Abstract
Stem cells do not all respond the same way, but the mechanisms underlying this heterogeneity are not well understood. Here, we found that expression of Hes1 and its downstream genes oscillate in mouse embryonic stem (ES) cells. Those expressing low and high levels of Hes1 tended to differentiate into neural and mesodermal cells, respectively. Furthermore, inactivation of Hes1 facilitated neural differentiation more uniformly at earlier time. Thus, Hes1-null ES cells display less heterogeneity in both the differentiation timing and fate choice, suggesting that the cyclic gene Hes1 contributes to heterogeneous responses of ES cells even under the same environmental conditions.
Keywords: ES cell heterogeneity, Hes1 oscillation, Notch signaling, Dll1, Gadd45g
Embryonic stem (ES) cells have the ability to differentiate into multiple cell types of all three germ layers, providing powerful tools to realize mammalian early development in culture dishes as well as to create specific types of cells for regenerative medicine (Smith 2001; Murry and Keller 2008). Many methods for triggering in vitro differentiation of ES cells have been established, but most of them require additional purification to collect homogeneous cell populations. One main reason for such heterogeneity is that ES cells asynchronously differentiate into diverse cell types, and thereby give rise to mixtures of differentiated and undifferentiated cells in a rather chaotic manner (Lowell et al. 2006). Recent studies have demonstrated that expression of the homeodomain factor Nanog and the zinc finger protein Rex1 fluctuates over several days in individual ES cells (Chambers et al. 2007; Singh et al. 2007; Toyooka et al. 2008). The phenotypic output of this heterogeneity appears during their differentiation: Nanog-negative ES cells are prone to differentiate into primitive endoderm-like cells (Chambers et al. 2007; Singh et al. 2007) while Rex1-negative ES cells correspond to epiblast and primitive ectoderm and more rapidly differentiate into somatic lineages (Toyooka et al. 2008). These studies indicate that ES cells are not homogeneous. Rather, they are heterogeneous in differentiation competency (Chambers et al. 2007; Toyooka et al. 2008). However, even Nanog-positive ES cells are not homogeneous, and the molecular mechanism underlying this heterogeneity is not well understood, although such heterogeneity is suggested to be important for dynamic differentiation process of ES cells, based on theoretical analysis of cell systems (Furusawa and Kaneko 2001; Kaneko 2006).
Hes1 is a member of the basic helix–loop–helix (bHLH) transcriptional repressors that regulate cell proliferation and differentiation in embryogenesis (Kageyama et al. 2007). We previously found that Hes1 is expressed in an oscillatory manner with a period of ∼2 h in various cells such as fibroblasts and neural progenitor cells (Hirata et al. 2002; Shimojo et al. 2008). This oscillatory expression is regulated by negative feedback and rapid degradation of the gene products: Hes1 can repress its own expression, but rapid degradation of both Hes1 mRNA and protein with half-lives of ∼20 min soon results in the release from negative regulation and allows the next round of expression (Hirata et al. 2002). We here found that Hes1 expression also oscillates in ES cells and that Hes1-null ES cells display less heterogeneity in both the differentiation timing and fate choice, suggesting that the cyclic gene Hes1 contributes to heterogeneous responses of ES cells even under the same environmental conditions.
Results and Discussion
Hes1 expression oscillates in ES cells
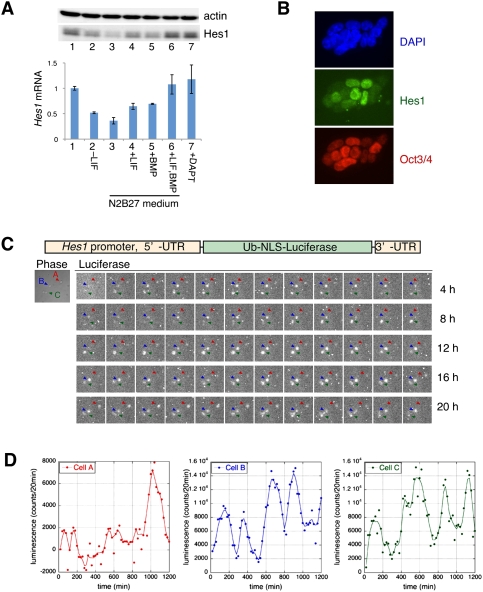
Hes1 protein was highly expressed by ES cells under the control of two essential factors, LIF and BMP (Fig. 1A, lanes 2–6; Ying et al. 2003), but not of Notch signaling (Fig. 1A, lane 7). Interestingly, Hes1 expression levels were variable in individual ES cells while Oct3/4 also seemed to be variable to some extent (Fig. 1B). To determine whether Hes1 expression oscillates in ES cells, we employed a real-time imaging method using a Hes1 promoter-driven ubiquitin-fused firefly luciferase reporter (Fig. 1C) that was shown previously to mimic the endogenous Hes1 expression (Masamizu et al. 2006). Indeed, the Hes1 promoter-driven reporter expression oscillated in individual ES cells (Fig. 1C,D; Supplemental Movie), whereas the human polypeptide chain elongation factor 1α (EF-1α) promoter-driven reporter expression did not (data not shown). These results indicated that Hes1 expression oscillates in ES cells.
