Abstract
Since the earliest stages of evolution, organisms have faced the challenge of sensing and adapting to environmental changes for their survival under compromising conditions such as food depletion or stress. Implicit in these responses are mechanisms developed during evolution that include the targeting of chromatin to allow or prevent expression of fundamental genes and to protect genome integrity. Among the different approaches to study these mechanisms, the analysis of the response to a moderate reduction of energy intake, also known as calorie restriction (CR), has become one of the best sources of information regarding the factors and pathways involved in metabolic adaptation from lower to higher eukaryotes. Furthermore, responses to CR are involved in life span regulation—conserved from yeast to mammals—and therefore have garnered major research interest. Herein we review current knowledge of responses to CR at the molecular level and their functional link to chromatin.
Keywords: Sirtuins, chromatin, calorie restriction, TOR, NAD+, metabolism
“‘You'll live longer and you'll be healthier too,’ he answered. ‘Because as we were saying today, there's nothing in the world like eating moderately to live a long life.’ ‘If that's the way things are,’ I thought to myself, ‘I never will die.’ Because I've always been forced to keep that rule, and with my luck I'll probably keep it all my life.”—Anonymous, The Life of Lazarillo de Tormes and of His Fortunes and Adversities (1554).
Since antiquity, human beings have associated immoderate levels of eating with disease and a shortened life span. This intuition, exemplified by the Renaissance era quote above, is only now starting to be truly understood. One of the most important laboratory tools for studying the effects of energy intake on eukaryotes is the intervention known as calorie restriction (CR), which was originally defined in mice as a reduction in food intake of 30%–50% as compared with animals fed with no control (ad libitum) (Masoro 2005). In a hallmark study in 1935, McCay et al. (1935) demonstrated that, compared with rats fed with a standard diet, rats fed under CR lived longer, weighed less, showed heart hypertrophy, and had smaller livers, which they explained at the time as an effect of growth retardation. Since then, considerable research efforts have revealed that, surprisingly, the effects of CR on life span are highly similar among diverse eukaryotes (e.g., yeast, insects, fish, and mammals) (Kennedy et al. 2007). Although we can only speculate on these findings, they suggest that the following general survival strategy has been conserved throughout eukaryotic evolution: In low-nutrient conditions, metabolism is adjusted to enable more efficient use of the energy available, which in turn protects the organism.
Studies of CR have not only provided a better understanding of aging, but also have generated considerable data on the mechanisms that control cellular responses to metabolic fluctuations and how these mechanisms work in concert to provide a coordinated response. One of the most important players in this adaptation is chromatin, which serves as a major hub whereby cell signals from different pathways come together to coordinate responses through gene expression. Indeed, many of these adaptive mechanisms are aimed at protecting chromatin itself.
CR research has become multidisciplinary and has yielded an overwhelming quantity of data, such that summarizing the relevant literature briefly is a difficult task. We do not purport to explore every aspect of the field, but rather to provide a current overview of the important and often underappreciated role of chromatin in the CR response.
Biological lean on CR
The mechanisms involved in the responses to CR are diverse and remain only partially understood. However, there is a growing body of evidence suggesting that these responses involve numerous pathways, which in some cases are intricately linked (Fig. 1). CR is associated with myriad changes at the cellular level and, in the case of higher eukaryotes, also at the organism level (Guarente and Picard 2005). In addition to increasing life span in rodents, CR has also been shown to delay a wide range of aging-associated diseases in mice, such as cancer, diabetes, artherosclerosis, cardiomyopathies, autoimmune diseases, neurodegenerative diseases, or respiratory and kidney diseases, among others (Hursting et al. 2003). Another remarkable feature of the CR response from lower to higher eukaryotes is the accompanying increase in resistance to different forms of stress (e.g., oxidative, genotoxic, or heat) (Guarente and Picard 2005).
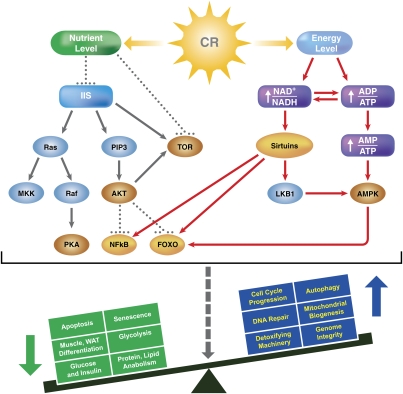
Figure 1.
Main pathways associated with response to CR. Metabolic fluctuations signal through a nutrient and energy level. A decrease in the nutrient levels down-regulates the IIS pathway and the nutrient-dependent kinases TOR, PKA, and AKT, which in turn up-regulate stress response organizers such as the transcription factors NF-κB and FOXOs. In parallel, the cell's energetic imbalance produced by CR up-regulates the NAD+-dependent family of Sirtuins and the nutrient-dependent kinase AMPK, which in turn activate and modulate the stress response. Sirtuins are also involved in the activation of the LKB1 kinase. Overall, as is shown in the bottom part of the figure, survival and life span increase is activated through the activation of several processes (in blue) and inhibition of others (in green).
Sensing the conditions of CR seems to occur at two different levels: the nutrient level and the energy level (Fig. 1). At the nutrient level, the main pathways involved include the endocrine insulin/IGF-1 pathway (Dilova et al. 2007) and the nutrient-responsive kinases target of rapamycin (TOR), cyclic AMP-dependent kinase (PKA), and Sch9/AKT. From the energetic standpoint, the two main cellular events during CR are a decrease in the ratios of NADH/NAD+ and ATP/ADP. This oxidative and hypoenergetic state of the cell is believed to trigger a response through the activation of different pathways, the best known of which involve the NAD+-dependent deacetylases called Sirtuins and the AMP-dependent kinase (AMPK). The elements involved do not work independently (Fig. 1). In fact, as we shall see, they seem to synergize for fine-tuning the regulation of cellular responses.
Heavyweights in the CR response
In higher organisms—particularly in mammals—the CR response implies a strong link to the endocrine system. CR is associated with a decrease in body temperature and size, low levels of blood glucose and insulin, an increase in insulin sensitivity, a decrease in levels of triodothyronine (T3) and growth hormone (GH), and inhibition of the insulin-like pathway (IGF-1) (Redman and Ravussin 2009). The CR response is ultimately reflected in many processes, such as inhibition of glycolysis and activation of gluconeogenesis (GNG) in the liver, inhibition of adipogenesis in WAT (white adipose tissue), myoblast differentiation in skeletal muscle, and others (Guarente and Picard 2005). The network formed by the insulin receptor and the IGF-1 pathway, known as insulin/IGF-1 signaling (IIS), is critical to the endocrine response to CR (Bartke 2008). Signal transduction through IIS is exerted mainly through the PI3K (phosphatidylinositol-3 kinase)/AKT and the RAS/MAPK (mitogen-activated protein kinase)/RAF pathways that signal downstream to a wide spectrum of transcription factors (Dilova et al. 2007). Among these, the FOXO subgroup of proteins that belongs to the family of forkhead factors seems to be very important for the functional link to CR in multicellular organisms. FOXO transcription factors control the response to different types of stress, promoting cell survival via activation of myriad genes involved in DNA repair, detoxification machinery, cell cycle progression, apoptosis inhibition, and other functions (see below; Puig and Tjian 2006). In normal nutrient conditions, the IIS pathway keeps the FOXO members inactive, whereas CR conditions down-regulate the IIS pathway and activate FOXO activity (Nakae et al. 2008). The IIS signaling components are important not only in CR responses, but are also the only components that are consistently involved in the control of life span from yeast to mammals (Dilova et al. 2007).
Another key element in the CR response is a group of functionally interrelated kinases involved in the adaptation to nutrient availability from yeast to humans: TOR, PKA, AMPK, and AKT/Sch9 kinase. TOR, AKT/Sch9, and PKA are activated under conditions of nutrient availability (Kennedy et al. 2007), whereas AMPK is activated under conditions of nutrient scarcity (such as CR) that generate a high AMP/ATP ratio (Dilova et al. 2007; Kennedy et al. 2007). One of the key mechanisms through which these cytoplasmic kinases exert their chromatin-related functions is by controlling the localization of certain effectors; phosphorylation of certain proteins—primarily transcription activators—keeps them sequestered in the cytoplasm, thereby preventing expression of their target genes (Raught et al. 2001). These nutrient-dependent kinase pathways are linked in a very complex network that ensures efficient and dynamic responses to environmental changes. For instance, TOR activity is positively modulated by IIS during glucose abundance, and negatively modulated by AMPK during glucose deprivation or conditions of stress (e.g., hypoxia) (Dilova et al. 2007). In addition, TOR regulates the activities of PKA and of AKT, which in turn also regulate TOR (Fig. 1; Bhaskar and Hay 2007). These kinases are functionally linked not only to each other, but also to other pathways. For instance, the kinase LKB1, a tumor suppressor that is required for AMPK activation under energy-deficient conditions, is regulated by SirT1, a member of the Sirtuin family (Lan et al. 2008).
The main signaling effect of these kinases is exerted from the cytoplasm. However, as we will see, TOR has a special relationship with chromatin, not only indirectly through a signaling cascade mechanism, but also via the direct regulation of chromatin structure.
The mitochondrion is another major player in CR responses (Guarente 2008), as a pivotal aspect of the CR response relates to the production of reactive oxygen species (ROS) including superoxide (O2−), hydrogen peroxide (H2O2) and hydroxyl radical (OH−) (Sohal and Weindruch 1996). Managing these reactive species is a survival challenge for both prokaryotes and eukaryotes in aerobic conditions, and requires specialized machinery and a complex regulatory network (Halliwell 1999). These molecules are generated mainly during electron transfer through the respiratory chain in which the final electron acceptor is O2, generating H2O. However, other sources can promote the formation of these species, such as ionizing radiation or certain enzymatic reactions such as demethylation of histone H3 Lys 4 (H3K4) by the mono-oxidase LSD1 (Forneris et al. 2008). Oxidative stress leads to increased production of ROS, thereby shifting the normal balance between the generation and elimination of these molecules. Overproduction of ROS leads to carbonylation (oxidation) of various proteins, lipid peroxidation, and general damage to both nuclear and mitochondrial DNA (see below).
CR in mammalian cells increases mitochondrial biogenesis and energy efficiency through the production of nitric oxide (NO) by endothelial NO synthase (eNOS) (Nisoli et al. 2005; Lopez-Lluch et al. 2006). During oxidative stress, CR induces a decrease in protein oxidation, lipid peroxidation, and ROS production, and this translates into a decrease in mitochondrial metabolism (Fig. 2; Sohal et al. 1994). Based on these observations, Sohal and Weindruch (1996) hypothesized that a CR-induced drop in ROS production could be the process most responsible for CR-dependent life span increase. Despite the evidence linking CR with decreased metabolism, new evidence suggests the opposite, at least in the cases of Saccharomyces cerevisiae and Caenorhabditis elegans, and cell culture studies suggest that the same might apply to mammals (Lin et al. 2002; Lopez-Lluch et al. 2006; Schulz et al. 2007). Perhaps this reflects a mechanism for increasing energy production in an efficient manner, probably by inhibiting those energy-producing reactions that are less efficient (Schulz et al. 2007).
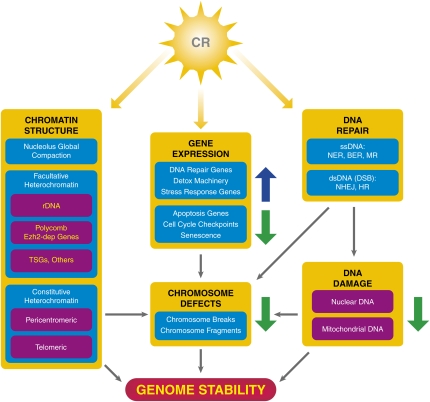
Figure 2.
CR response on chromatin. CR has an effect on chromatin at three different levels—chromatin structure, gene expression (up-regulation of certain genes and down-regulation of others as indicated by the arrows), and DNA repair—which in general produce an increase in genome stability and can explain, at least partially, the life span increase effect of CR (see the text).
The final key element of the response to CR and the focus of this review is chromatin. Historically perceived as a passive target of the damaging effects of ROS, chromatin is now being appreciated as a key and active participant in the response to CR. As we will see, chromatin is not only an important mediator of the CR response mechanism, but also the specific target of many of the events taking place in the process.
