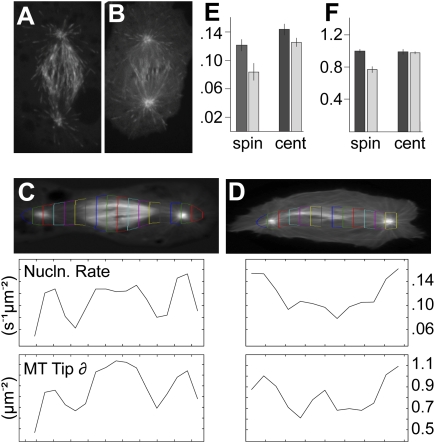
MTs have been reported to be nucleated by γ-tubulin present on pre-existing MTs in a variety of organisms. For example, in interphase tobacco BY-2 cells, γ-tubulin localizes to punctae along cortical MTs, regulating the growth of new MTs nucleated from branch points (Murata et al. 2005), while in Schizosaccharomyces pombe, γ-tubulin is required for the formation of MT bundles via MT-dependent MT nucleation during interphase (Janson et al. 2005). A similar mechanism has been proposed in Drosophila, where γ-tubulin is thought to contribute to the increase in density of MTs constituting the mitotic spindle (Goshima et al. 2008). This model stems from the observations that (1) the MT plus-end-binding protein EB1 labels MTs emerging from throughout the mitotic spindle in addition to those nucleated from both centrosomes and around chromatin (Mahoney et al. 2006), and (2) the decrease in spindle density that occurs when the Augmin complex is silenced in S2 cells corresponds to a decrease in spindle-associated γ-tubulin (Goshima et al. 2008). We therefore asked whether reducing Msd1 levels results in a decrease in EB1-GFP-labeled MTs emerging from within the mitotic spindle. In control-treated S2 cells expressing EB1-GFP, punctae are clearly visible throughout the mitotic spindle, including areas distinct from either the centrosomes or chromatin (Fig. 3A; Supplemental Movie S4). Strikingly, however, in cells treated with msd1 dsRNA, EB1-GFP punctae can be seen originating from around the poles and in the center of the spindle (presumably from around the chromatin), but very few are present within the mitotic spindle itself (Fig. 3B; Supplemental Movie S5). To quantify this effect, we designed software to track the EB1-GFP punctae over time. Initial points of the trajectories were collected and their spatio-temporal distribution analyzed, as well as the distribution of all points in the trajectories, giving both the MT growth initiation rate density and the density of MT plus ends in selected regions (see the Materials and Methods). As expected, RNAi against msd1 significantly reduced both the density and nucleation rate (the latter defined as appearance of new EB1-GFP ends per unit area over time) of MT plus ends in the region of the mitotic spindle, in comparison with control-treated cells, but had little effect on density and nucleation rate at centrosomes (Fig. 3C–F). Thus we conclude that Msd1 is required for mitotic spindle-templated MT nucleation.
Figure 3.
Msd1 regulates intraspindle MT nucleation. (A,B) Frames taken from time-lapse images of control (A) and msd1 dsRNA-treated (B) S2 cells, expressing EB1-GFP. (C,D) Images of movie-averaged real-time localization of EB1-GFP in control (C) and msd1 dsRNA-treated (D) cells. Regions of interest (RoIs) were selected around each spindle and segmented into 1.26-μm strips along the axis of the spindle. The growing MT tip density (MT Tip δ) and MT nucleation (MT Nucln.) in each strip was calculated (see the Materials and Methods) and are represented as graphs. (E,F) Bar charts showing the growing MT tip density (E) and MT nucleation (F) values for each strip averaged over all time frames of each data set.
msd1 mutant flies are viable but female sterile
The above results demonstrate the importance of mitotic spindle-templated MT nucleation in Drosophila tissue culture cells. To assess the role of Msd1 and therefore this mechanism of MT nucleation in a living organism, we generated mutants of msd1 (Fig. 4A). Flies homozygous or hemizygous (over a deficiency chromosome that uncovers the msd1 gene) for the msd1 mutation msd1ex51 are viable but female sterile (Supplemental Fig. S3). Perhaps surprisingly, sequencing demonstrated that the resultant mutation is due to a deletion internal to the P-element, rather than affecting the surrounding msd1 sequence (Fig. 4A). However, three pieces of evidence confirmed that the deletion at this locus affects levels of Msd1 in the fly: (1) the female sterility was fully rescued by maternal expression of Msd1-GFP (Supplemental Fig. S3); (2) mutant embryos laid by hemizygous mothers arrested development at an earlier stage than those laid by homozygote msd1ex51 mothers (Supplemental Fig. S4); and (3) the expression of msd1 was shown to be greatly reduced using RT–PCR in msd1ex51 third-instar larvae (Fig. 4B).