Figure 1.
Oscillatory expression of Hes1 in mouse ES cells. (A) Hes1 protein (top panel) and mRNA (bottom panel) expression in normal (lane 1) or other culture conditions, after the removal of LIF in normal ES medium (lane 2), in N2B27 serum-free medium (lane 3), in the presence of LIF (lane 4), BMP (lane 5), or both LIF and BMP (lane 6) in N2B27 medium, and in the presence of DAPT, a γ-secretase inhibitor (Notch signaling inhibitor), in normal ES medium (lane 7). Hes1 mRNA levels were analyzed by Q-PCR, normalized by GAPDH and given in ratios to ES cells cultured in normal ES medium. (B) Immunocytochemistry using anti-Hes1 (green) and anti-Oct4 antibodies (red) with DAPI (blue). (C) Bioluminescence images were taken by 20-min exposures for 20 h from three ES cells that express the ubiquitin (Ub)-fused and nuclear localization signal (NLS)-fused firefly luciferase reporter under the control of the Hes1 promoter (see the Supplemental Movie). (D) Quantification of the bioluminescence activity of cells A (red arrowhead), B (blue arrowhead), and C (green arrowhead) shown in C.
The oscillations continued throughout the cell cycle, but the Hes1 tended to be expressed at higher levels during S–G2 phases compared with G1 phase (data not shown). The oscillations seemed to be synchronized between neighboring daughter cells after cell division, although they easily became out of synchrony in large colonies (data not shown). The period of Hes1 oscillations was variable from cycle to cycle and from cell to cell (Fig. 1D). The power spectrum of Hes1 oscillations after Fourier transform showed that the periodicity was ∼3–5 h, although it included many noises of short periodicity (Supplemental Fig. S1). The period of 3–5 h in ES cells was longer than in other cell types (2–3 h) (Hirata et al. 2002; Masamizu et al. 2006; Shimojo et al. 2008). The half-life of Hes1 protein was ∼16 min in ES cells, which was similar to fibroblasts, whereas that of Hes1 mRNA was about two to four times longer in ES cells (∼46 min) than in fibroblasts (data not shown), implying that the stabilization of Hes1 mRNA contributes to a longer periodicity in ES cells.
Hes1 represses expression of Notch ligand and cell cycle regulator genes
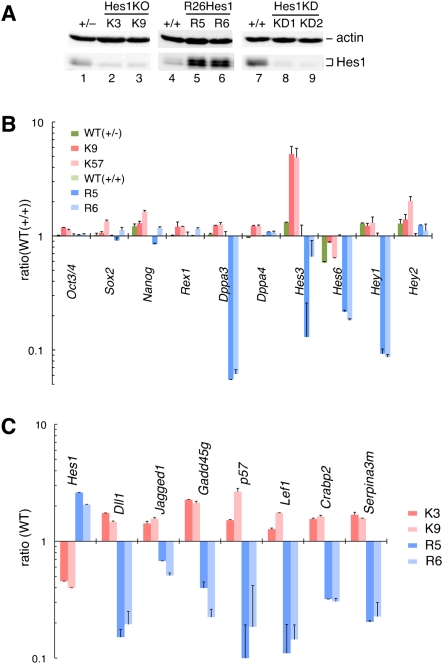
To elucidate the role of Hes1 in ES cells, we next sought to determine the downstream target genes. To this end, we established two types of ES cells (two or three independent lines for each type), Hes1-null and Hes1-sustained cells. Hes1-null (K3, K9, and K57) cells were made by transient Cre expression in ES cells carrying Hes1-floxed and Hes1-null alleles (Fig. 2A, lanes 2,3; Supplemental Fig. S2). Hes1-sustained (R5 and R6) cells were made by knocking Hes1 cDNA into the Rosa26 locus (Supplemental Fig. S3), and these cells expressed Hes1 in a sustained manner, whose levels