Abstract
The target of rapamycin complex 1 (TORC1) is an essential multiprotein complex conserved from yeast to humans. Under favorable growth conditions, and in the absence of the macrolide rapamycin, TORC1 is active, and influences virtually all aspects of cell growth. Although two direct effectors of yeast TORC1 have been reported (Tap42, a regulator of PP2A phosphatases and Sch9, an AGC family kinase), the signaling pathways that couple TORC1 to its distal effectors were not well understood. To elucidate these pathways we developed and employed a quantitative, label-free mass spectrometry approach. Analyses of the rapamycin-sensitive phosphoproteomes in various genetic backgrounds revealed both documented and novel TORC1 effectors and allowed us to partition phosphorylation events between Tap42 and Sch9. Follow-up detailed characterization shows that Sch9 regulates RNA polymerases I and III, the latter via Maf1, in addition to translation initiation and the expression of ribosomal protein and ribosome biogenesis genes. This demonstrates that Sch9 is a master regulator of protein synthesis.
Keywords: Maf1, Sch9, TOR, phosphoproteomics, rapamycin, ribosome biogenesis
The target of rapamycin complex 1 (TORC1) and TORC2 are large essential multiprotein assemblies structurally and functionally conserved throughout eukaryotic evolution. At their structural core are the TOR proteins, which are large Ser/Thr kinases belonging to the phosphatidylinositol kinase-like kinase family (Wullschleger et al. 2006). It is generally believed that TORC1 is active when sufficient nutrients are present and noxious stressors are absent (DH Kim et al. 2002; Urban et al. 2007). TORC1 also appears to monitor intracellular cues: Treatment with cycloheximide, a translation elongation inhibitor, causes a potent increase in TORC1 activity (Beugnet et al. 2003; Urban et al. 2007).
Rapamycin has been an invaluable tool to study pathways downstream from TORC1. This hydrophobic macrolide easily crosses cell membranes to rapidly and specifically inhibit TORC1. Application of rapamycin to yeast cells demonstrated that TORC1 signals promote cell growth through both the stimulation of anabolic processes, such as protein synthesis and ribosome biogenesis, and the inhibition of catabolic processes, such as autophagy, and stress-responsive transcription programs. For a comprehensive review on TORC1 signaling in yeast, see De Virgilio and Loewith (2006).
The molecular pathways linking TORC1 to its distal readouts are presently only partially characterized. At least two direct effectors downstream from TORC1 have been described: Tap42 and Sch9 (Di Como and Arndt 1996; Jiang and Broach 1999; Urban et al. 2007). Tap42 binds and regulates PP2A and PP2A-like protein phosphatases (Nanahoshi et al. 1998). Tap42 also interacts with and is directly phosphorylated by TORC1 (Jiang and Broach 1999). Genetic evidence clearly shows that Tap42 mediates TORC1 signals to a number of distal readouts. This is based on the observation that tap42-11, a temperature-sensitive allele of TAP42, confers semidominant resistance to rapamycin at the permissive temperature of 25°C (Di Como and Arndt 1996; Duvel et al. 2003). Indeed, in tap42-11 cells, the activation of the transcription factors Gcn4, Gln3, Gat1, and Msn2/4, and the kinase Npr1, normally observed after inhibition of TORC1 with rapamycin, is partially or, in some cases, completely blocked (Schmidt et al. 1998; Cherkasova and Hinnebusch 2003; Duvel et al. 2003; Santhanam et al. 2004).
Sch9 is a Ser/Thr protein kinase of the AGC family. It is directly phosphorylated by TORC1 at its C terminus on at least five residues, and these phosphorylation events are required for catalytic activity (Urban et al. 2007). Replacing some or all of these residues with acidic amino acids (SCH93E and SCH9DE alleles) yields versions of Sch9 that retain activity even in the absence of upstream signals from TORC1 (Urban et al. 2007). Sch9 mediates TORC1 signals to a number of distal readouts: Sch9 blocks the induction of genes required for entry into G0 by directly phosphorylating, and thereby antagonizing, the nuclear accumulation of the Ser/Thr kinase Rim15 (Wanke et al. 2008; Wei et al. 2008); Sch9 is critical for TORC1 to antagonize eIF2α phosphorylation and thus maintain efficient translation initiation (Urban et al. 2007); and Sch9 plays important roles in the regulated expression of RNA polymerase II (Pol II)-dependent genes required for ribosome biogenesis (Jorgensen et al. 2004; Urban et al. 2007). Except for Rim15, the substrates of Sch9 involved in these processes are not known.