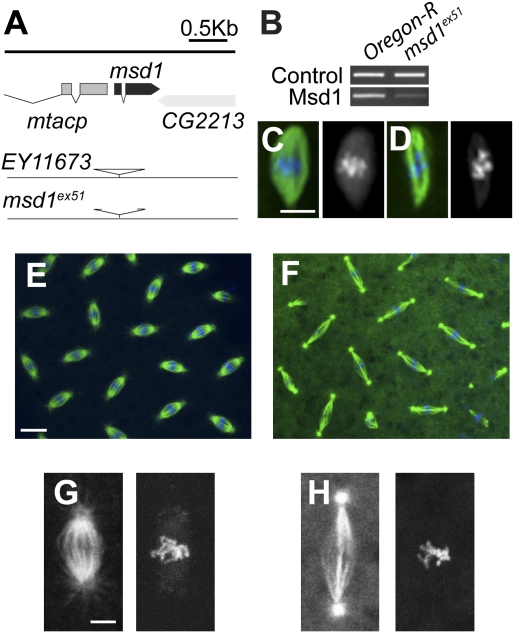
Figure 4.
msd1 mutants are viable, but female sterile. (A) Diagram of msd1 gene region. msd1 (shown in dark gray) spans a small genomic region at 61F6 on the third chromosome. The P-element in stock EY11673 inserted in the 5′ untranslated region (UTR) of msd1 was remobilized to generate the msd1ex51 mutation, which has a deletion internal to the P-element. (B) RT–PCR to compare expression of msd1 in wild-type (WT) and msd1 mutants. Control primers show equal amounts of template RNA extracted from wild-type and msd1 homozygous third instar larvae. Msd1 RNA-specific primers demonstrate a reduction of msd1 expression in msd1ex51 mutants. (C,D) Acentrosomal female meiosis I spindle from eggs laid by wild-type (C) or homozygous msd1 mutant mothers (D), α-tubulin (green), DNA (blue). Bar, 5 μm. (E,F) Wild-type (E) and msd1 mutant (F) embryo during metaphase of cycle 10; α-tubulin (green), DNA (blue). Bar, 10 μm. (G,H) Close-ups of metaphase spindles from wild-type (G) and msd1 mutant (H) embryos. Bar, 2 μm.
To understand why msd1 mutant females are sterile, we stained 0- to 3-h embryos laid by wild-type or msd1ex51 mutant mothers (hereafter msd1 mutant embryos) for DNA and MTs. Following female meiosis and fertilization, Drosophila embryos undergo 13 rounds of synchronous mitoses in the absence of cytokinesis. Female meiotic spindles in msd1 mutant embryos appeared to show qualitative differences to wild type, both in terms of apparent spindle density and/or length, and in chromosome alignment (Fig. 4C,D). However, bipolar spindle organization was not affected, and many msd1 mutant embryos underwent a variable number of mitotic divisions, before accumulating defects and arresting prior to cellularization (Fig. 4E,F; Supplemental Fig. S4). Similarly to the original phenotype in S2 cells (Hughes et al. 2008), the primary defect observed in msd1 mutant embryos was an increase in mitotic spindle length during metaphase, and a reduction in mitotic spindle density, when compared with wild-type spindles (Fig. 4G,H).
Although the above results show that Msd1 has a crucial role in the rapid mitotic divisions of the early embryo, the viability of msd1ex51 homozygous and hemizygous mutants suggests that it is not essential. Although this may be due to the hypomorphic nature of the allele, it is in good agreement with the phenotype of null mutants of another recently described Drosophila Augmin gene, wac (Meireles et al. 2009). Together, these mutants suggest that the Augmin complex is dispensable for viability, and that mitotic spindle-templated MT nucleation is not essential for most somatic mitoses or for the formation of acentrosomal polarized MT arrays in Drosophila.