were similar to the endogenous maximal level (Fig. 2A, lanes 5,6; Supplemental Fig. S4). Both Hes1-null and Hes1-sustained cells expressed similar levels of Oct3/4, Sox2, Nanog, and Rex1, markers of ES cells (Fig. 2B), and proliferated on feeder cells at similar levels to wild-type ES cells (data not shown). Furthermore, both types of ES cells were able to form three germ layers in embryoid body (Supplemental Fig. S5) and chimeric embryo formation assays (Supplemental Fig. S6), indicating that the proliferative and multipotential activities are maintained in both Hes1-null and Hes1-sustained ES cells. We searched the downstream genes of Hes1 by comparing mRNA expression levels between wild-type, R5/R6 and K3/K9 cells using a microarray method. We found that 36 genes displayed more than twofold higher expression in K3/K9 (Hes1-null) cells than in R5/R6 (Hes1-sustained) cells on microarray analysis, suggesting that these genes are repressed by Hes1 (Supplemental Table S1). Among them, seven genes were confirmed as Hes1 targets by real-time PCR, including two Notch ligands, Dll1 and Jagged1 (Jag1), and two cell cycle inhibitory genes, Gadd45g (Vairapandi et al. 2002) and p57 (Fig. 2C). We also performed chromatin immunoprecipitation (ChIP) by using Hes1 antibody and subjected the DNA precipitates to oligonucleotide tiling arrays (ChIP–chip) (Wendt et al. 2008) to identify direct target genes for Hes1. We used Hes1 knockdown ES cells as a negative control (Fig. 2A, lanes 8,9). Five-hundred-six Hes1-binding regions were identified, and the list contained many gene loci for transcription factors that regulate cell differentiation as well as Notch signaling, suggesting that Hes1 is involved in ES cell differentiation (Supplemental Table S2). Among the genes identified by the expression array, Dll1, Jag1, Gadd45g, Lef1, Crabp2, and Hes1 were also identified by ChIP–chip analysis.
Figure 2.
Comparison of gene expression between Hes1-null and Hes1-sustained ES cells. (A) Hes1 protein levels in Hes1-null (K3 and K9; lanes 2,3), Hes1-sustained (R5 and R6; lanes 5,6), Hes1 knockdown (KD1 and KD2; lanes 8,9), and parental cells (+/−, +/+; lanes 1,4,7). Hes1 knockdown cells were made by two different shRNA constructs (KD1, KD2). Actin is a loading control for Western blotting. (B) The mRNA levels of ES cell marker genes (Oct3/4, Sox2, Nanog, Rex1, Dppa3, and Dppa4) and Hes family genes (Hes3, Hes6, Hey1, and Hey2). They were analyzed by Q-PCR and given in ratios to wild-type (+/+) ES cell line (lane 4 in A). Most of ES cell markers, except Dppa3, were expressed at similar levels in all cell lines. Hes5 expression was very low in all ES cells (data not shown). (C) The mRNA levels of Hes1 target genes in ES cell lines (+/−, +/+, K3, K9, R5, and R6) were analyzed by Q-PCR and given in ratios to their parental wild-type cells.