The skinny on CR and chromatin
The evidence suggests that the biological effects of CR are closely related to chromatin function. In fact, given the parallels between aging and loss of chromatin integrity, one of the models for explaining the delaying effect of CR on aging is based on increased genomic stability (Fig. 2; Heydari et al. 2007). This has been shown clearly in yeast where, as we will see, Sirtuin Sir2p has been linked to aging control through the modulation of chromatin structure (Guarente 2000).
The consequences of CR on chromatin historically have been interpreted as antagonistic to the well-known effects produced by ROS in chromatin during oxidative stress (see below). While this is partially true, it is becoming increasingly clear that ROS are not the only consideration in this process. Current data suggest that at least part of the CR effect on life span could be elicited through a long-term activation of protective machinery under low-intensity stressors (e.g., activation of detoxifying machinery, cell cycle progression, DNA repair, and inhibition of apoptosis and senescence) (Masoro 2005). This would entail a tighter and more efficient global control on the cell. This theory, known as the Hormesis theory, was proposed by Masoro in 1998 (Masoro 1998), and is currently under study (Masoro 2005).
The evidence suggests that responses to CR involving chromatin are twofold: mediating cellular adaptation to changes in metabolism through the control of gene expression, and promoting the protection of genome integrity and chromatin structure (Fig. 2). The former occurs not only in the nucleus but also in mitochondrial DNA, as addressed above. CR induces major changes in gene expression patterns, which in mammals are apparently highly tissue-specific (Fu et al. 2006). The genes targeted can be classified generally as those encoding proteins involved in (1) metabolic pathways, (2) stress responses, (3) DNA damage repair, (4) chromatin structure regulation, (5) detoxification, and (6) mitochondria-related processes. The mechanisms that mediate these changes include modulation of the activity of certain key transcription factors that regulate a specific set of genes (e.g., FOXO transcription factors) (Guarente and Picard 2005).
Also implicit to the CR response is gene inactivation, which seems to occur via two different mechanisms: regulation of individual genes by modulation of specific transcription factors, and heterochromatinization of large parts of the genome that might include many genes, as has been observed in the case of nucleolar rDNA with either TOR inhibition or yeast Sir2p activation (Guarente 2000; Tsang and Zheng 2007). The former may occur through changes in DNA-binding affinity, enzymatic activity, or cellular localization of key transcription factors such as stress-responsive transcription factors, or via recruitment of silencing machinery (e.g., histone deacetylases [HDACs] or methyltransferases) (see below).
CR leads to protection of genome integrity and preservation of chromatin structure from yeast to mammals (Heydari et al. 2007). This effect is closely related to that described above, since part of the machinery involved in genomic protection depends on appropriate gene expression. This protection occurs via two series of mechanisms. The first encompasses activation of DNA repair (Cabelof et al. 2003), ROS detoxification machinery (Xia et al. 1995), and increased fidelity of DNA replication (Srivastava et al. 1993). The second comprises direct modulation of chromatin structure, which has positive effects on DNA recombination, DNA replication, and cell cycle progression, among other processes (Vaquero 2009).
ROS and chromatin
The link between ROS and nuclear chromatin is supported by an overwhelming amount of evidence. ROS production is associated with genomic instability through an increase in DNA damage and chromosome degradation (Barzilai and Yamamoto 2004). In addition to their well-characterized effect on ssDNA damage, ROS have also been linked to production of double-strand breaks (DSBs) that result in chromosome breaks (Karanjawala et al. 2002). Thus, overexpression of superoxide dismutase (SOD1), the enzyme responsible for processing superoxide radicals (O2−), in transgenic mice results in increased H2O2 and OH− and increased frequency of chromosome breaks in primary fibroblasts, from 9% to 23% (Karanjawala et al. 2002). Another interesting observation associated with ROS production in the cell is the accumulation of chromosomal fragments, including the so-called giant fragments (>1 Mb) and high-molecular-weight (HMW) ones (50–800 kb) (Higuchi 2003). This phenomenon is H2O2-dependent, linked to specific nucleases, and believed to arise from the breakdown of higher-ordered chromatin organization, probably as a part of the events involved in cell death (Konat 2003).
Telomeres present a clear example of how ROS are linked to chromatin structure. Oxidative stress is linked to the replicative senescence phenotype, and both are interrelated with telomere uncapping and shortening (von Zglinicki 2002). Although these connections are not completely understood, they appear to be due to a progressive decrease in effective telomerase activity through its export to the cytoplasm under stress conditions (Haendeler et al. 2004), and to an increase in telomeric DNA damage that correlates with telomere shortening during DNA replication (Petersen et al. 1998). However, this cumulative damage is likely a function of the special structural features of telomeres resulting in their less efficient repair, rather than their being subjected to a higher rate of damage, per se (Petersen et al. 1998).
The link between oxidative stress and DNA damage is evidenced by other findings: ROS production correlates with an increased frequency of certain post-translational chromatin modifications related to DNA damage, such as phosphorylation of histones H2A (γH2A) and H3, poly-ADP-ribosylation of histones, and certain acetylation and methylation patterns of histones H3 and H4 (Monks et al. 2006). Consistent with this, mice deficient in the DNA repair signaling kinase ATM exhibited abnormally high levels of ROS, among many other phenotypes (Ito et al. 2006).
As mentioned previously, CR increases cellular resistance to oxidative stress by reducing the accumulation of both nuclear and mitochondrial DNA damage produced by ROS (Fig. 2). However, the level of DNA protection in each cell compartment is not equivalent among different tissues (Stuart et al. 2004). CR increases DNA repair activity and reverses the decreased levels of such activity observed in aged mice (Heydari et al. 2007). The DNA repair pathways up-regulated by CR are the three main mechanisms involving ssDNA, NER (nucleotide excision repair) (Guo et al. 1998), BER (base excision repair) (Cabelof et al. 2003), S-phase-dependent mismatch repair (Tsao et al. 2002), and the most common form of DSB repair, NHEJ (nonhomologous end joining) (Um et al. 2003). Interestingly, CR not only increases expression of the DNA repair machinery (Heydari et al. 2007), but also amplifies the activity and fidelity of critical enzymes such as DNA polymerases α and β (Cabelof et al. 2003). The effects of CR on NHEJ are tissue-specific, being most pronounced in kidneys and lungs, but less evident in testes or liver. These effects seem to occur via altered levels of the NHEJ modulation complex Ku70/86 (Um et al. 2003).
Personal trainers of CR effects on chromatin
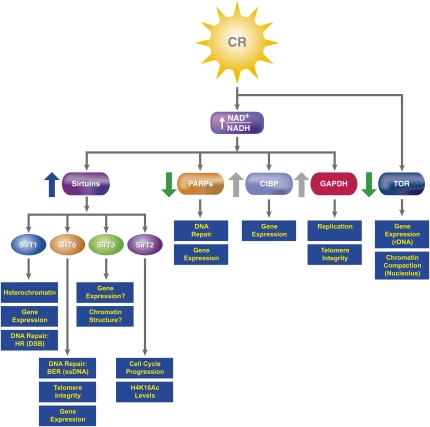
Several groups of factors have been postulated as being mediators of the CR response on chromatin (Fig. 3; Table 1). The candidates with the best credentials for such a role are the Sir2 NAD+-dependent deacetylases known as Sirtuins that have emerged recently as major players in signaling to chromatin during responses to CR (Guarente and Picard 2005; Saunders and Verdin 2007; Vaquero et al. 2007b). Another interesting case is that of TOR. In addition to its signaling pathway, TOR participates directly in the regulation of nucleolar chromatin. Additionally, given the strong relationship between CR and the NAD+/NADH levels, it is likely that other chromatin-related factors whose activity is strongly influenced by the redox state of the cell might be involved in CR responses (Fig. 3). Below we review these factors and discuss the nature of their link to CR and to chromatin functions (Table 1).
Figure 3.
CR signaling mediators to chromatin. CR induces an energetic imbalance through changes in the NAD+/NADH ratio that activates Sirtuins and down-regulates PARPs. Because of their capacity to respond to changes in the NAD+/NADH ratio, CtBP and GAPDH (and other metabolic enzymes such as LDH) are likely to be activated by CR response (gray arrows), but the link between their activity and CR has not been formally demonstrated yet. Interestingly, the inhibition of the TOR pathway by CR has dramatic effects on chromatin nucleolar structure and expression (see the text). In blue boxes are represented the roles that these mediators perform or are likely to perform (indicated by ?) in chromatin.
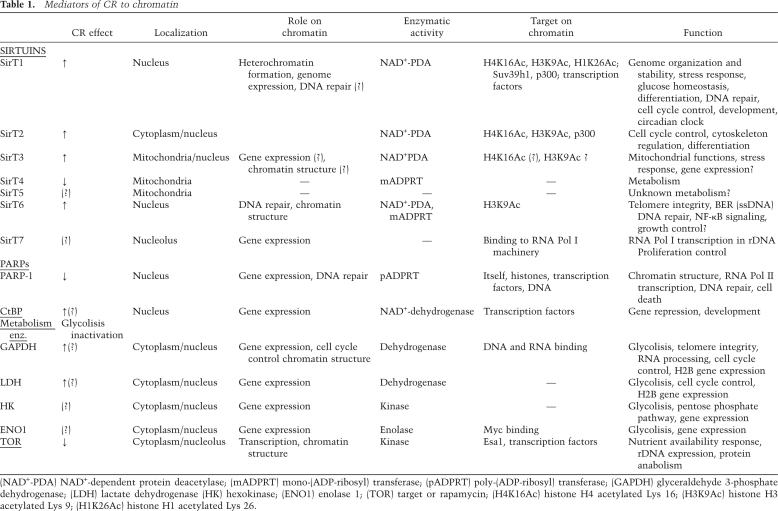
Table 1.
Mediators of CR to chromatin
(NAD+-PDA) NAD+-dependent protein deacetylase; (mADPRT) mono-(ADP-ribosyl) transferase; (pADPRT) poly-(ADP-ribosyl) transferase; (GAPDH) glyceraldehyde 3-phosphate dehydrogenase; (LDH) lactate dehydrogenase (HK) hexokinase; (ENO1) enolase 1; (TOR) target or rapamycin; (H4K16Ac) histone H4 acetylated Lys 16; (H3K9Ac) histone H3 acetylated Lys 9; (H1K26Ac) histone H1 acetylated Lys 26.
The exertion of Sirtuins
The Sirtuin family is defined by its homology with the budding yeast silencing factor Sir2p, which participates in different chromatin-associated functions, and has members in organisms from bacteria to mammals (Vaquero 2009). Remarkably, Sirtuins have a role in regulating metabolism since early evolution, and their diversification from one member in bacteria to five in Drosophila and seven in mammals suggests that they have acquired new, specialized functions to match the increasing complexity of the organism (Vaquero 2009). This is evidenced by the diverse localization of the mammalian Sirtuins (SirT1-7): SirT1 and SirT6 in the nucleoplasm, SirT7 in the nucleolus, SirT2 in the cytoplasm, SirT3 in the mitochondria and the nucleus, and SirT4 and SirT5 in the mitochondria (Michishita et al. 2005). Despite the diversity of their localization, current evidence has unveiled a certain degree of functional redundancy between Sirtuins, although this is not yet well understood. For instance, among the targets of SirT1 are the histone modifications H4K16Ac and H3K9Ac, p53, and the transcription factors FOXO and NF-κB (Luo et al. 2001; Vaziri et al. 2001; Brunet et al. 2004; Motta et al. 2004; Vaquero et al. 2004; Yeung et al. 2004). SirT2 deacetylates H4K16Ac, H3K9Ac, p53, and FOXO, whereas SirT6 targets H3K9Ac and regulates the activity of NF-κB (Vaquero et al. 2006; Wang et al. 2007; Jin et al. 2008; Michishita et al. 2008; Kawahara et al. 2009). Sirtuins show two associated enzymatic activities that likely diverged from an original one: protein deacetylation and mono[ADP-ribosyl] transferase (mADPRT) (Tanny et al. 1999; Imai et al. 2000). Interestingly, while most Sirtuins seem to exhibit at least weak mADPRT activity (Tanny et al. 1999), only SirT1, SirT2, SirT3, and SirT6 have been shown to be active, NAD+-dependent deacetylases of both histones and nonhistone proteins (North et al. 2003; Saunders and Verdin 2007; Vaquero et al. 2007b). The deacetylase SirT6 and one of the nondeacetylase Sirtuins, SirT4, both show strong mADPRT activities (Saunders and Verdin 2007). However, whether all Sirtuins can be both deacetylases and mADPRTs when presented with an appropriate substrate is still not clear and needs to be addressed.