Msd1 contributes to mitotic progression and is essential for spindle-associated γ-tubulin, in vivo
As in S2 cells, γ-tubulin is normally present on both centrosomes and the mitotic spindles of dividing neuroblasts (Fig. 5A). However, in msd1 mutant cells, γ-tubulin localizes solely to the centrosomes (Fig. 5B). This confirms that levels of Msd1 in these mutant larval neuroblasts are at least sufficiently reduced to disrupt the localization of γ-tubulin to the mitotic spindle, and thus the ability of γ-tubulin to facilitate MT nucleation from within the spindle. Intriguingly, however, immunofluorescence of wild-type and msd1 mutant third-instar larval neuroblasts, fixed and stained to reveal α-tubulin, failed to uncover any visible abnormalities in mitotic spindle formation (Fig. 5C,D). No quantifiable difference in MT spindle density was observed, nor was there any significant increase of monopolar spindles (data not shown). However, DAPI-stained squashes of third instar larvae showed that msd1 mutants have an increased mitotic index and a decrease in the proportion of cells in anaphase (wild-type MI 1.12, 392 fields, n = 4; msd1 MI 2.17, fields 379, n = 4) (Fig. 5G), suggesting that reduction of Msd1 levels results in a prolonged prometaphase/metaphase.
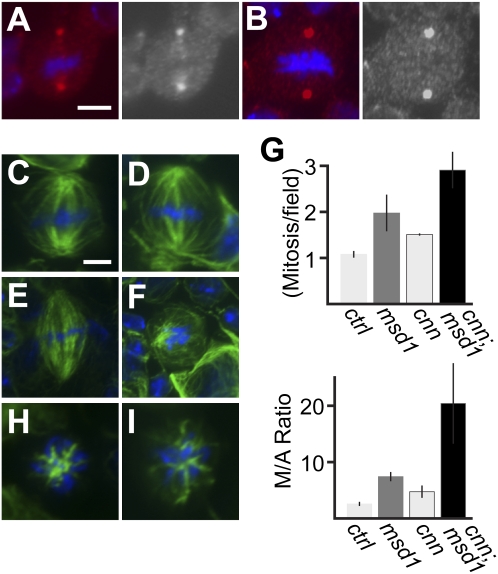
Figure 5.
Msd1 is essential for neuroblast mitosis in the absence of functional centrosomes. (A,B). Wild-type (A) and msd1 mutant (B) mitotic neuroblasts showing γ-tubulin (red) and DNA (blue). Bar, 5 μm. (C,D) Wild-type (C) and msd1 mutant (D) neuroblast showing α-tubulin (green) and DNA (blue). Bar, 5 μm. (E,F) Mitotic larval neuroblast from a cnn mutant (E) and cnn;msd1 double mutants (F) showing α-tubulin (green) and DNA (blue). (G) Bar chart showing mitotic index (Metaphase/field) and metaphase-to-anaphase ratio (M/A Ratio) in wild-type, msd1, cnn, and cnn; msd1 mutants. (H,I) MTs (green) and DNA (blue) during prometaphase in cnn mutants (H) and cnn;msd1 double mutants (I), showing MT nucleation around mitotic chromatin.
The differences observed between the phenotypes of msd1 dsRNA treatment in S2 cells, msd1 mutant embryos, and msd1 mutant neuroblasts are intriguing. Perhaps the simplest explanation is that different cell types rely on the same underlying mechanisms for spindle assembly, but that they do so to different extents. In addition to Augmin-driven mitotic spindle-templated MT nucleation, two other pathways are known to contribute to spindle assembly in animal cells: the centrosome-directed pathway that predominates in most cell types and the Ran-GTP-driven chromatin-mediated pathway that is essential for acentrosomal divisions such as female meiosis and cells in which centrosomes have been inactivated (for review, see Walczak and Heald 2008). It is possible that MTs nucleated from centrosomes and chromatin contribute more to mitotic spindle assembly in neuroblasts than in tissue culture cells or the syncytial blastoderm, placing less reliance on mitotic spindle-templated MT nucleation. In support of this, γ-tubulin on the spindle in neuroblasts, although very consistent, is subtle compared with that seen in S2 cells (cf. Figs. 1F and 5A), and embryos (A Wainman, unpubl.). Furthermore, Drosophila blastoderm embryos undergo mitosis very rapidly. Although these embryos possess an intact spindle checkpoint, the presence of a proportion of abnormal spindles within a normal population is insufficient to arrest the entire embryo. Thus, many mutations affecting spindle formation in this tissue result in asynchrony, nuclear fallout, and a variety of aberrant phenotypes (e.g., Gonzalez et al. 1990; Glover et al. 1995; Megraw et al. 1999). It is therefore likely that the perturbation in spindle organization seen in msd1 mutant embryos leads to an accumulation of defects during the following divisions and subsequent female sterility.