Expression of some downstream genes also oscillates in ES cells
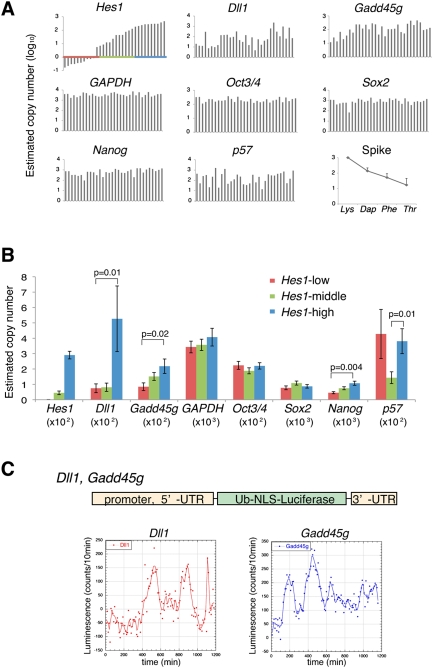
Because Hes1 expression oscillated, some downstream genes might be also expressed in an oscillatory manner. To determine the relationship of Hes1 expression with the downstream gene expression, we randomly picked up 32 single ES cells and made cDNAs from each cell to perform quantitative real-time PCR (single-cell Q-PCR) (Kurimoto et al. 2006, 2007). These ES cells expressed variable levels of Hes1 and were classified into three groups according to the Hes1 expression level (Fig. 3A). When Hes1 expression was high, both Dll1 and Gadd45g expression was also high and vice versa, whereas the other downstream genes p57, Lef1, Jag1, and Crabp2 displayed different patterns (Fig. 3B; data not shown). Thus, expression levels of Hes1, Dll1, and Gadd45g changed in a similar manner in individual ES cells, suggesting that these genes oscillate in phase. When the Hes1 protein level is high, transcription of all Hes1, Dll1, and Gadd45g genes may be repressed, but they may be activated when the Hes1 protein level is low. Due to this delayed negative regulation by Hes1, expression of all these genes probably oscillated in phase. To obtain direct evidence that both Dll1 and Gadd45g expression also oscillate in ES cells, we further analyzed their expression dynamics by a real-time imaging method using a ubiquitin-fused luciferase reporter under the control of Dll1 or Gadd45g promoter. The expression of both Dll1 and Gadd45g dynamically changed in many individual ES cells (Fig. 3C). These results suggest that the differentiation competency of ES cells can be changed rapidly by oscillations of Dll1, a ligand of Notch signaling that induces neural differentiation (Lowell et al. 2006), and Gadd45g, which inhibits cell cycle progression (Vairapandi et al. 2002). Interestingly, while Nanog was mostly expressed by picked-up ES cells (Fig. 3A), its expression level seemed to be variable and became higher when Hes1 was highly expressed (Fig. 3B), whereas there was no such relationship with Oct3/4 and Sox2 (Fig. 3B), suggesting that Hes1 oscillations may have some correlation with Nanog fluctuation (Chambers et al. 2007) but not with Oct3/4 or Sox2.
Figure 3.
Dynamic expression of Hes1 target genes in ES cells. (A) Single-cell Q-PCR analysis with 32 randomly picked up ES cells. The results were aligned along the x-axis according to the Hes1 expression levels. Graphs show the putative number (log10) of transcript of Hes1, Hes1 downstream genes (Dll1, Gadd45g, and p57), a housekeeping gene (GAPDH), and ES cell marker genes (Oct3/4, Sox2, and Nanog) in each single cell. (Bottom right) Control spike RNAs (Lys, Dap, Phe, and Thr for 1000, 100, 20, and 5 copies, restrictively) were quantified by Q-PCR and plotted by the average of all cDNA samples with standard errors. (B) Cells were classified into three groups according to the Hes1 expression levels: Hes1-low (red), Hes1-middle (green) and Hes1-high (blue), shown in A. The average transcript number in each population was plotted with standard errors (normal scale). P-value was calculated by Mann-Whitney U-test. (C) Real-time imaging of Dll1 and Gadd45g expression in ES cells was performed by using Dll1 promoter-driven or Gadd45g promoter-driven ubiquitin-fused luciferase reporter. Bioluminescence images were taken by 10-min exposures for 20 h, and representative plots of luminescence in each single cell were shown.
Hes1-high and Hes1-low ES cells display different responses
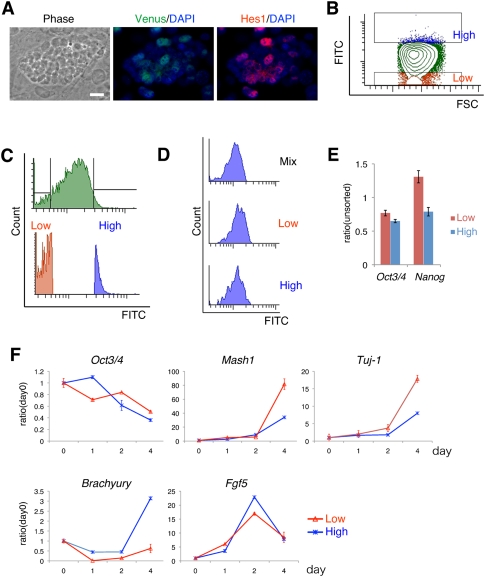
We showed that Hes1 expression oscillates in ES cells and that this Hes1 oscillation leads to oscillatory expression of some of the downstream genes that regulate Notch signaling and the cell cycle. We next asked whether different Hes1 expression levels contribute to heterogeneous responses in the differentiation of ES cells. We reasoned that ES cells could respond differently to the differentiation signal according to the Hes1 expression levels at a given time. To address this issue, the Venus (a YFP variant) fragment was knocked into the Hes1 locus of ES cells so that the Venus-Hes1 fusion protein was expressed under the control of the endogenous Hes1 promoter. These cells were found to express variable levels of the Venus-Hes1 fusion protein, and the Venus fluorescence intensity well correlated with the endogenous Hes1 protein levels (Fig. 4A).