The link between Sirtuins and chromatin functions is apparently as old as eukaryotes, probably reflecting the intimate relationship between environmental changes and DNA regulation from early evolution (Vaquero 2009). In fact, members of the family in archeobacteria that already contain a genome organized into a chromatin-like structure are involved in the control of genome compaction through their regulation of the histone-like protein Alba (Bell et al. 2002). Interestingly, eukaryotic Sirtuins share a functional link with the modifications H4K16Ac and H3K9Ac, which are critical to the regulation of chromatin structure and epigenetic phenomena throughout evolution (Vaquero et al. 2007b; Vaquero 2009).
Of all Sirtuins, yeast Sir2p is the best characterized and most clearly related to CR. It is involved in epigenetic silencing through the formation of heterochromatin-like compacted regions in three different loci of the yeast genome: the mating type loci, rDNA in the nucleolus, and telomeres (Steinkraus et al. 2008). Sir2p has also been linked to cell cycle control, DNA replication, DNA repair, and meiosis (Guarente 2000).
Yeast life span can be measured at two levels: the replicative life span, which represents the number of times a cell can replicate during its lifetime before reaching senescence, and the chronological life span, which refers to the time that nondividing cells can maintain viability. CR produces an increase in both the replicative and the chronological life spans of yeast. Extra copies of SIR2 increase yeast replicative life span by 30%–40%, whereas ΔSIR2 decreases it by 50% and CR treatment is ineffectual in replicative life span in such mutant cells (Steinkraus et al. 2008). These observations, together with the evidence that CR up-regulates Sir2p, led to speculation that Sir2p could also be a mediator of CR effects in aging (Guarente 2000). In fact, further evidence has demonstrated that other factors (e.g., nutrient-responsive kinases) are also mediators of the CR response, and to a greater extent than Sir2p. Whereas Sir2p is not involved in CR-mediated chronological life span (Kaeberlein et al. 2004), Sch9 and TOR participate in both the chronological and replicative CR-dependent increases in life span (Fabrizio et al. 2001).
The most accepted explanation for the effects of Sir2p on life span is that Sir2p-dependent silencing of rDNA copies inhibits, by fivefold to 10-fold, homologous recombination events that generate extrachromosomal rDNA circles (ERCs). Accumulation of ERCs in the mother cell has been associated with aging, although the details of this relationship remain the subject of debate (Steinkraus et al. 2008). Some recent studies further support a chromatin link to life span control in yeast: Levels of H4K16Ac increase during yeast aging, which correlates with a loss of Sir2p protein levels. Interestingly, the mutation of K16 to Q, which mimics acetylated lysine, is associated with a shorter life span. Aging in yeast is accompanied by hyperacetylation of H4K16 at different Sir2p-binding loci, particularly in telomeric regions (Dang et al. 2009). Actually, ERCs do not seem to exist outside of yeast. Thus, this inverse relationship between Sir2 and H4K16Ac levels during aging appears to be the model most relevant to the possible regulatory role of Sirtuins in higher eukaryotic life span.
In terms of multicellular organisms, the orthologs of SirT1 have been shown to increase life span in both C. elegans (Sir-2.1) and Drosophila (dSir2) and improve many aging-associated parameters in mammals (Tissenbaum and Guarente 2001; Rogina and Helfand 2004; D Chen et al. 2005; Boily et al. 2008). However, whether these effects are due to a mechanism dependent on or independent of CR is unknown. In fact, Drosophila dSir2 has been functionally linked to CR-dependent life span through a mechanism linked to the HDAC Rpd1 (Rogina and Helfand 2004), but C. elegans Sir-2.1 increases life span through a CR-independent mechanism linked to the IIS pathway (Tissenbaum and Guarente 2001). Nevertheless, there is no doubt that Sirtuins are functionally linked to the CR response in mammals, since CR up-regulates SirT1, SirT2, SirT3, and SirT6 and down-regulates SirT4 (Cohen et al. 2004; Shi et al. 2005; Wang et al. 2007; Kanfi et al. 2008). In fact, recent and controversial data have suggested that CR might not induce SirT1 up-regulation, but its down-regulation in liver (Chen et al. 2008a). However, the nature of the discrepancy between this study and several others that showed CR-mediated SirT1 up-regulation in liver tissues from mice and rats is not understood. It might indicate that SirT1 levels are tightly controlled and can changedrastically under certain conditions.
This link with CR has only been clearly established and developed with SirT1. Extensive research is necessary to clarify the role of the other Sirtuins and determine if mammalian Sirtuins participate in CR-related life span effects.
SIRT1.
Among mammalian Sirtuins, SirT1 is the closest homolog to yeast Sir2p and the best studied by far. It plays multiple roles in the targeting of myriad proteins (Feige and Auwerx 2007). Although SirT1 is localized mainly to the nucleus (Vaquero et al. 2004; Michishita et al. 2005), in certain cell types (e.g., pancreatic β cells or cardiomyocytes) it can be localized to the cytoplasm (Moynihan et al. 2005).
SirT1 has been linked to the major players in CR response and life span control. First, SirT1 is functionally linked to the IIS pathway through inhibition of the IGF signaling pathway and to activation of the FOXO transcription factor upon CR or stress (Brunet et al. 2004; Motta et al. 2004). SirT1 is also linked to other components of the endocrine network, such as the nuclear receptors LXR, GR, or AR (Amat et al. 2007; Li et al. 2007). Second, SirT1 regulates the activity and localization of the main AMPK kinase LKB1 (Lan et al. 2008). In fact, AMPK has also been implicated in SirT1 activation upon CR in certain tissues such as skeletal muscle, in which it inhibits myoblast differentiation (Fulco et al. 2008). Third, SirT1 is also involved in autophagy activation under nutrient starvation, a mechanism that seems to be linked to the inactivation of the TOR pathway. Consistent with this, the loss of SirT1 inhibits autophagy, an important response during starvation and probably in CR as well (Lee et al. 2008). Fourth, an increase in CR-induced activity requires SirT1 in mice (D Chen et al. 2005).
SirT1 is activated via up-regulation of the nicotinamide phosphoribosyltransferase Nampt, a functional homolog of yeast PNC1 that is involved in the nuclear NAD+ salvage pathway. Nampt is an NAD+ biosynthetic enzyme that increases the NAD+/NADH ratio and decreases the levels of nicotinamide (Anderson et al. 2003), the latter being both a product and a competitive inhibitor of Sirtuin enzymatic activity. Further evidence of the connection between SirT1 in CR responses and life span control has been provided by studies in which SirT1 is shown to be specifically activated by the synthetic molecule SRT1720 (Feige et al. 2008). Among other effects, SRT1720 mimics CR by increasing diet-associated metabolic defects through the inhibition of adipogenesis and induction of lipid oxidation. Moreover, moderate overexpression of ectopic SirT1 in mice winds up optimizing energy reserves by increasing insulin sensitivity, thereby inhibiting diabetes (Banks et al. 2008; Feige et al. 2008).
SirT1 serves as a metabolic sensor at both the cellular and organismal levels, performing general and tissue-specific dependent functions. The majority of these functions enable cell survival and growth under compromised conditions through optimization of available resources (Feige and Auwerx 2007; Saunders and Verdin 2007). They can be divided into four classes: cell survival under stress, glucose homeostasis, cell differentiation and development, and chromatin organization and stability.
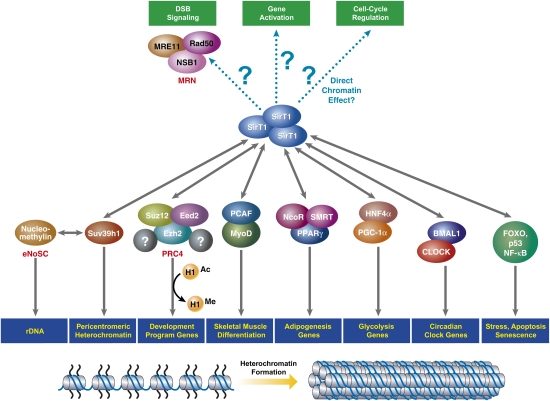
The first noteworthy feature about the mechanism of action of SirT1 is that while the majority of its associated functions are exerted through chromatin, its role is far more expansive (Feige and Auwerx 2007; Vaquero et al. 2007b). As opposed to other HDACs, for example, SirT1 is a coordinator of simultaneous events. The scope of this is evidenced by its capacity to bind myriad factors and to target for deacetylation the histone marks H4K16Ac and H3K9Ac (Vaquero et al. 2004), as well as transcription factors, and even other enzymes such as the histone acetyltransferase (HAT) p300 (Bouras et al. 2005) or the histone methyltransferase (HMT) Suv39h1 (Fig. 4; see below; Vaquero et al. 2007a).
Figure 4.
SirT1 regulates multiple processes through coordination of heterochromatin formation. To perform its specific functions, SirT1 interacts with a wide variety of factors that seem to provide specificity and localization to specific targets (in blue boxes). However, SirT1 is involved in other functions such as DNA repair signaling of DSBs, gene activation, or cell cycle regulation that have not been shown to be due to a direct effect of this sirtuin on chromatin.
Sirtuins in general, and SirT1 in particular, do not seem to bind directly to chromatin, but are instead recruited by specific factors (Fig. 4). The arrival of SirT1 to chromatin is associated with its compaction and gene silencing, as has been described for yeast Sir2p (Vaquero et al. 2004). However, as a consequence of its many specialized targets, SirT1 can interact with and deacetylate specific transcription factors, thereby modulating their ability to activate or repress genes, as a function of tissue context. For instance, under stress, SirT1 binds to stress-dependent transcription factors (e.g., FOXO transcription factors [FOXO1, FOXO3, and FOXO4], NF-κB, and p53) and induces the expression of their target genes involved in stress protection, while inhibiting those target genes involved in cell cycle arrest, senescence, or apoptosis. The underlying mechanisms of SirT1-induced activation seem to be related to deacetylation of the transcription factor itself, such as in the case of FOXO4, and they seem to be apart from direct deacetylation of chromatin components (Fig. 4). Another interesting case is the relationship between SirT1 and transcription factors involved in metabolic control such as PGC1-α, which is key to glucose homeostasis and mitochondria regulation in the liver (Rodgers et al. 2005). Under low-glucose conditions, SirT1 in the liver binds to and deacetylates PGC-1α and activates the transcription of GNG-dependent genes while silencing glycolysis-related genes. In contrast, SirT1 in white adipocytes inhibits adipogenesis by interfering with the activity of PPAR-γ by binding to the corepressors NCoR and SMRT (Picard et al. 2004).
SirT1 is involved in the formation of two different forms of heterochromatin: facultative and constitutive. Facultative heterochromatin refers to chromatin regions that become heterochromatinized (tightly packed) during certain processes, such as development or differentiation, or in response to certain stimuli, but its structure can be restored to the more open form of euchromatin as required. These regions may encompass one gene, a few genes, or even an entire chromosome, such as the inactive X chromosome. In contrast, constitutive heterochromatin (CH) describes those chromatin regions that, once formed, never decompact again. CH has a very low density of genes and is located mainly in the pericentromeric and telomeric chromosomal regions, and its role is chiefly structural (Trojer and Reinberg 2007).
SirT1 promotes formation of facultative chromatin via several coordinated events, the first of which is deacetylation of histones, particularly H4K16Ac and H3K9Ac residues (Vaquero et al. 2004). In the case of facultative heterochromatin, this seems to be restricted to the regulatory regions of genes. There is evidence suggesting a close relationship between SirT1 and these two residues, since SirT1−/− mouse embryonic fibroblasts (MEFs) or cells depleted of SirT1 by RNAi exhibit hyperacetylation of both H4K16 and H3K9 (Vaquero et al. 2004, 2007a; Wang et al. 2008). Interestingly, brain sections of SirT1−/− embryonic day 11 (E11) embryos show hyperacetylation of H3K9 but not of H4K16, suggesting that the SirT1-regulated levels of H4K16Ac are tissue-specific or related to certain stages of differentiation (Wang et al. 2008).