Msd1 is required for the viability of flies in the absence of functioning centrosomes
Analysis of null mutants in genes expressing core centrosomal or centriolar proteins, such as Centrosomin (Cnn) and D-Sas4, demonstrates unequivocally that Drosophila can develop to adulthood without functioning centrosomes (Megraw et al. 2001; Basto et al. 2006). This is thought to be due to compensatory MT nucleation via the Ran-GTP-driven chromatin-mediated pathway. However, it could also be due, at least in part, to mitotic spindle-templated MT nucleation. To address this, we compared spindle formation in larval neuroblasts carrying mutations in either cnn alone or both cnn and msd1. Third instar larval neuroblasts from cnn mutants have mitotic spindles with broad poles, lacking astral MTs (Fig. 5E; Megraw et al. 2001). However, these spindles are capable of orchestrating chromosome segregation, and cnn mutants are viable (Megraw et al. 2001). In contrast, cnn; msd1 double mutants are homozygous lethal, dying at an early pupal stage. α-Tubulin staining of third-instar larval neuroblasts from these cnn; msd1 mutants showed “metaphase-like” cells with weak, unorganized MT arrays (Fig. 5F); very few robust spindles were observed. DAPI staining confirmed an increased mitotic index in double mutants, greater than the increase seen in either cnn or msd1 single mutants, and a dramatic increase in metaphase-to-anaphase ratio (Fig. 5G). Interestingly, MT nucleation around chromatin in cnn; msd1 mutants was still observed (Fig. 5I), similar to that seen during prometaphase in cnn mutants (Fig. 5H), suggesting that nucleation of MTs from around mitotic chromatin is insufficient for viability of Drosophila in the absence of a functional centrosomes and mitotic spindle-templated MT nucleation.
In summary, the above study demonstrates the importance of Msd1, the Augmin complex, and mitotic spindle-templated MT nucleation in the development of the fly. It suggests that spindle formation in vivo occurs via at least three independent mechanisms and that mitotic spindle-templated MT nucleation, although not essential on its own, functions in concert with centrosome-driven MT nucleation to ensure faithful mitosis. Interestingly, although acentrosomal flies have been shown to be viable, the lack of astral MTs during the asymmetric cell divisions of stem cells in these mutants results in a randomization of the plane of cytokinesis with regard to asymmetrically localized fate determinants (Giansanti et al. 2001; Megraw et al. 2001; Basto et al. 2006). Larval brains from such acentrosomal mutants often develop tumors in an allograft transplantation assay, suggesting a link between loss of centrosomes and tumor formation (Castellanos et al. 2008). Our study therefore raises the intriguing possibility that global inhibition of Augmin function would prevent cell division only in cells possessing tumorogenic activity brought about through an absence of functioning centrosomes, while having minimal effect on all other cells. Given that at least some components of the Augmin complex are conserved to humans (Zhu et al. 2008; G Goshima, pers. comm.), it may, therefore, present an exciting potential future drug target.
Materials and methods
Fly stocks
The P-element line EY11673, Df(3L)Bab-PG, mat-α-tubulin-VP16 GAL4, and cnnhk21 were obtained from Bloomington (Indiana University, Bloomington, IN). The original P-element insertion line is viable and fertile. Remobilization events were selected and analyzed over Df(3L)Bab-PG. To follow Msd1 localization in vivo, full-length msd1 was cloned into the Gateway expression vector pPWG (Drosophila Genome Resource Center) via pENTR/D/TOPO (Invitrogen). The plasmid was injected into w1118 embryos by Bestgene, Inc. Rescue of msd1ex51/Df(3L)Bab-PG mutants was performed by driving Msd1-GFP in early embryos using mat-α-tubulin-VP16 GAL4. We used Oregon-R flies as controls.