Figure 4.
Hes1-high and Hes1-low ES cells are prone to mesodermal and neural differentiation, respectively. (A) Venus-Hes1 knock-in ES cells on feeder cell layer. Detailed information about these ES cells will be described elsewhere. Phase contrast view and merged images of immunofluorescence using anti-GFP (green) and anti-Hes1 antibodies (red) with DAPI (blue). Bar, 20 μm. (B) Venus-high cells (blue) and Venus-low cells (red) were sorted by flow cytometory. FSC, forward scatter. (C) Histogram of whole cells (green, top panel) and two fractions of Venus-high and Venus-low cells (blue and red, bottom panel). (D) Hes1-high and Hes1-low ES cells returned to the original distribution of variable expression levels within 1 d when they were cultured in ES medium. (E) mRNA levels of Oct3/4 and Nanog were analyzed by Q-PCR of Hes1-high and Hes1-low ES cells just after sorting into the N2B27 medium. Each value was given in the ratio to unsorted cells. (F) Hes1-high and Hes1-low cells were sorted and directly transferred into the N2B27 differentiation medium and cultured for 4 d. Gene expression was quantified by Q-PCR. Each value was given in the ratio to unsorted cells (day 0).
Using fluorescence-activated cell sorting (FACS), Hes1-high and Hes1-low ES cells were isolated and further characterized (Fig. 4B,C). These cells returned to the original distribution of variable expression levels within 1 d (Fig. 4D), indicating that Hes1-high and Hes1-low cells represent different phases of Hes1 oscillations rather than different subpopulations of ES cells. We next transferred Hes1-high and Hes1-low cells to the neural differentiation medium (Ying and Smith 2003) immediately after sorting. Both sorted cells initially expressed Oct3/4 and Nanog at comparable levels (Fig. 4E). Although there was no significant difference in Oct3/4 decrease and Fgf5 increase, a primitive-ectoderm marker (Kunath et al. 2007), associated with differentiation (Fig. 4F), Hes1-low cells expressed a higher level of the neural marker Mash1 and the neuronal marker Tuj-1 than Hes1-high cells at day 4 (Fig. 4F), suggesting that Hes1-low cells more efficiently differentiated into neurons. In contrast, Hes1-high cells expressed a higher level of the mesodermal marker Brachyury than Hes1-low cells at day 4 (Fig. 4F), suggesting that Hes1-high cells more efficiently differentiated into mesodermal cells. These results indicated that different Hes1 expression levels contribute to heterogeneous responses in the cell fate choice. Interestingly, Hes1 oscillations were arrested after induction of differentiation but resumed later in somatic stem/progenitor cells (data not shown).
Inactivation of Hes1 leads to accelerated and more uniform differentiation into the neural fate
To obtain further evidence that Hes1 is involved in heterogeneous responses of ES cells, we compared the differentiation timings and cell fate choice of Hes1-null and wild-type ES cells. We measured the expression kinetics of marker genes during differentiation after removal of both LIF and feeder cells, a condition known to induce all three germ layers (Rathjen and Rathjen 2001). Expression of all differentiation marker genes was activated in wild-type ES cells within 10 d (+/+ and +/− in Supplemental Fig. S5B), but up-regulation of Mash1 was more pronounced in Hes1-null (K9 and K57) cells than in wild-type cells (Supplemental Fig. S5B).