Second, SirT1 interacts with the linker histone H1b, deacetylates its K26 residue, and recruits it to chromatin, thereby facilitating the formation of higher-order chromatin organization (Vaquero et al. 2004). Although the role of H1K26Ac remains poorly understood, evidence suggests that it functions similarly to acetylated core histones in that its presence correlates with RNA polymerase II (Pol II) as evidenced by chromatin immunoprecipitation (ChIP)-on-chip results, and it is excluded from heterochromatin regions as evidenced in immunofluorescence (IF) experiments (A Vaquero and D Reinberg, unpubl.). Interestingly, the H1K26 residue can also be methylated by the Polycomb group protein Ezh2, an ortholog of Drosophila E(Z) (Kuzmichev et al. 2004)—a finding that pointed to a possible functional link between SirT1 and Ezh2. Indeed, both proteins are components of the PRC4 complex that, while only partially purified thus far, is detectable only during specific stages of development in Drosophila and mammals (Furuyama et al. 2004; Kuzmichev et al. 2005). Interestingly, despite the fact that Ezh2 usually targets H3K27 for methylation with the resultant H3K27me3 mark being involved in Ezh2-dependent silencing throughout development, PRC4 specifically methylates histone H1K26, not H3. To date, the role of PRC4 and the implications of H1K26me3 are not completely understood, but several of the PRC4 components (e.g., SirT1 and a specific isoform of the subunit EED) have been localized to a variety of Ezh2-dependent target genes (Kuzmichev et al. 2005).
Third, arrival of SirT1 to chromatin results in the loss of certain marks associated with active transcription, such as H3K79me2 (Vaquero et al. 2004). The loss of H3K79me2 spreads a few kilobases away from the original site and this hints at the existence of a histone demethylase or histone exchange activity, since SirT1-dependent loss of H3K79me2 does not seem to require passage through S phase (A Vaquero, unpubl.).
Fourth, SirT1 promotes the arrival of heterochromatin-associated marks such as H3K9me3 and H4K20me1, the former of which is particularly interesting because it is present in all forms of heterochromatin and is a hallmark of compacted chromatin from early stages of eukaryote evolution (Trojer and Reinberg 2007). Moreover, SirT1 depletion through RNAi renders global hyperacetylation of H3K9 and hypomethylation of H3K9me3 (Vaquero et al. 2007a). We recently identified the means by which SirT1 promotes H3K9me3. This entails the intimate functional relationship between SirT1 and the main H3K9me3 HMT activity, Suv39h1 (Peters et al. 2001), through which SirT1 exerts three mechanisms. First, SirT1 interacts with and recruits Suv39h1 to genomic loci and deacetylates H3K9Ac, thereby setting the stage for the subsequent methylation of this residue by Suv39h1. Interaction between SirT1 and Suv39h1 involves the N-terminal region of SirT1 and the first 89 residues of Suv39h1. Second, SirT1 binding to Suv39h1 induces a conformational change in the methyltransferase that increases the specific activity of SirT1 by more than fivefold. This has been demonstrated in vitro and in vivo and depends exclusively on the N-terminal domain of SirT1. Third, SirT1 up-regulates Suv39h1 activity directly by deacetylating residue K266 present in its catalytic SET domain. This residue lies within a loop that seems to be involved in the folding of the SET domain into the post-SET domain (Min et al. 2002), and when acetylated renders Suv39h1 50% less active (Vaquero et al. 2007a). This is the first evidence of the means by which such methyltransferase activity is regulated and might very well be applicable to other HKMTs in which the K266 residue is conserved (e.g., Ezh2 or Suv4-20). Further studies should clarify not only the features of this modification, but also the conditions under which SirT1 exerts its regulatory function.
The involvement of SirT1 in the formation/maintenance of CH remains unclear. SirT1−/− MEFs exhibit a loss in H3K9me3 and in structural proteins (e.g., HP1) at pericentromeric heterochromatin foci, indicating a loss of heterochromatin structure in >50% of the cells tested (Vaquero et al. 2007a). Supporting this observation, an identical phenotype was shown in brain cells from SirT1−/− E11 embryos, suggesting that the role of SirT1 in CH is not tissue-specific (Wang et al. 2008). This relationship seems to be conserved through evolution, since previous studies in Schizosaccharomyces pombe revealed that spSir2 was required for deacetylation of H3K9Ac in pericentromeric regions to allow methylation of the same residue by Clr4, the ortholog of Suv39h1 (Shankaranarayana et al. 2003). Why half of the SirT1−/− MEF population showed this phenotype whereas the other half looked normal is still not understood. During development, these mice may have adapted to the loss of SirT1 and, in some cases, found a way to overcome it. Another possibility is that this phenotype is related to a particular stage of the cell cycle. Interestingly, this effect of SirT1 on CH seems to be direct, since transfection of SirT1 recovered the wild-type phenotype completely. Yet transfection of either a catalytically dead point mutant of SirT1 or a ΔNSirT1 mutant led to only partial recovery, suggesting that both its activity and its N-terminal domain are required for SirT1 to exert its function with respect to Suv39h1 and H3K9me3 (Vaquero et al. 2007a).
Another curiosity stemming from the link between CH and Suv39h1 is the unexpected presence of H4K16Ac in the CH loci that lost H3K9me3 and HP1 in Suv39h1−/− MEFs (Fig. 4; Vaquero et al. 2007a). Interestingly, this is the only mark tested to date that relocalizes to CH under these conditions. While not yet understood, it does not seem to relate to SirT1 or any other Sirtuin given that SirT1 localization was only slightly altered to a more granular distribution in IF experiments (Vaquero et al. 2007a). One possibility is that H4K16Ac is somehow related to replication or DNA repair of CH regions, and that loss of H3K9me3 perhaps compromises or even prevents DNA repair. Further characterization of H4K16Ac and its relationship with Sirtuins (particularly SirT1 and SirT2) are required to resolve the implications of these findings.
This relationship between SirT1 and Suv39h1 has been further confirmed by the identification of the complex eNoSC, which contains both proteins along with the H3K9me2-binding factor nucleomethylin (Murayama et al. 2008). eNoSC is involved in repressing rDNA transcription under conditions of low energy, such that cells can maintain tight control over ribosome biogenesis (Fig. 4).
Another interesting facet regarding SirT1 and Suv39h1 (and, in fact, also Ezh2) is the apparent discrepancies they exhibit in the context of DNA methylation. Studies have shown that SirT1 colocalizes in rDNA regions with the DNA methyltransferase Dnmt1 (Espada et al. 2007). Although the presence of SirT1 in these regions is dependent on Dnmt1, the levels of DNA methylation are unaffected by its absence. On the other hand, Suv39h1 (and Ezh2) has been shown to be part of a complex together with DNA methyltransferases (Fuks et al. 2003), and its loss is associated with a marked redistribution of DNA methylation in certain regions (Lehnertz et al. 2003). This bespeaks of Suv39h1 involvement in two different stages of gene silencing: one with Dnmts to establish DNA methylation, and another with SirT1 to participate in the actual silencing process, independent of DNA methylation. This possibility is supported by studies showing that loss of SirT1 induces reactivation of certain tumor suppressor genes without affecting the methylation levels of their promoters (Pruitt et al. 2006). Yet this raises a very important question: What, then, is the role of such DNA methylation in these promoters if not for the silencing process? Further studies will be required in the future to understand this puzzling evidence and the involvement of SirT1 in these processes.
Another remarkable role of SirT1 in chromatin that is directly linked to its protective effect under conditions of CR and stress is its direct involvement with DNA repair. SirT1−/− thymocytes show increased sensitivity to certain forms of irradiation (Cheng et al. 2003), whereas SirT1−/− embryonic stem (ES) cells show chromosomal aberrations upon H2O2 treatment. The idea that SirT1 has a role in chromosome stability is bolstered by the observation that SirT1−/− E10.5 embryos exhibit aneuploidy in 37% of the cells (Wang et al. 2008). Evidence suggests that SirT1 is linked to DSB repair through interaction with the MRE11–RAD50–NBS1 (MRN) complex, which is involved in the signaling of S-phase-dependent repair of DSBs through homologous recombination. SirT1 deacetylates NBS1 (Nijmegen breakage syndrome protein 1), thereby facilitating the proper response to certain insults such as ionic radiation (IR) and enabling NBS1 phosphorylation (Yuan et al. 2007). Interestingly, SirT1 localizes to sites of DSB repair, similar to the case of Sir2p in yeast (Oberdoerffer et al. 2008; O'Hagan et al. 2008). This relocation has been interpreted as a survival response when DSBs arise, promoting genome stability, but also changes in the expression pattern of many genes that might have implications in the process of aging (Oberdoerffer et al. 2008). SirT1 also interacts with and deacetylates Ku70, which is a component of the NHEJ pathway for DSB repair. This results in repressing Ku70-dependent apoptosis and Ku70-dependent DNA repair, suggesting a role for SirT1 in NHEJ (Cohen et al. 2004). Whether the function of SirT1 in DNA repair is limited strictly to regulating NBS1 and Ku70 or whether other factors are also targeted remains unknown and will require further studies.
In keeping with the scope of its targets, which span so many processes, SirT1 is also associated with circadian clock regulation (Asher et al. 2008; Nakahata et al. 2008). Its deacetylase activity is regulated in a circadian manner, and its binding to the CLOCK–BMAL1 chromatin complex that regulates circadian-associated genes contributes to tight regulation of the oscillation cycles associated with these processes (Asher et al. 2008; Nakahata et al. 2008). SirT1 appears to counteract CLOCK-associated HAT activity toward histone H3 in these genes, and to deacetylate BMAL1 (Nakahata et al. 2008). Interestingly, the mechanism involving SirT1 activation in these processes is related to oscillations of nuclear NAD+ levels, with a cycle of ∼24 h. This cycling seems to be dependent on Nampt and the NAD+ salvage pathway (Nakahata et al. 2009). However, although circadian clock regulation has been suggested to be a mediator of CR in life span control, the functional mechanism is still not well understood and will require extensive work in the future.
Finally, the link of SirT1 with chromatin is also borne out by its regulation of tissue differentiation. Levels of SirT1 are very high in early stages of development and decrease progressively upon differentiation (Kuzmichev et al. 2005). Studies in different tissue types have shown that SirT1 negatively affects differentiation, particularly in those tissues with high energy expenditure (e.g., WAT or skeletal muscle, in which SirT1, once activated during energy deprivation, inhibits adipogenesis or myogenesis, respectively) (Fulco et al. 2003; Picard et al. 2004). A particularly interesting case is that of the brain, where SirT1 has a major protective role in injuries or axon degeneration (Araki et al. 2004). This observation has important implications in neurodegenerative disorders such as Parkinson's, Alzheimer's, Huntington's, and prion-related diseases (Kim et al. 2007; Chen et al. 2008b) Interestingly, differentiation and proliferation in brain cells is modulated by mild changes in redox conditions. Upon activation, SirT1 favors differentiation of mouse neural progenitor cells (NPCs) into the astroglial lineage in lieu of the neuronal one. SirT1 binds to and represses the transcription factor Hes1, thereby silencing at least one Hes1 target, the proneuronal factor Mash1 (Prozorovski et al. 2008) On the other hand, SirT1 seems to induce differentiation in the case of spermatogenesis, although the mechanism involved is not understood and might be indirect (McBurney et al. 2003)
SIRT2.
SirT2 is one of the most conserved Sirtuins: Its orthologs can be traced from yeast to humans. Albeit much about SirT2 functions remains unknown, evidence suggests that cytoplasmic SirT2 is involved in cell cycle control in proliferating cells. Contrary to most of the chromatin-related Sirtuins such as SirT1 and SirT6, SirT2 does not seem to induce cell survival, but instead participates in a mitotic checkpoint during the G2/M transition (Inoue et al. 2007a). This is supported by various findings. First, overexpression of SirT2 is associated with inhibition of cell division in starfish oocytes and mitosis delay in cultured mammalian cells (Borra et al. 2002; Dryden et al. 2003) Second, SirT2 blocks cell cycle progression at G2/M under conditions of mitotic stress or uncontrolled growth (Inoue et al. 2007b), which is in accord with the down-regulation of its activity in different types of tumors (e.g., gliomas, gastric carcinomas, and melanomas) (Hiratsuka et al. 2003; Lennerz et al. 2005) Third, SirT2 activity and localization is controlled by cyclin-dependent kinases (Cdks) throughout the cell cycle (North and Verdin 2007). Fourth, SirT2 has been found to associate with the microtubule-specific HDAC6 and to deacetylate α-tubulin (North et al. 2003). The implications of acetylation and deacetylation of α-tubulin are not completely understood, but these processes have been hypothesized to regulate microtubule organization and assembly. However, the lack of α-tubulin-associated defects in SirT2−/− MEFs or mice (AVaquero and D Reinberg, unpubl.), combined with the fact that lower eukaryotes do not show acetylation in α-tubulin (Polevoda and Sherman 2002), suggests that there are other substrates that could justify SirT2 conservation through evolution.