Ribosome biogenesis is a highly coordinated process requiring the concerted activity of the three nuclear RNA polymerases (RNA Pol I–III) (Planta 1997; Venema and Tollervey 1999). As it consumes a high amount of cellular energy (Warner 1999), it is not surprising that ribosome biogenesis is tightly coupled to environmental growth conditions. Much of this regulation is mediated by TORC1 (Zaragoza et al. 1998; Mayer and Grummt 2006).
RNA Pol I is dedicated to the transcription of 35S pre-rRNA, which is subsequently processed to 25S, 18S, and 5.8S rRNAs (Venema and Tollervey 1999). Among other models, RNA Pol I was proposed to be regulated by TORC1 via recruitment of the essential initiation factor Rrn3 (Claypool et al. 2004).
RNA Pol II transcription is required for expression of ribosomal protein (RP) genes and ribosome biogenesis (ribi) genes that encode proteins required for nucleolar rRNA processing and assembly of ribosomal subunits (Jorgensen et al. 2004). TORC1 regulates the expression of both RP and ribi genes by controlling the activities of several transcription factors. Some of this regulation is mediated by Sch9 (Jorgensen et al. 2004; Urban et al. 2007).
RNA Pol III transcribes the 5S rRNA, tRNAs, and other stable noncoding RNAs (Geiduschek and Kassavetis 2001). TORC1 regulates RNA Pol III via its conserved repressor, Maf1 (Upadhya et al. 2002). Under favorable growth conditions, Maf1 is highly phosphorylated and is shuttled out of the nucleus. Inactivation of TORC1 results in the rapid dephosphorylation and nuclear accumulation of Maf1 (Oficjalska-Pham et al. 2006; Roberts et al. 2006). TORC1 has been proposed to maintain Maf1 phosphorylation by antagonizing the activity of PP2A family phosphatases, while PKA has been proposed to be the Maf1 kinase (Moir et al. 2006; Oficjalska-Pham et al. 2006). Dephosphorylated Maf1 binds RNA Pol III and, consequently, blocks RNA Pol III transcription via poorly defined mechanisms (Desai et al. 2005; Oficjalska-Pham et al. 2006; Roberts et al. 2006).
Although many distal readouts downstream from TORC1 are known, undoubtedly more remain to be identified. Additionally, the signaling cascades that couple TORC1 to its known readouts remain incompletely understood. To better characterize these pathways we wished to define the TORC1-regulated phosphoproteome. Several liquid chromatography tandem mass spectrometry (LC-MS/MS) approaches have been developed recently to quantify protein phosphorylation (Domon and Aebersold 2006; Olsen et al. 2006). Most prominent among these are protocols based on the differential isotopic labeling of phosphopeptides enriched from protein digests that enables relative quantification between biological samples (Zhou et al. 2001; Olsen et al. 2006). Beyond the restricted number of biological comparisons possible, these methods suffer additional limitations: Currently, only peptide ions identified using MS/MS can be quantified, and the doubling of peptide ion signals in isotopically labeled samples makes the spectra more complex (Mueller et al. 2008). These issues can be largely overcome by label-free LC-MS-based quantitative proteomic strategies, particularly if high mass accuracy MS is used (Rinner et al. 2007). Thereby, every detectable phosphopeptide ion signal can be tracked across multiple LC-MS feature maps, and each tracked peptide can be quantified (Mueller et al. 2007). As a result, the quantification process is applicable across a high number of samples.