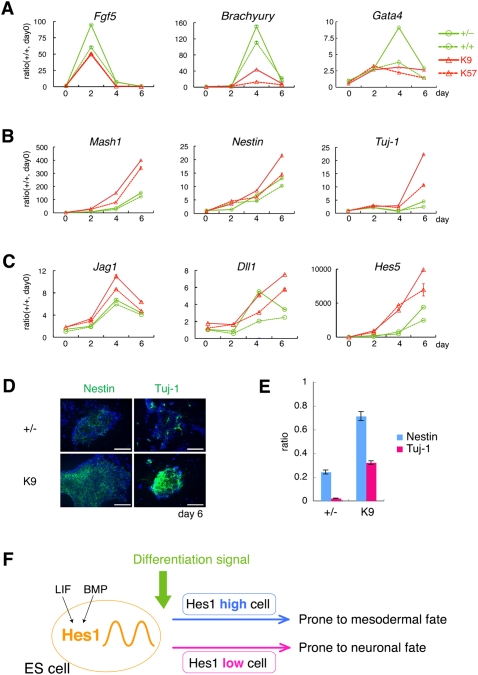
To further characterize the role of Hes1 in ES cells, we focused on neural differentiation because inactivation of Hes1 promotes neuroectoderm differentiation (Supplemental Fig. S5B). Under a neural differentiation condition, Hes1-null ES cells (K9 and K57) expressed higher levels of Mash1, Nestin, Tuj-1 (Fig. 5B), and Notch signaling molecules Jag1, Dll1, and Hes5 (Fig. 5C), which are involved in neural induction (Lowell et al. 2006; Nemir et al. 2006), while they expressed lower levels of other marker genes—Fgf5, Brachyury, and Gata4—than wild-type cells (Fig. 5A). This preference to the neural fate agreed well with that of Hes1-low cells (Fig. 4F). Importantly, while wild-type ES cells only randomly differentiated into neural cells even under the neural differentiation condition, Hes1-null ES cells (K9) more uniformly differentiated into neural cells at earlier timings (Fig. 5D,E). At day 6, almost all Hes1-null ES cells differentiated into neural cells (either Nestin+ neural progenitor cells or Tuj-1+ neurons), whereas only subsets of wild-type ES cells did so (Fig. 5D,E). Thus, inactivation of Hes1 reduces heterogeneous responses and leads to more uniform and preferential differentiation of ES cells into the neural fate.
Figure 5.
Hes1-null ES cells display less heterogeneity in differentiation timing and cell fate choice. Cells were cultured in the N2B27 medium. (A–C) mRNA levels of marker genes in the control (+/− and +/+; green) and Hes1-null ES cells (K9 and K57; red) during differentiation were analyzed by Q-PCR. Each value was given in the ratio to wild-type cells (+/+) at day 0. (D) Hes1-null (K9) and parental control cells (+/−) cultured in N2B27 medium were analyzed at day 6 by immunocytochemistory using anti-Nestin and anti-Tuj-1 antibodies (green) with DAPI (blue). Bars, 100 μm. (E) Quantification of the ratio of Nestin-positive (blue) and Tuj-1-positive cells (red) of parental (+/−) and Hes1-null cells (K9) at day 6. Averages with standard errors were calculated from five fields of confocal images for each cell type. (F) Proposed model. Hes1-high and Hes1-low ES cells tend to differentiate into mesodermal and neural cells, respectively.
Recent reports showed that reversible changes in gene expression occur slowly, over several days, in ES cells and hematopoietic progenitor cells, resulting in different potentials for differentiation, although the mechanism for such slow changes remains to be elucidated (Chambers et al. 2007; Chang et al. 2008; Graf and Stadtfeld 2008; Hayashi et al. 2008; Toyooka et al. 2008). We here showed that Hes1 and its downstream gene expression dynamically change much faster, over several hours, in ES cells. Hes1-high cells tend to adopt the mesodermal fate whereas Hes1-low cells tend to differentiate into neural cells (Fig. 5F). Furthermore, virtually all Hes1-null ES cells differentiated into the neural cells within 6 d under the neural differentiation condition, whereas only subsets of wild-type ES cells did so. Thus, in the absence of Hes1, ES cells display less heterogeneity in both the differentiation timing and fate choice, suggesting that the cyclic gene Hes1 contributes to heterogeneous responses of ES cells. Such rapid cycling of gene expression might be suitable to make multiple cell types even under a single differentiation condition. Hes1 expression also oscillates in neural progenitor cells, but this oscillation seems to contribute to maintenance of the undifferentiated state rather than the diversity in responses (Shimojo et al. 2008). Thus, it is likely that Hes1 oscillations have different functions in different cell types.