An important clue as to its basic role has come from biochemical studies demonstrating that both SirT2 and its yeast ortholog HST2p have a strong NAD+-dependent HDAC activity with a striking specificity for H4K16Ac (Vaquero et al. 2006). In agreement with this finding, cells depleted of SirT2 by RNAi or Δhst2 yeast cells exhibit hyperacetylated levels of H4K16, suggesting that SirT2 and HsT2p are involved in H4K16Ac regulation during the cell cycle (Vaquero et al. 2006). During the G2/M transition, SirT2 is hyperphosphorylated and transported to the nucleus, where it localizes to chromatin (Dryden et al. 2003; Vaquero et al. 2006; Wilson et al. 2006) This G2/M translocation to chromatin coincides with a massive deacetylation of H4K16Ac, suggesting that SirT2 is responsible for the drop in global H4K16Ac levels before mitosis, ensuring proper chromosome condensation. Consistent with this, loss of SirT2 correlates with hyperacetylation of H4K16Ac in mitotic cells. Interestingly, SirT2−/− MEFs do not show any marked changes in mitosis length, but do undergo a delayed G1/S transition (Vaquero et al. 2006). Given that H4K16Ac hyperacetylation is associated with high levels of DNA damage (Taipale et al. 2005; Gupta et al. 2008), this G1/S delay might accommodate DNA repair, requisite to the onset of DNA replication. However, it remains unclear whether the effects on G1/S resulting from SirT2 loss are actually due to H4K16Ac or to other, unknown substrates.
Despite the link between SirT2 and cell cycle control, the consequences of SirT2 up-regulation by CR on H4K16Ac levels or chromatin integrity are completely unknown. Recent studies have demonstrated that SirT2 also might be localized in the nucleus under certain conditions and interact with—and in some cases, deacetylate—different chromatin factors such as the HAT p300, p53, FOXO transcription factors, and the homeobox transcription factor HOXA10 (Bae et al. 2004; Wang et al. 2007; Black et al. 2008; Wang and Tong 2009) This indicates that SirT2 might also be involved in gene silencing and stress control, providing another possible link between CR and SirT2.
Interestingly, SirT2 also has tissue-specific roles. For instance, SirT2 is involved in cardiac stress tolerance—specifically, in anoxia–reoxygenation stress—through its regulation of the protein chaperone 14–3–3 ζ, which results in cell death (Lynn et al. 2008). SirT2 is also present at high levels in the brain where it seems to participate in oligodendrocyte differentiation, and has been associated with neurodegenerative diseases (Outeiro et al. 2007). The presence of SirT2 in both cardiac and brain tissues is related to inhibition of cell survival under stress situations, supporting its negative role in the cell cycle. However, whether the role of SirT2 in these organs is even somewhat related to chromatin is unknown. A very interesting link between SirT2 and CR has been shown in adipocytes. SirT2 inhibits adipocyte differentiation through its deacetylation of FOXO1, but only under CR conditions (Wang and Tong 2009). Hence, studies are needed to clarify the role of SirT2 during stress, and its possible role as a mediator of CR.
SIRT3–5.
SirT3–5 would appear to be perfect candidates as participants in the CR response in the mitochondria in which they reside (Onyango et al. 2002; Schwer et al. 2002; Saunders and Verdin 2007). Although their role is largely unknown, they seem to be intimately linked to regulation of mitochondrial metabolism and energy. Of all three, SirT3 is the only one that might also be linked to chromatin regulation, since it is also present in the nucleus (Scher et al. 2007; Nakamura et al. 2008; Sundaresan et al. 2008). Mitochondrial SirT3 has been shown to regulate mitochondrial metabolism, adaptive thermogenesis, energy homeostasis, and apoptosis (Shi et al. 2005; Allison and Milner 2007) through deacetylation of numerous mitochondrial proteins (e.g., acetyl-CoA synthetase 2 [AceCS2], the NDUFA9 component of Complex I of the respiratory chain, isocitrate dehydrogenase 2, glutamate dehydrogenase, and Ku70) (Hallows et al. 2006; Schwer et al. 2006; Lombard et al. 2007; Ahn et al. 2008; Schlicker et al. 2008; Sundaresan et al. 2008). SirT3 exists in two forms: a 44-kDa full-length protein and a 28-kDa processed version (Schwer et al. 2002). Some contradictory data have been published regarding the localization of each among different cell types: In 293 and HeLa cells, the 44-kDa form is nuclear and the 28-kDa form is present in both the nucleus and mitochondria (Scher et al. 2007), whereas in mouse cardiomyocytes the 44-kDa form is found in the nucleus, mitochondria, and cytoplasm, and the 28-kDa form is exclusively mitochondrial (Sundaresan et al. 2008). These differences are not surprising though, given that in rat cardiomyocytes SirT1—a nuclear protein in most cell lines—localizes to the nucleus in the embryo, to the cytoplasm in newborns, and to both the nucleus and cytoplasm in adults (Chen et al. 2006). Nuclear SirT3 localizes to certain presently uncharacterized foci, and under stress conditions (e.g., UV irradiation, genotoxic stress, or SirT3 overexpression), relocalizes completely to mitochondria (Scher et al. 2007). Interestingly, SirT3 has been shown to induce gene silencing by histone deacetylation of reporter genes in transfected human 293 cells, which suggests that nuclear SirT3 might be involved in gene regulation. In fact, like SirT2, SirT3 has a strict in vitro specificity for H4K16Ac, and to a lesser extent H3K9Ac. Given that cells depleted of SirT3 by RNAi and SirT3−/− MEFs do not exhibit any hyperacetylation of H4K16 or H3K9, SirT3 might affect only a very specific subset of genes—perhaps nuclear-encoded mitochondrial factors (Scher et al. 2007). Interestingly, overexpression of SirT5 also induces the relocalization of SirT3 to the nucleus while SirT5 stays in the mitochondria, suggesting that SirT3 and SirT5 are functionally linked (Nakamura et al. 2008). Identification of the nuclear foci and the gene targets of SirT3 in the near future should clarify whether SirT3 is a mediator of chromatin during the CR response.
SIRT6.
Among the Sirtuins, SirT6, like SirT1, is one of the best candidates for mediating the CR response to chromatin. In fact, SirT6 is actually the only Sirtuin that upon deletion of its gene produces an aging-like phenotype in mice, including severe metabolic defects such as abnormal levels of serum IGF-1 and glucose, lymphopenia, loss of subcutaneous fat, and lordokyphosis (Mostoslavsky et al. 2006). SirT6−/− MEFs or ES cells exhibit a broad range of defects associated with genomic instability—including chromosomal breaks and gaps, and detached chromosomes—with defects in the BER pathway responsible for repairing ssDNA damage that arises from oxidative or genotoxic stress (Mostoslavsky et al. 2006). However, the mechanism by which SirT6 is functionally linked to BER is not understood. Indeed, some data suggest that SirT6 is not part of the pathway, whereas other data suggest that overexpression of DNA polymerase β, the main DNA polymerase involved in mammalian BER, rescues the sensitivity of SirT6−/− MEFs to stress (Mostoslavsky et al. 2006).
Recent evidence supports an additional role for SirT6 in telomeric chromatin, not linked to the BER pathway. SirT6−/− MEFs show premature cellular senescence and associated telomere dysfunction, a phenotype that can be reverted by overexpression of telomerase (hTERT) (Michishita et al. 2008). SirT6 was detected by ChIP experiments in telomeric chromatin, where it is required for deacetylating H3K9Ac (Michishita et al. 2008). Interestingly, before this evidence, SirT6 was considered to be mainly a mADPRT whose only known target was itself (Liszt et al. 2005). In fact, and in contrast to SirT1–3, SirT6 shows strict specificity for H3K9Ac in vitro. The telomeric role of SirT6 remains poorly understood, but its H3K9Ac-specific deacetylase activity seems to have important consequences during telomere replication, since hyperacetylation of H3K9 in telomeres is associated with a loss of telomeric maintenance factors such as the helicase WRN RecQ (Michishita et al. 2008). Interestingly, given that WRN is reported to be a substrate of SirT1 (Vaitiekunaite et al. 2007), SirT6 and SirT1 might be functionally related.
Overall, current findings suggest that SirT6 protects the genome. However, its reported interaction with the putative tumor suppressor GCIP, which negatively regulates cell proliferation, suggests that SirT6 is itself tightly regulated by cell cycle checkpoint pathways (Ma et al. 2007). A recent report has also linked SirT6 with NF-κB-dependent regulation through deacetylation of H3K9Ac in NF-κB target genes by binding to the NF-κB subunit RelA. This observation seems to explain the aging-related phenotype of SirT6−/− mice (Kawahara et al. 2009). Interestingly, SirT1 has also been identified as a modulator of NF-κB transcription through deacetylation of RelA (Yeung et al. 2004). However, whether SirT1 and SirT6 exert similar overlapping functions or are involved in different processes is completely unknown. Array studies between SirT6 and NF-κB suggest that the involvement of SirT6 in NF-κB signaling is global, whereas the role of SirT1 seems to be related to inducing cell survival under conditions of stress. Further studies should clarify not only the link of SirT6 to CR, but also the mechanisms through which SirT6 is involved in survival pathways and chromatin integrity.
SIRT7.
Nucleolar SirT7 is detected in the promoter and coding regions of the rDNA genes, where it associates with RNA Pol I and up-regulates both its activity at and its association with these genes (Ford et al. 2006). In fact, depletion of SirT7 by RNAi or overexpression of a catalytically inactive point mutant of SirT7 correlates with a loss of Pol I in these rDNA regions. However, the mechanism through which SirT7 affects Pol I activity is unknown. Recent evidence suggest that SirT7 interacts with the Pol I transcription factor UBF and is involved in the reactivation of rDNA gene expression after mitosis (Grob et al. 2009). Although findings clearly support the idea that SirT7 has enzymatic activity toward the Pol I machinery, a target has yet to be identified. Consistent with an active role on ribosomal biogenesis, SirT7 mRNA levels are high in metabolic tissue (Vakhrusheva et al. 2008b), and its depletion by RNAi has been reported to inhibit proliferation and induce apoptosis in cell culture (Ford et al. 2006). However, studies on SirT7−/− knockout mice suggest that SirT7 has more intricate functions. Although the primary cardiomyocytes in these mice show abnormally high levels of apoptosis and low capacity for stress response, the proliferative AKT- and Ras-dependent pathways are hyperactivated and the heart is hypertrophied (Vakhrusheva et al. 2008b). Consistent with these findings, SirT7 has been shown to be up-regulated in different types of tumors and SirT7−/− MEFs do not show proliferation defects, while SirT7 overexpression significantly inhibits cell growth (Vakhrusheva et al. 2008a). Perhaps SirT7 functions differently according to tissue type. Interestingly, its purported role in Pol I transcription is one of antagonism with respect to SirT1 in the nucleolus (Espada et al. 2007; Murayama et al. 2008), and this might also indicate a functional link between SirT7 and TOR function in chromatin rDNA. Hence, studies are needed to clarify this role. Additionally, although SirT7 has a close relationship with chromatin, as corroborated by its localization to chromosomes during mitosis (Grob et al. 2009), it is yet unknown whether SirT7 levels are modulated under conditions of CR.
TOR and chromatin
TOR is a serine/threonine kinase from the phosphatidylinositol kinase-related kinase (PIKK) family and is responsible for sensing certain nutrients, particularly nitrogen sources such as amino acids (Hay and Sonenberg 2004). TOR senses glutamine, which is the preferred source of nitrogen in yeast and is crucial for the biosynthesis of purine and pyrimidine nucleotides and other related molecules such as NAD+. TOR pathway activation induces cell growth and proliferation by promoting protein anabolism through the activation of different mechanisms that include amino acid transport, ribosome biogenesis including activation of ribosome gene expression, and protein translation (Hay and Sonenberg 2004).
In addition to its link with the cytoplasmic signaling network described above, studies in yeast and mammals have shown that TOR also is involved directly in the regulation of chromatin in the nucleus, particularly the nucleolar chromatin, both at the transcriptional and structural levels (Tsang and Zheng 2007). Under optimal nutrient conditions, part of the TOR population shuttles from the cytoplasm to the nucleus, where it localizes in the nucleolus (Li et al. 2006). TOR then induces ribosome biogenesis by activating transcription of ribosomal genes through the three RNA polymerases (Pol I, Pol II, and Pol III). In fact, TOR seems to up-regulate Pol I and Pol III genes directly, whereas its effect on the expression of Pol II-transcribed ribosomal genes is exerted from the cytoplasm through its control of the localization of transcription factors SFP1 and CRF1 (Li et al. 2006). In the case of Pol III, TOR is localized to the promoters of the 5S genes, but its mechanism of action is unknown (Tsang and Zheng 2007).