Cell culture and RNAi
The culture of S2 cells and RNAi against msd1 was performed as described previously (Buster et al. 2007; Hughes et al. 2008) using dsRNA corresponding to Escherichia coli β-lactamase as a control.
Biochemistry and Western blotting
The MT-cosedimentation assay was carried out as described previously (Hughes et al. 2008), using 0.15 g of 0- to 3-h-old Msd1-GFP embryos homogenized in 1 mL of C Buffer (50 mM HEPES at pH 7.4, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1% NP-40, protease inhibitors [Roche]).
For coimmunoprecipitation of the Augmin complex, 0.15 g of 0- to 3-h-old Msd1-GFP embryos were homogenized in 1 mL of C Buffer, clarified by repeated centrifugation at 14,000 rpm, and divided into two. Fractions were incubated with either 20 μg of mouse monoclonal anti-GFP antibody (Roche), or 20 μg of mouse monoclonal anti-β-gal (Sigma) for 2 h at room temperature, followed by addition of 40 μL of Protein G beads for a further 2 h. Samples of immunodepleted extract were diluted in 2× protein sample buffer (Bio-Rad), while beads were washed extensively with C buffer before resuspension in 1× protein sample buffer.
Samples were subjected to standard SDS-PAGE and Western blotting. Membranes were probed with 1:1000 anti-GFP of 0.8 mg/mL stock (Roche); 1:1000 anti-α-tubulin (DM1A; Sigma); 1:500 anti-Wac (a gift from H. Ohkura, University of Edinburgh, UK); 1:500 anti-Dgt4, 1:500 anti-Dgt5, 1:500 anti-Dgt6 (all gifts from G. Goshima); and 1:1000 anti-Pnut (Hughes et al. 2008).
Cytological analysis
Third-instar larval brains were dissected, fixed, and stained as described (Bonaccorsi et al. 2000). S2 cells were methanol-fixed and stained as described (Hughes et al. 2008). The antibodies used were α-tubulin (DM1A; Sigma), γ-tubulin (GTU-88; Sigma), Dgt6, and Dgrip71 (a gift from Y. Zheng, Carnegie Institution of Washington, USA), all at 1:500. Preparations were examined under oil at 25°C using an Eclipse TE2000-U microscope with a Plan APO VC 60× 1.4 NA objective (Nikon) and a 1.5× integrated zoom and camera (c8484-056; Hamamatsu). Pictures were captured using IPlab software (BD Biosciences) merged in Photoshop CS2 (Adobe), and pseudocolored. Relative intensity of α-tubulin, γ-tubulin, and Dgrip71 at the centrosomes and on the mitotic spindle were analyzed as described in Goshima et al. (2007), with the exception of γ-tubulin intensity, where cytoplasmic background levels were subtracted from spindle and centrosome intensities. The spindle intensity of Dgt6 was measured as a region of interest (RoI) for each half-spindle. Live embryos were prepared as described (Buttrick et al. 2008), and images collected every 10 sec using a laser scanning confocal microscope (Radiance 2000; Bio-Rad Laboratories).
Live analysis in S2 cells and EB1-GFP analysis
S2 cells were maintained and visualized as described previously (Buster et al. 2007). EB1 punctae were tracked using an in-house MATLAB particle tracking algorithm. RoIs were selected around each spindle and segmented into 1.26-μm strips along the axis of the spindle. The growing MT tip density in each strip was calculated by counting the number of points and dividing by the area of each region. These numbers were averaged over all frames of each data set to provide the movie-average growing MT end density for each region. The mean density in the spindle region for each movie was determined by averaging over all RoI segments within the spindle area of the cell, while the centrosomal densities were calculated using circular RoIs centered on a centrosome.
Acknowledgments
We thank Maurizio Gatti, Patrizia Somma, Ana Meireles, Hiro Okhura, Gohta Goshima, and Katherine Fisher for providing reagents and for discussing unpublished results. We thank members of the Wakefield laboratory for stimulating discussion and critical reading of the manuscript, and the reviewers for helpful comments. This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) PhD studentship (to T.D.) and a lectureship associated with the Engineering and Physical Sciences Research Council (EPSRC) Oxford Life Sciences Interface/Doctoral Training Centre (to J.G.W). D.J.S was supported by the NIH (grant R01-GM65940) and is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.532209.