Materials and methods
ES cell lines, culture condition, and real-time imaging
The MG1.19 cell line (Gassmann et al. 1995; Takahashi et al. 2003) was used for the quantification of oscillatory expression and Hes1 knockdown, and the TT2 cell line was used for genetic manipulation. Construction of Hes1 reporter cell lines derived from MG1.19 are described in the Supplemental Material. For real-time imaging, cells were cultured on glass-based dishes in ES medium supplemented with 1 mM luciferin. Luminescence images were captured with 20-min or 10-min exposure and by binning of pixels 8 × 8 or 4 × 4 to increase the signal-to-noise ratio. Strong white dots are derived from cosmic rays. Images collected from CCD camera were analyzed with IMAGE-PRO, as described previously (Masamizu et al. 2006).
Hes1 knockdown
Plasmid vector encoding shRNA under the 7SK promoter (Yoshiura et al. 2007) with polyoma ori for replication (Gassmann et al. 1995) and the puromycin-resistant gene was transfected into MG1.19 ES cells by lipofection, and puromycin-resistant cells were selected. The sequences for Hes1 knockdown were 5′-GCCAATTTGCCTTTCTCATCC-3′ for KD1 and 5′-GTAGAGAGCTGTATTAAGTGA-3′ for KD2. Randomly scrambled sequences were used for control.
Microarray analysis
Total RNA was collected from each cell line after removal of feeder cells and subjected to microarray analysis using GeneChip Mouse Genome 430 2.0 Array (Affimetrix). Data were analyzed by using GCOS (Affimetrix) and were normalized to the parental cells by using Gene Spring (Agilent Technologies). The average of two cell lines are shown in Supplemental Table S1. For ChIP–chip analysis, we prepared two Hes1 knockdown cells for negative control and performed ChIP using rabbit anti-Hes1 antibody as described previously (Ishii et al. 2008). ChIP samples were amplified and hybridized to the mouse promoter 1.0R array and mouse tiling 2.0R array (Affimetrix), and analyzed as described previously (Wendt et al. 2008).
Real-time PCR and immunostaining
Quantification by real-time PCR (Q-PCR) and Western blotting was performed as described previously (Yoshiura et al. 2007). For all real-time PCR analysis, a standard curve was drawn for each primer set using mixtures of cDNA samples. Quantified values of RNA were normalized with those of GAPDH. Primers were listed in Supplemental Table S3. Quantitative analyses were performed at least three times and shown with standard errors. For immunocytochemistry, cells were fixed at 4% PFA on ice, blocked with 0.1% Triton-2% skim milk in PBS, and stained with specific antibodies as described in the Supplemental Material.
Q-PCR of single ES cells
Single cells of the wild-type MG1.19 ES cell line were randomly picked up manually by a glass capillary mouse pipet, and cDNA production and amplification were performed as described previously (Kurimoto et al. 2007). After single-cell cDNA amplification, we performed Q-PCR using specific primers as described previously (Kurimoto et al. 2006; Kawaguchi et al. 2008). Primers are listed in Supplemental Table S3. The copy number of each gene was calculated using Coefficient A and B, which were measured for all primer pairs by using plasmid or mouse genome, and normalized with the spike RNA Lys (1000 copies per cell) as described previously (Kurimoto et al. 2006).
Construction of Dll1 and Gadd45g reporters
For Dll1 reporter plasmid, the promoter region of pDll1-Ub1-Luc (Shimojo et al. 2008) was replaced with a 6-kb Dll1 promoter including Hes1-binding sites cloned from mouse genome. For Gadd45g reporter plasmid, 3-kb Gadd45g promoter including Hes1-binding sites and 5′-UTR were placed upstream of and 3′-UTR of Gadd45g was placed downstream from ubiquitin (Ub)-nuclear localization signal (NLS)-luc2. Lentiviral vector construction (Miyoshi 2004) is described in the Supplemental Material.
Cell sorting by flow cytometory
ES cells were dissociated by 0.25% trypsin-EDTA, suspended in ES medium and passed through a cell strainer with a pore size of 35μm. Cells were sorted by a FACSAria Cell Sorter (BD Bioscience). MG1.19 wild-type cells were used for negative control. Single cells were gated by forward scatter (FSC), side scatter (SSC), a pulse width parameter of both FSC and SSC. Sorted cells were collected in normal ES medium or N2B27 medium, and cultured on gelatin-coated dish.
Acknowledgments
We thank Max Gassmann, Shinya Yamanaka, Hiroyuki Miyoshi, and Atsushi Miyawaki for materials; Kazuki Kurimoto, Michinori Saitou, and Siok Lay Tan for methods; Hitoshi Miyachi for technical help; and Kazutoshi Takahashi and Hiroshi Kiyonari for discussion. This work was supported by the Genome Network Project and the Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Mitsubishi Foundation, and the Uehara Memorial Foundation. I.I. was supported by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1823109.