TOR localizes to rDNA promoters, particularly in the case of the 35S gene where it regulates Pol I activity via phosphorylation of the Pol I cofactor TIF-IA, resulting in increased Pol I binding to rDNA regions. Additionally, yeast TOR is involved in recruiting the H4-specific HAT Esa1p, part of the NuA4 complex that seems to specifically target H4K5 and H4K12 in the nucleolus (Rohde and Cardenas 2003). Consistent with this, incubation of NIH3T3 mouse embryonic cells with the HDAC inhibitor TSA leads to hyperacetylation of histone H4 and up-regulation of rDNA genes, highlighting the critical role of H4 acetylation in rDNA control. Interestingly, in yeast that are either exposed to nutrient scarcity or whose TOR pathway has been inactivated with rapamycin, TOR, Esa1p, and Pol I are depleted in nucleolar regions and this correlates with the arrival of the HDAC Sin3/Rpd3 complex, which silences these rDNA genes through deacetylation (Tsang et al. 2003). Strikingly, inhibition of the TOR pathway in yeast and mammals is also associated with compaction of the nucleolus, which results in condensation of the nucleolar chromatin to one-fifth of its original size. In other words, nutrient starvation induces general transcriptional shutdown of rDNA by compaction of the entire nucleolus structure. This mechanism seems to imply heterochromatinization within the nucleolus, based on the activity of HDAC Rpd3 as well as on local accumulation of condensin, a structural factor involved in control of mitotic chromosome structures (Tsang et al. 2003, 2007a). When nutrients are available and/or TOR is activated, condensin is released from the nucleolus, enabling decompaction or euchromatinization of the nucleolus in keeping with transcriptional activation (Tsang et al. 2007b). Although this reversible compaction/decompaction event is linked to TOR, whether it is directly related to its activity is unknown. A functional relationship between the TOR pathway and Sirtuins cannot be excluded given that, upon CR, the former is deactivated whereas yeast Sir2p or SirT1—involved in the repression of rDNA copies—is up-regulated. In fact, some studies have suggested an activation of Sir2p activity through the up-regulation of the nicotinamidase PNC1 (Anderson et al. 2003) upon TOR pathway inhibition (Medvedik et al. 2007). However, yeast TOR inhibits phosphorylation of the Sir2p-associated factor Sir3p in subtelomeric heterochromatin via inhibition of the MAPK Mpk1p (Ai et al. 2002). Thus, phosphorylation of Sir3p under conditions of stress seems to be associated with decompaction and derepression of the subtelomeric regions. This suggests that TOR activity modulates chromatin structure: In subtelomeric regions, TOR favors heterochromatin structures, whereas in the nucleolus it induces euchromatinization and gene activation. Therefore, the TOR pathway and the Sirtuins represent a direct link between CR and large-scale control of chromatin structure.
CtBP
Another interesting factor that might be involved in CR is CtBP, a transcription factor that interacts with the adenovirus oncoprotein E1A (Boyd et al. 1993). The main function of CtBP is to corepress several transcription factors (e.g., Snail, Kruppel, Knips, and Hairless), many of them key in developmental regulation (Chinnadurai 2007). There is a considerable body of information on CtBP stemming from work on Drosophila, wherein this factor is encoded by a single gene, dCtBP (Chinnadurai 2007). In contrast, the mammalian CtBP gene has diversified into two partially redundant genes—CtBP1 and CtBP2—each of which encodes different isoforms. CtBP1 and CtBP2 appear to repress transcription factors via recruitment of chromatin silencing enzymes (e.g., HDAC1, HDAC2, and the H3K9 methyltransferases G9a and EuHMT) to targeted genes (Shi et al. 2003). Interestingly, both CtBP1 and CtBP2 can bind to either NADH or NAD+ (Kumar et al. 2002). This binding modulates dimerization of the proteins and their subsequent binding to specific transcription factors (Kumar et al. 2002). In vitro and in vivo studies suggest that CtBP binds preferentially to NADH over NAD+ (Fjeld et al. 2003). In some in vivo studies, conditions that led to a higher NADH/NAD+ ratio, such as hypoxia, also led to increased CtBP-repressive activity via increased binding to transcriptional repressors (e.g., ZEB) (Zhang et al. 2002). However, the amount of NADH does not always positively correlate with CtBP activity. In other studies, increased levels of NADH gave rise to the opposite effect. For example, hypoxia resulted in decreased binding in vivo between CtBP and the transcription factor Hdm2—a CtBP-dependent repressor of p53-dependent transcription—releasing CtBP and consequently relieving p53 inhibition (Mirnezami et al. 2003).
Interestingly, CtBP was found recently to be indirectly involved in the CR response through its regulation of SirT1 levels, and this provides a mechanistic explanation for the up-regulation of SirT1 during CR. CtBP is present at the SirT1 promoter, where it interacts with the transcription factor HIC1 (Zhang et al. 2007). CR conditions can be mimicked by inhibiting glycolysis using 2-deoxyglucose (2-DG), resulting in decreased NADH and hence a decreased NADH/NAD+ ratio. This inhibits CtBP binding to HIC1, and in turn SirT1 expression is up-regulated at the transcriptional level (WY Chen et al. 2005; Zhang et al. 2007). Whether CR conditions induce other CtBP-dependent events is currently unknown.
Poly-(ADP-ribosyl) transferases (PARPs)
The superfamily of PARPs is evolutionarily linked to the Sirtuins and represents the other large NAD+-dependent group of cellular chromatin factors (Ame et al. 2004). PARP-1 is the member best characterized, being responsible for >90% of PARP activity (Kim et al. 2005). PARP-1 is also the member most clearly involved in chromatin regulation, followed by PARP-2 and the telomeric tankyrases. The ADP-ribose donor in the PARP catalytic reaction is NAD+, which is processed as in the case of Sirtuins via breakage of the dinucleotide in nicotinamide and ADP-ribose. The (ADP-ribosyl)ation occurs in glutamic and arginine residues forming a heterogeneous branched polymeric structure that can contain >100 ADP-ribose molecules. The targets of PARP-1 described to date comprise the enzyme itself, histones, and various other nuclear proteins such as transcription factors and chromatin structural proteins (e.g., high-mobility group [HMG] proteins) (Kim et al. 2005). PARP-1 is involved in numerous processes including DNA repair, transcriptional regulation, genome instability, and cell death (Kraus 2008). PARP-1 auto(ADP-ribosyl)ation seems to alter its capacity to bind to factors involved in DNA repair, transcription, and chromatin structure. Among the different stimuli that activate PARP-1 activity is oxidative stress. Specifically, PARP-1 recognizes DNA damaged by oxidation, alkylation, or IR and has been implicated in multiple DNA repair pathways. Moreover, under conditions of low DNA damage, cell survival is promoted by PARP-1 as it recruits DNA repair machinery and regulates the functions of p53, NF-κB, and other transcription factors crucial to the stress response (Kim et al. 2005). In contrast, massive DNA damage produces cell death through necrosis and apoptosis, mainly through the depletion of NAD+ (Kraus 2008).
Given the functional parallels between PARPs and Sirtuins, it is reasonable to speculate that PARPs might have a role in the response to CR and its chromatin signaling. This idea is supported by various findings. First, PARPs are clearly linked to life span control and aging. Poly-(ADP-ribosyl)ation decreases with age, and increased PARP activity in blood mononuclear cells correlates with life span increase in several mammals (Hiona and Leeuwenburgh 2004). In fact, this link is supported not only by the functions of PARPs in DNA repair, but also by their role in protecting insulator regions and in conserving telomere length and structure (Tong et al. 2001; Yu et al. 2004). Second, under conditions of oxidative stress, PARP-1 promotes autophagy through activation of AMPK, leading to down-regulation of the TOR pathway (Huang et al. 2009). Since CR activates both AMPK activity and autophagy, and can restore the age-dependent decline in autophagy in mouse liver cells, it is possible that part of this CR response is mediated through PARP activity. Third, PARP-1 interacts with the histone variant mH2A1.1, which seems to inhibit PARP-1 enzymatic activity (Ouararhni et al. 2006). mH2A1.1 binds to O-acetyl-ADP-ribose (AADPR), the product of Sirtuin-catalyzed deacetylation (Kustatscher et al. 2005); thus, it is possible that activation of Sirtuins by CR would increase AADPR levels, which in turn could modulate PARP-1 activity. Fourth, there is evidence suggesting that SirT1 might control PARP-1 activity under DNA damage conditions, and this would link PARP1 to CR through SirT1. Activation of SirT1 by the polyphenol resveratrol reduces PARP1 activity, decreasing the levels of poly-(ADP-ribose) in the cell. In contrast, SirT1−/− cells show higher levels of poly-(ADP-ribose) (Kolthur-Seetharam et al. 2006). Other studies in Drosophila have shown that the enzyme responsible for degrading poly-(ADP-ribose) polymers, poly-(ADP-ribose) glycohydrolase (PARG), colocalizes with dSir2 at several genomic loci, and its loss induces mislocalization of the Sirtuin (Tulin et al. 2006). Finally, other findings, although not well understood, indicate that PARP1 and/or PARG might also regulate ADP-ribosylation of dSir2, which has led to the suggestion that PARP and PARG might contribute to modulation of dSir2 function (Tulin et al. 2006). Although likely, a role for PARPs in the CR response has yet to be clearly established and therefore requires extensive study.
The glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-containing complexes
Among the most surprising findings in the transcription field in the last decade is the discovery that metabolic enzymes previously thought to be exclusively cytoplasmic have nuclear functions. The best-known case is that of the cytoplasmic enzyme GAPDH, involved in DNA repair, telomere length control, transcriptional regulation, nuclear RNA export, apoptosis, and nuclear membrane fusion (Chuang et al. 2005; Sirover 2005). GAPDH catalyzes the conversion of GAP into 1,3 bisphosphoglycerate in glycolysis, with concomitant reduction of NAD+ to NADH. This enzyme is present in the nucleus under conditions such as S phase, stress, NO signaling, and apoptosis (Sirover 2005).
Given that GAPDH and other glycolytic oxidoreductases are de facto metabolic sensors that operate via changes in NADH/NAD+, among other mechanisms, and that stress response and DNA containing fraudulent nucleosides (e.g., thioguanosine, cytosine arabinoside, or 5-fluorouridine) might trigger localization of GAPDH and other such proteins to the nucleus (Chuang et al. 2005; Sirover 2005), it is feasible to envision a link between CR signaling and the role of nuclear GAPDH functions. GAPDH composes different nuclear complexes and has been shown to bind to DNA and RNA. However, the mechanisms that it employs in these processes are generally poorly understood. It is noteworthy that in several of these complexes, GAPDH is not present in its active homotetrameric form, suggesting that its catalytic dehydrogenase activity might not be relevant for many of its nuclear functions.
A good example of the nuclear functions of GAPDH is its role in the S-phase-dependent expression of the H2B gene. GAPDH is part of OCA-S, an S-phase-specific coactivator of the transcription factor Oct-1, a key regulator of many S-phase genes (Fletcher et al. 1987; Zheng et al. 2003). There are three other components of the complex that are closely linked to metabolic regulation, indicating a strong link between OCA-S and cell metabolism: lactate dehydrogenase (LDH), which catalyzes the conversion of pyruvate to lactate and oxidation of NADH to NAD+; uracil-DNA glycosylase (UDG), involved in DNA repair and recombination, metabolism, and transcription; and nucleoside diphosphate kinase (NDP), involved in nucleotide synthesis and histone H4 phosphorylation (Zheng et al. 2003; Shi 2004). Interestingly, S-phase-dependent expression of the H2B gene is determined by a specific range of NADH/NAD+ values and requires both Oct-1 and OCA-S. Increased NAD+ levels lead to increased expression of H2B in a GAPDH-dependent manner, whereas increased levels of NADH have the opposite effect (Zheng et al. 2003). This finding, together with data from in vitro competition experiments, suggests that binding of GAPDH to NAD+ is required for OCA-S functioning and H2B expression. Additionally, GAPDH binds directly to Oct-1 and is responsible for OCA-S binding to Oct-1.