Supplemental material is available at http://www.genesdev.org.
References
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Bonaccorsi S, Giansanti MG, Gatti M. Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nat Cell Biol. 2000;2:54–56. doi: 10.1038/71378. [DOI] [PubMed] [Google Scholar]
- Buster DW, Zhang D, Sharp DJ. Poleward tubulin flux in spindles: Regulation and function in mitotic cells. Mol Biol Cell. 2007;18:3094–3104. doi: 10.1091/mbc.E06-11-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttrick GJ, Beaumont LM, Leitch J, Yau C, Hughes JR, Wakefield JG. Akt regulates centrosome migration and spindle orientation in the early Drosophila melanogaster embryo. J Cell Biol. 2008;180:537–548. doi: 10.1083/jcb.200705085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos E, Dominguez P, Gonzalez C. Centrosome dysfunction in Drosophila neural stem cells causes tumors that are not due to genome instability. Curr Biol. 2008;18:1209–1214. doi: 10.1016/j.cub.2008.07.029. [DOI] [PubMed] [Google Scholar]
- Giansanti MG, Gatti M, Bonaccorsi S. The role of centrosomes and astral microtubules during asymmetric division of Drosophila neuroblasts. Development. 2001;128:1137–1145. doi: 10.1242/dev.128.7.1137. [DOI] [PubMed] [Google Scholar]
- Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- Gonzalez C, Saunders RD, Casal J, Molina I, Carmena M, Ripoll P, Glover DM. Mutations at the asp locus of Drosophila lead to multiple free centrosomes in syncytial embryos, but restrict centrosome duplication in larval neuroblasts. J Cell Sci. 1990;96:605–616. doi: 10.1242/jcs.96.4.605. [DOI] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: A protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JR, Meireles AM, Fisher KH, Garcia A, Antrobus PR, Wainman A, Zitzmann N, Deane C, Ohkura H, Wakefield JG. A microtubule interactome: Complexes with roles in cell cycle and mitosis. PLoS Biol. 2008;6:e98. doi: 10.1371/journal.pbio.0060098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson ME, Setty TG, Paoletti A, Tran PT. Efficient formation of bipolar microtubule bundles requires microtubule-bound γ-tubulin complexes. J Cell Biol. 2005;169:297–308. doi: 10.1083/jcb.200410119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mahoney NM, Goshima G, Douglass AD, Vale RD. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr Biol. 2006;16:564–569. doi: 10.1016/j.cub.2006.01.053. [DOI] [PubMed] [Google Scholar]
- Megraw TL, Li K, Kao LR, Kaufman TC. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 1999;126:2829–2839. doi: 10.1242/dev.126.13.2829. [DOI] [PubMed] [Google Scholar]
- Megraw TL, Kao LR, Kaufman TC. Zygotic development without functional mitotic centrosomes. Curr Biol. 2001;11:116–120. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- Meireles AM, Fisher KH, Colombie N, Wakefield JG, Ohkura H. Wac: A new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J Cell Biol. 2009;184:777–784. doi: 10.1083/jcb.200811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Sonobe S, Baskin TI, Hyodo S, Hasezawa S, Nagata T, Horio T, Hasebe M. Microtubule-dependent microtubule nucleation based on recruitment of γ-tubulin in higher plants. Nat Cell Biol. 2005;7:961–968. doi: 10.1038/ncb1306. [DOI] [PubMed] [Google Scholar]
- Raynaud-Messina B, Merdes A. γ-Tubulin complexes and microtubule organization. Curr Opin Cell Biol. 2007;19:24–30. doi: 10.1016/j.ceb.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Verollet C, Colombie N, Daubon T, Bourbon HM, Wright M, Raynaud-Messina B. Drosophila melanogaster γ-TuRC is dispensable for targeting γ-tubulin to the centrosome and microtubule nucleation. J Cell Biol. 2006;172:517–528. doi: 10.1083/jcb.200511071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Zhu H, Coppinger JA, Jang CY, Yates JR, III, Fang G. FAM29A promotes microtubule amplification via recruitment of the NEDD1–γ-tubulin complex to the mitotic spindle. J Cell Biol. 2008;183:835–848. doi: 10.1083/jcb.200807046. [DOI] [PMC free article] [PubMed] [Google Scholar]