Supplemental material is available at http://www.genesdev.org.
References
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa C, Kaneko K. Theory of robustness of irreversible differentiation in a stem cell system: Chaos hypothesis. J Theor Biol. 2001;209:395–416. doi: 10.1006/jtbi.2001.2264. [DOI] [PubMed] [Google Scholar]
- Gassmann M, Donoho G, Berg P. Maintenance of an extrachromosomal plasmid vector in mouse embryonic stem cells. Proc Natl Acad Sci. 1995;92:1292–1296. doi: 10.1073/pnas.92.5.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, Stadtfeld M. Heterogeneity of embryonic and adult stem cells. Cell Stem Cell. 2008;3:480–483. doi: 10.1016/j.stem.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Lopes SM, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, Kageyama R. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- Ishii A, Kobayashi T, Kageyama R. Requirement of multiple lysine residues for the transcriptional activity and the instability of Hes7. Biochem Biophys Res Commun. 2008;372:142–146. doi: 10.1016/j.bbrc.2008.05.015. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: Repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- Kaneko K. Life: An introduction to complex systems biology. Springer-Verlag; New York: 2006. [Google Scholar]
- Kawaguchi A, Ikawa T, Kasukawa T, Ueda HR, Kurimoto K, Saitou M, Matsuzaki F. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Ono Y, Uno KD, Yamada RG, Ueda HR, Saitou M. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res. 2006;34:e42. doi: 10.1093/nar/gkl050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Saitou M. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protocols. 2007;2:739–752. doi: 10.1038/nprot.2007.79. [DOI] [PubMed] [Google Scholar]
- Lowell S, Benchoua A, Heavey B, Smith AG. Notch promotes neural lineage entry by pluripotent embryonic stem cells. PLoS Biol. 2006;4:805–818. doi: 10.1371/journal.pbio.0040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masamizu Y, Ohtsuka T, Takashima Y, Nagahara H, Takenaka Y, Yoshikawa K, Okamura H, Kageyama R. Real-time imaging of the somite segmentation clock: Revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc Natl Acad Sci. 2006;103:1313–1318. doi: 10.1073/pnas.0508658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H. Gene delivery to hematopoietic stem cells using lentiviral vectors. Methods Mol Biol. 2004;246:429–438. doi: 10.1385/1-59259-650-9:429. [DOI] [PubMed] [Google Scholar]
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Nemir M, Croquelois A, Pedrazzini T, Radtke F. Induction of cardiogenesis in embryonic stem cells via downregulation of Notch1 signaling. Circ Res. 2006;98:1471–1478. doi: 10.1161/01.RES.0000226497.52052.2a. [DOI] [PubMed] [Google Scholar]
- Rathjen J, Rathjen PD. Mouse ES cells: Experimental exploitation of pluripotent differentiation potential. Curr Opin Genet Dev. 2001;11:587–594. doi: 10.1016/s0959-437x(00)00237-9. [DOI] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R. Oscillations in Notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Singh AM, Hamazaki T, Hankowski KE, Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- Smith AG. Embryo-derived stem cells: Of mice and men. Annu Rev Cell Dev Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Mitsui K, Yamanaka S. Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. Nature. 2003;423:541–545. doi: 10.1038/nature01646. [DOI] [PubMed] [Google Scholar]
- Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Vairapandi M, Balliet AG, Hoffman B, Liebermann DA. GADD45b and GADD45g are cdc2/cyclinB1 kinase inhibitors with a role in S and G2/M cell cycle checkpoints induced by genotoxic stress. J Cell Physiol. 2002;192:327–338. doi: 10.1002/jcp.10140. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Ying Q-L, Smith AG. Defined conditions for neural commitment and differentiation. Methods Enzymol. 2003;365:327–341. doi: 10.1016/s0076-6879(03)65023-8. [DOI] [PubMed] [Google Scholar]
- Ying Q-L, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Yoshiura S, Ohtsuka T, Takenaka Y, Nagahara H, Yoshikawa K, Kageyama R. Ultradian oscillations of Stat, Smad, and Hes1 expression in response to serum. Proc Natl Acad Sci. 2007;104:11292–11297. doi: 10.1073/pnas.0701837104. [DOI] [PMC free article] [PubMed] [Google Scholar]