Another interesting nuclear function of GAPDH is its role in telomere length regulation. GAPDH binds to ssDNA and dsDNA in vitro and associates with telomeric regions in a cell cycle-dependent manner in vivo (Sundararaj et al. 2004). Interestingly, in vitro analysis reveals that binding of NAD+ to GAPDH inhibits its telomeric-binding activity, suggesting that, contrary to the OCA-S case, a decrease in NADH/NAD+ (e.g., under CR) might alter GAPDH localization at the telomere. The importance of GAPDH to telomere integrity is demonstrated by the finding that treatment of cells with the sphingolipid ceramide leads to shortening of telomeres through inhibition of nuclear GAPDH localization, among other effects (Sundararaj et al. 2004). The implications of these findings are completely unknown and should require the characterization of the mechanism of GAPDH at the telomere, and any possible link to changes in the metabolic status of the cell, such as CR conditions.
In addition to GAPDH and LDH, glycolytic enzymes such as hexokinase (HK) and enolase 1 (ENO1) have also been found in the nucleus, where they have been associated with transcription regulation in yeast (HK), plants (ENO1), and mammals (ENO1) (Kim and Dang 2005). Given the close relationship between CR and glycolysis inhibition, studies are required to determine if these alternative nuclear functions are somehow linked.
Gaining perspectives
Understanding the mechanisms that dictate cellular responses to CR might help elucidate the path to controlling longevity in mammals. As we discussed in this review, the response to these compromised conditions implies a more efficient use of the available energy and a protective effect on genomic stability. Although its active role in the CR response has been generally underappreciated, chromatin is actually at the front line since it is a key response mediator of this signaling and the main objective of the protective role of most of the triggered measures. Chromatin factors are the targets of signaling cascades that modulate their ability to elicit the requisite changes in chromatin that give rise to the global response to CR. A full understanding of CR and its effect will require a complete characterization of the role of chromatin in these processes. Although our knowledge about the mechanisms and the factors underlying this signaling is relatively limited at present, this will likely change through extensive efforts in the future. What we already know suggests that, as in the case of Sirtuins, these mechanisms and associated factors might provide the key to improve life for human beings in the not too distant future.
Acknowledgments
We apologize to the many colleagues whose work could not be cited due to space limitations. We are grateful to Lynne Vales for critical reading of the manuscript and helpful comments. The work of D.R.'s laboratory is supported by grants from NIH and the HHMI. The Chromatin Biology Laboratory (A.V.) is funded by the Spanish Ministry of Science and Innovation (MICINN), the European Union (EU)-Marie Curie Actions, and Fundación Sandra Ibarra. A.V. is an ICREA researcher.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1807009.
References
- Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai W, Bertram PG, Tsang CK, Chan TF, Zheng XF. Regulation of subtelomeric silencing during stress response. Mol Cell. 2002;10:1295–1305. doi: 10.1016/s1097-2765(02)00695-0. [DOI] [PubMed] [Google Scholar]
- Allison SJ, Milner J. SIRT3 is pro-apoptotic and participates in distinct basal apoptotic pathways. Cell Cycle. 2007;6:2669–2677. doi: 10.4161/cc.6.21.4866. [DOI] [PubMed] [Google Scholar]
- Amat R, Solanes G, Giralt M, Villarroya F. SIRT1 is involved in glucocorticoid-mediated control of uncoupling protein-3 gene transcription. J Biol Chem. 2007;282:34066–34076. doi: 10.1074/jbc.M707114200. [DOI] [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Bae NS, Swanson MJ, Vassilev A, Howard BH. Human histone deacetylase SIRT2 interacts with the homeobox transcription factor HOXA10. J Biochem (Tokyo) 2004;135:695–700. doi: 10.1093/jb/mvh084. [DOI] [PubMed] [Google Scholar]
- Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke A. Insulin and aging. Cell Cycle. 2008;7:3338–3343. doi: 10.4161/cc.7.21.7012. [DOI] [PubMed] [Google Scholar]
- Barzilai A, Yamamoto K. DNA damage responses to oxidative stress. DNA Repair (Amst) 2004;3:1109–1115. doi: 10.1016/j.dnarep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Bell SD, Botting CH, Wardleworth BN, Jackson SP, White MF. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science. 2002;296:148–151. doi: 10.1126/science.1070506. [DOI] [PubMed] [Google Scholar]
- Bhaskar PT, Hay N. The two TORCs and Akt. Dev Cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Black JC, Mosley A, Kitada T, Washburn M, Carey M. The SIRT2 deacetylase regulates autoacetylation of p300. Mol Cell. 2008;32:449–455. doi: 10.1016/j.molcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borra MT, O'Neill FJ, Jackson MD, Marshall B, Verdin E, Foltz KR, Denu JM. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J Biol Chem. 2002;277:12632–12641. doi: 10.1074/jbc.M111830200. [DOI] [PubMed] [Google Scholar]
- Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, Hay RT, Gu W, Pestell RG. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- Boyd JM, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Cabelof DC, Yanamadala S, Raffoul JJ, Guo Z, Soofi A, Heydari AR. Caloric restriction promotes genomic stability by induction of base excision repair and reversal of its age-related decline. DNA Repair (Amst) 2003;2:295–307. doi: 10.1016/s1568-7864(02)00219-7. [DOI] [PubMed] [Google Scholar]
- Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Chen IY, Lypowy J, Pain J, Sayed D, Grinberg S, Alcendor RR, Sadoshima J, Abdellatif M. Histone H2A.z is essential for cardiac myocyte hypertrophy but opposed by silent information regulator 2α. J Biol Chem. 2006;281:19369–19377. doi: 10.1074/jbc.M601443200. [DOI] [PubMed] [Google Scholar]
- Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes & Dev. 2008a;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Steele AD, Hutter G, Bruno J, Govindarajan A, Easlon E, Lin SJ, Aguzzi A, Lindquist S, Guarente L. The role of calorie restriction and SIRT1 in prion-mediated neurodegeneration. Exp Gerontol. 2008b;43:1086–1093. doi: 10.1016/j.exger.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol. 2007;39:1593–1607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Chuang DM, Hough C, Senatorov VV. Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2005;45:269–290. doi: 10.1146/annurev.pharmtox.45.120403.095902. [DOI] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilova I, Easlon E, Lin SJ. Calorie restriction and the nutrient sensing signaling pathways. Cell Mol Life Sci. 2007;64:752–767. doi: 10.1007/s00018-007-6381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol. 2003;23:3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada J, Ballestar E, Santoro R, Fraga MF, Villar-Garea A, Nemeth A, Lopez-Serra L, Ropero S, Aranda A, Orozco H, et al. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res. 2007;35:2191–2198. doi: 10.1093/nar/gkm118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- Feige JN, Auwerx J. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007;17:292–301. doi: 10.1016/j.tcb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Fjeld CC, Birdsong WT, Goodman RH. Differential binding of NAD+ and NADH allows the transcriptional corepressor carboxyl-terminal binding protein to serve as a metabolic sensor. Proc Natl Acad Sci. 2003;100:9202–9207. doi: 10.1073/pnas.1633591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher C, Heintz N, Roeder RG. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell. 1987;51:773–781. doi: 10.1016/0092-8674(87)90100-0. [DOI] [PubMed] [Google Scholar]
- Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes & Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F, Binda C, Battaglioli E, Mattevi A. LSD1: Oxidative chemistry for multifaceted functions in chromatin regulation. Trends Biochem Sci. 2008;33:181–189. doi: 10.1016/j.tibs.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Fu C, Hickey M, Morrison M, McCarter R, Han ES. Tissue specific and non-specific changes in gene expression by aging and by early stage CR. Mech Ageing Dev. 2006;127:905–916. doi: 10.1016/j.mad.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Banerjee R, Breen TR, Harte PJ. SIR2 is required for polycomb silencing and is associated with an E(Z) histone methyltransferase complex. Curr Biol. 2004;14:1812–1821. doi: 10.1016/j.cub.2004.09.060. [DOI] [PubMed] [Google Scholar]
- Grob A, Roussel P, Wright JE, McStay B, Hernandez-Verdun D, Sirri V. Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J Cell Sci. 2009;122:489–498. doi: 10.1242/jcs.042382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes & Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- Guarente L. Mitochondria—A nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–176. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L, Picard F. Calorie restriction—The SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Guo Z, Heydari A, Richardson A. Nucleotide excision repair of actively transcribed versus nontranscribed DNA in rat hepatocytes: Effect of age and dietary restriction. Exp Cell Res. 1998;245:228–238. doi: 10.1006/excr.1998.4269. [DOI] [PubMed] [Google Scholar]
- Gupta A, Guerin-Peyrou TG, Sharma GG, Park C, Agarwal M, Ganju RK, Pandita S, Choi K, Sukumar S, Pandita RK, et al. The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol Cell Biol. 2008;28:397–409. doi: 10.1128/MCB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haendeler J, Hoffmann J, Diehl JF, Vasa M, Spyridopoulos I, Zeiher AM, Dimmeler S. Antioxidants inhibit nuclear export of telomerase reverse transcriptase and delay replicative senescence of endothelial cells. Circ Res. 2004;94:768–775. doi: 10.1161/01.RES.0000121104.05977.F3. [DOI] [PubMed] [Google Scholar]
- Halliwell B. Antioxidant defence mechanisms: From the beginning to the end (of the beginning) Free Radic Res. 1999;31:261–272. doi: 10.1080/10715769900300841. [DOI] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes & Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Heydari AR, Unnikrishnan A, Lucente LV, Richardson A. Caloric restriction and genomic stability. Nucleic Acids Res. 2007;35:7485–7496. doi: 10.1093/nar/gkm860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y. Chromosomal DNA fragmentation in apoptosis and necrosis induced by oxidative stress. Biochem Pharmacol. 2003;66:1527–1535. doi: 10.1016/s0006-2952(03)00508-2. [DOI] [PubMed] [Google Scholar]
- Hiona A, Leeuwenburgh C. Effects of age and caloric restriction on brain neuronal cell death/survival. Ann N Y Acad Sci. 2004;1019:96–105. doi: 10.1196/annals.1297.018. [DOI] [PubMed] [Google Scholar]
- Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, Watanabe T, Ohama E, Tahimic CG, Kurimasa A, et al. Proteomics-based identification of differentially expressed genes in human gliomas: Down-regulation of SIRT2 gene. Biochem Biophys Res Commun. 2003;309:558–566. doi: 10.1016/j.bbrc.2003.08.029. [DOI] [PubMed] [Google Scholar]
- Huang Q, Wu YT, Tan HL, Ong CN, Shen HM. A novel function of poly(ADP-ribose) polymerase-1 in modulation of autophagy and necrosis under oxidative stress. Cell Death Differ. 2009;16:264–277. doi: 10.1038/cdd.2008.151. [DOI] [PubMed] [Google Scholar]
- Hursting SD, Lavigne JA, Berrigan D, Perkins SN, Barrett JC. Calorie restriction, aging, and cancer prevention: Mechanisms of action and applicability to humans. Annu Rev Med. 2003;54:131–152. doi: 10.1146/annurev.med.54.101601.152156. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Inoue T, Hiratsuka M, Osaki M, Oshimura M. The molecular biology of mammalian SIRT proteins: SIRT2 in cell cycle regulation. Cell Cycle. 2007a;6:1011–1018. doi: 10.4161/cc.6.9.4219. [DOI] [PubMed] [Google Scholar]
- Inoue T, Hiratsuka M, Osaki M, Yamada H, Kishimoto I, Yamaguchi S, Nakano S, Katoh M, Ito H, Oshimura M. SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene. 2007b;26:945–957. doi: 10.1038/sj.onc.1209857. [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Takubo K, Matsuoka S, Miyamoto K, Ohmura M, Naka K, Hosokawa K, Ikeda Y, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- Jin YH, Kim YJ, Kim DW, Baek KH, Kang BY, Yeo CY, Lee KY. Sirt2 interacts with 14-3-3 β/γ and down-regulates the activity of p53. Biochem Biophys Res Commun. 2008;368:690–695. doi: 10.1016/j.bbrc.2008.01.114. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfi Y, Shalman R, Peshti V, Pilosof SN, Gozlan YM, Pearson KJ, Lerrer B, Moazed D, Marine JC, de Cabo R, et al. Regulation of SIRT6 protein levels by nutrient availability. FEBS Lett. 2008;582:543–548. doi: 10.1016/j.febslet.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanjawala ZE, Murphy N, Hinton DR, Hsieh CL, Lieber MR. Oxygen metabolism causes chromosome breaks and is associated with the neuronal apoptosis observed in DNA double-strand break repair mutants. Curr Biol. 2002;12:397–402. doi: 10.1016/s0960-9822(02)00684-x. [DOI] [PubMed] [Google Scholar]
- Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Steffen KK, Kaeberlein M. Ruminations on dietary restriction and aging. Cell Mol Life Sci. 2007;64:1323–1328. doi: 10.1007/s00018-007-6470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Dang CV. Multifaceted roles of glycolytic enzymes. Trends Biochem Sci. 2005;30:142–150. doi: 10.1016/j.tibs.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes & Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolthur-Seetharam U, Dantzer F, McBurney MW, de Murcia G, Sassone-Corsi P. Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle. 2006;5:873–877. doi: 10.4161/cc.5.8.2690. [DOI] [PubMed] [Google Scholar]
- Konat GW. H2O2-induced higher order chromatin degradation: A novel mechanism of oxidative genotoxicity. J Biosci. 2003;28:57–60. doi: 10.1007/BF02970132. [DOI] [PubMed] [Google Scholar]
- Kraus WL. Transcriptional control by PARP-1: Chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Carlson JE, Ohgi KA, Edwards TA, Rose DW, Escalante CR, Rosenfeld MG, Aggarwal AK. Transcription corepressor CtBP is an NAD+-regulated dehydrogenase. Mol Cell. 2002;10:857–869. doi: 10.1016/s1097-2765(02)00650-0. [DOI] [PubMed] [Google Scholar]
- Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14:183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Cacicedo JM, Ruderman N, Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J Biol Chem. 2008;283:27628–27635. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Lennerz V, Fatho M, Gentilini C, Frye RA, Lifke A, Ferel D, Wolfel C, Huber C, Wolfel T. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci. 2005;102:16013–16018. doi: 10.1073/pnas.0500090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Tsang CK, Watkins M, Bertram PG, Zheng XF. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 2006;442:1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo MV, Allard J, Ingram DK, Navas P, et al. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci. 2006;103:1768–1773. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Lynn EG, McLeod CJ, Gordon JP, Bao J, Sack MN. SIRT2 is a negative regulator of anoxia-reoxygenation tolerance via regulation of 14–3–3 ζ and BAD in H9c2 cells. FEBS Lett. 2008;582:2857–2862. doi: 10.1016/j.febslet.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Stafford LJ, Li D, Luo J, Li X, Ning G, Liu M. GCIP/CCNDBP1, a helix–loop–helix protein, suppresses tumorigenesis. J Cell Biochem. 2007;100:1376–1386. doi: 10.1002/jcb.21140. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. Hormesis and the antiaging action of dietary restriction. Exp Gerontol. 1998;33:61–66. doi: 10.1016/s0531-5565(97)00071-5. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. Overview of caloric restriction and ageing. Mech Ageing Dev. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2α protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size. J Nutr. 1935;10:63–79. [PubMed] [Google Scholar]
- Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e261. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Zhang X, Cheng X, Grewal SI, Xu RM. Structure of the SET domain histone lysine methyltransferase Clr4. Nat Struct Biol. 2002;9:828–832. doi: 10.1038/nsb860. [DOI] [PubMed] [Google Scholar]
- Mirnezami AH, Campbell SJ, Darley M, Primrose JN, Johnson PW, Blaydes JP. Hdm2 recruits a hypoxia-sensitive corepressor to negatively regulate p53-dependent transcription. Curr Biol. 2003;13:1234–1239. doi: 10.1016/s0960-9822(03)00454-8. [DOI] [PubMed] [Google Scholar]
- Monks TJ, Xie R, Tikoo K, Lau SS. Ros-induced histone modifications and their role in cell survival and cell death. Drug Metab Rev. 2006;38:755–767. doi: 10.1080/03602530600959649. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S. Increased dosage of mammalian Sir2 in pancreatic β cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- Nakae J, Oki M, Cao Y. The FoxO transcription factors and metabolic regulation. FEBS Lett. 2008;582:54–67. doi: 10.1016/j.febslet.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Ogura M, Tanaka D, Inagaki N. Localization of mouse mitochondrial SIRT proteins: Shift of SIRT3 to nucleus by co-expression with SIRT5. Biochem Biophys Res Commun. 2008;366:174–179. doi: 10.1016/j.bbrc.2007.11.122. [DOI] [PubMed] [Google Scholar]
- Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- North BJ, Verdin E. Mitotic regulation of SIRT2 by cyclin-dependent kinase 1-dependent phosphorylation. J Biol Chem. 2007;282:19546–19555. doi: 10.1074/jbc.M702990200. [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci. 2002;99:13653–13658. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouararhni K, Hadj-Slimane R, Ait-Si-Ali S, Robin P, Mietton F, Harel-Bellan A, Dimitrov S, Hamiche A. The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes & Dev. 2006;20:3324–3336. doi: 10.1101/gad.396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, et al. Sirtuin 2 inhibitors rescue α-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Petersen S, Saretzki G, von Zglinicki T. Preferential accumulation of single-stranded regions in telomeres of human fibroblasts. Exp Cell Res. 1998;239:152–160. doi: 10.1006/excr.1997.3893. [DOI] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B, Sherman F. The diversity of acetylated proteins. Genome Biol. 2002;3:reviews0006. doi: 10.1186/gb-2002-3-5-reviews0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prozorovski T, Schulze-Topphoff U, Glumm R, Baumgart J, Schroter F, Ninnemann O, Siegert E, Bendix I, Brustle O, Nitsch R, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Tjian R. Nutrient availability and growth: Regulation of insulin signaling by dFOXO/FOXO1. Cell Cycle. 2006;5:503–505. doi: 10.4161/cc.5.5.2501. [DOI] [PubMed] [Google Scholar]
- Raught B, Gingras AC, Sonenberg N. The target of rapamycin (TOR) proteins. Proc Natl Acad Sci. 2001;98:7037–7044. doi: 10.1073/pnas.121145898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman LM, Ravussin E. Endocrine alterations in response to calorie restriction in humans. Mol Cell Endocrinol. 2009;299:129–136. doi: 10.1016/j.mce.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde JR, Cardenas ME. The tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol Cell Biol. 2003;23:629–635. doi: 10.1128/MCB.23.2.629-635.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders LR, Verdin E. Sirtuins: Critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes & Dev. 2007;21:920–928. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SI. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol. 2003;13:1240–1246. doi: 10.1016/s0960-9822(03)00489-5. [DOI] [PubMed] [Google Scholar]
- Shi Y. Metabolic enzymes and coenzymes in transcription—A direct link between metabolism and transcription? Trends Genet. 2004;20:445–452. doi: 10.1016/j.tig.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar EB, Whetstine JR, Lan F, Ogawa H, Luke MP-S, Nakatani Y, Shi Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- Sirover MA. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J Cell Biochem. 2005;95:45–52. doi: 10.1002/jcb.20399. [DOI] [PubMed] [Google Scholar]
- Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal RS, Ku HH, Agarwal S, Forster MJ, Lal H. Oxidative damage, mitochondrial oxidant generation and antioxidant defenses during aging and in response to food restriction in the mouse. Mech Ageing Dev. 1994;74:121–133. doi: 10.1016/0047-6374(94)90104-x. [DOI] [PubMed] [Google Scholar]
- Srivastava VK, Miller S, Schroeder MD, Hart RW, Busbee D. Age-related changes in expression and activity of DNA polymerase α: Some effects of dietary restriction. Mutat Res. 1993;295:265–280. doi: 10.1016/0921-8734(93)90025-x. [DOI] [PubMed] [Google Scholar]
- Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: The means to the end. Annu Rev Cell Dev Biol. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JA, Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA. Mitochondrial and nuclear DNA base excision repair are affected differently by caloric restriction. FASEB J. 2004;18:595–597. doi: 10.1096/fj.03-0890fje. [DOI] [PubMed] [Google Scholar]
- Sundararaj KP, Wood RE, Ponnusamy S, Salas AM, Szulc Z, Bielawska A, Obeid LM, Hannun YA, Ogretmen B. Rapid shortening of telomere length in response to ceramide involves the inhibition of telomere binding activity of nuclear glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 2004;279:6152–6162. doi: 10.1074/jbc.M310549200. [DOI] [PubMed] [Google Scholar]
- Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Rea S, Richter K, Vilar A, Lichter P, Imhof A, Akhtar A. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol. 2005;25:6798–6810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Tong WM, Hande MP, Lansdorp PM, Wang ZQ. DNA strand break-sensing molecule poly(ADP-Ribose) polymerase cooperates with p53 in telomere function, chromosome stability, and tumor suppression. Mol Cell Biol. 2001;21:4046–4054. doi: 10.1128/MCB.21.12.4046-4054.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Reinberg D. Facultative heterochromatin: Is there a distinctive molecular signature? Mol Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Tsang CK, Zheng XF. TOR-in(g) the nucleus. Cell Cycle. 2007;6:25–29. doi: 10.4161/cc.6.1.3675. [DOI] [PubMed] [Google Scholar]
- Tsang CK, Bertram PG, Ai W, Drenan R, Zheng XF. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003;22:6045–6056. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang CK, Li H, Zheng XS. Nutrient starvation promotes condensin loading to maintain rDNA stability. EMBO J. 2007a;26:448–458. doi: 10.1038/sj.emboj.7601488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang CK, Wei Y, Zheng XF. Compacting DNA during the interphase: Condensin maintains rDNA integrity. Cell Cycle. 2007b;6:2213–2218. doi: 10.4161/cc.6.18.4733. [DOI] [PubMed] [Google Scholar]
- Tsao JL, Dudley S, Kwok B, Nickel AE, Laird PW, Siegmund KD, Liskay RM, Shibata D. Diet, cancer and aging in DNA mismatch repair deficient mice. Carcinogenesis. 2002;23:1807–1810. doi: 10.1093/carcin/23.11.1807. [DOI] [PubMed] [Google Scholar]
- Tulin A, Naumova NM, Menon AK, Spradling AC. Drosophila poly(ADP-ribose) glycohydrolase mediates chromatin structure and SIR2-dependent silencing. Genetics. 2006;172:363–371. doi: 10.1534/genetics.105.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JH, Kim SJ, Kim DW, Ha MY, Jang JH, Chung BS, Kang CD, Kim SH. Tissue-specific changes of DNA repair protein Ku and mtHSP70 in aging rats and their retardation by caloric restriction. Mech Ageing Dev. 2003;124:967–975. doi: 10.1016/s0047-6374(03)00169-6. [DOI] [PubMed] [Google Scholar]
- Vaitiekunaite R, Butkiewicz D, Krzesniak M, Przybylek M, Gryc A, Snietura M, Benedyk M, Harris CC, Rusin M. Expression and localization of Werner syndrome protein is modulated by SIRT1 and PML. Mech Ageing Dev. 2007;128:650–661. doi: 10.1016/j.mad.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Vakhrusheva O, Braeuer D, Liu Z, Braun T, Bober E. Sirt7-dependent inhibition of cell growth and proliferation might be instrumental to mediate tissue integrity during aging. J Physiol Pharmacol. 2008a;59:201–212. [PubMed] [Google Scholar]
- Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008b;102:703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- Vaquero A. The conserved role of sirtuins in chromatin regulation. Int J Dev Biol. 2009;53:303–322. doi: 10.1387/ijdb.082675av. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes & Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007a;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007b;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem Sci. 2002;27:339–344. doi: 10.1016/s0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1’s repressive interaction with PPARγ. Mol Biol Cell. 2009;20:801–808. doi: 10.1091/mbc.E08-06-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JM, Le VQ, Zimmerman C, Marmorstein R, Pillus L. Nuclear export modulates the cytoplasmic Sir2 homologue Hst2. EMBO Rep. 2006;7:1247–1251. doi: 10.1038/sj.embor.7400829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia E, Rao G, Van Remmen H, Heydari AR, Richardson A. Activities of antioxidant enzymes in various tissues of male Fischer 344 rats are altered by food restriction. J Nutr. 1995;125:195–201. doi: 10.1093/jn/125.2.195. [DOI] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet. 2004;36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang SY, Fleuriel C, Leprince D, Rocheleau JV, Piston DW, Goodman RH. Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc Natl Acad Sci. 2007;104:829–833. doi: 10.1073/pnas.0610590104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zheng L, Roeder RG, Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114:255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]