Employing these advances, we established a novel, integrated experimental and computational pipeline for the label-free quantification of cellular phosphorylation patterns between a theoretically unlimited number of related samples. We applied this technique to compare the protein phosphorylation patterns in yeast cells upon cycloheximide or rapamycin treatment. This led to the identification of many phosphorylation sites that are presumably directly or indirectly targeted by TORC1. Repeating these assays using cells expressing TORC1-insensitive alleles of TAP42 or SCH9 we were able to accurately partition rapamycin-sensitive phosphoproteins to these two main TORC1 effector branches. These studies led to the observation that Maf1 is directly phosphorylated by Sch9, and that Sch9 regulates both Maf1 localization and binding to RNA Pol III. During the course of these experiments it became apparent that Sch9 also regulates RNA Pol I activity. Altogether this work reveals new effectors downstream from TORC1 and positions Sch9 as a central coordinator of protein synthesis.
Results
Sch9 and Tap42 act in parallel to mediate TORC1 signals
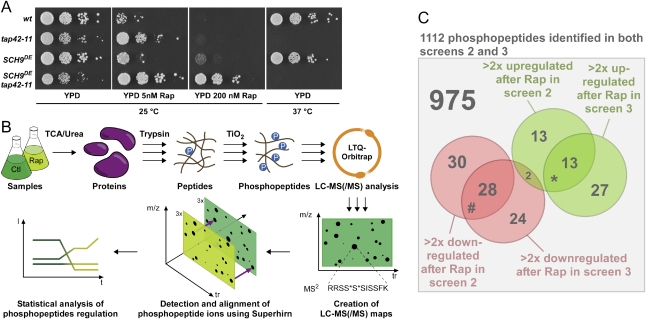
Two direct TORC1 effectors have been reported: Sch9 and Tap42. In order to test their functional relationship, we took advantage of the SCH9DE and tap42-11 alleles that uncouple the encoded protein from upstream regulation by TORC1 (Di Como and Arndt 1996; Urban et al. 2007). In the TB50 yeast background SCH9DE or tap42-11 alone conferred only very slight resistance to rapamycin but, together, the two alleles showed strong synthetic rapamycin resistance with cells still growing, albeit slowly, in the presence of 200 nM rapamycin (∼20 times the minimal lethal concentration for this strain; Fig. 1A). This argues that Sch9 and Tap42 function in parallel downstream from TORC1. The observation that the growth of SCH9DE tap42-11 cells is still slowed by the presence of rapamycin suggests the existence of additional direct TORC1 substrates like Sfp1 (Lempiainen et al. 2009). Beyond Tap42 and Sch9, the molecular events that couple TORC1 to its diverse range of downstream readouts remain largely uncharacterized.
Figure 1.
Label-free quantitative phosphoproteomic screens. (A) TAP42 and SCH9 act in parallel downstream from TORC1. Ten-fold serial dilutions of sch9 tap42 cells complemented with indicated alleles of SCH9 and TAP42 and made prototroph with pAH149 were spotted onto the indicated media and incubated for 2–5 d at 25°C or 37°C. (Rap) Rapamycin. (B) Strategy for label-free quantitative phosphoproteomics. Triple arrows indicate steps performed in triplicate. (C) Venn diagram of phosphopeptides identified in both screens 2 and 3. Subsets of phosphopeptides found to be up-/down-regulated by rapamycin in each screen and their overlaps are shown. The overlap of phosphopeptides predicted to be down-regulated in screen 3 and up-regulated in screen 2 is not statistically significant (P = 0.25). P-values associated with the overlaps enrichment. (*) P < 10−12; (#) P < 10−24.
Label-free quantitative phosphoproteomic screens
To tackle this challenge on a broad scale we employed a novel label-free quantitative phosphoproteomic approach as follows (Fig. 1B): Exponentially dividing cells were either treated with drug or mock-treated with drug vehicle. Subsequently, biochemical reactions were quenched. Proteins were extracted, enzymatically digested, and phosphopeptides were enriched using titanium dioxide-based affinity chromatography (Pinkse et al. 2004; Bodenmiller et al. 2007b). LC-MS feature maps, called phosphorylation patterns, were generated from each sample, and these phosphorylation patterns were aligned using the Superhirn algorithm (Mueller et al. 2007). Analysis of these alignments revealed phosphorylation events that were significantly regulated by a given treatment in a given genetic background.
Our specific strategy employed three different screens. First, we compared protein phosphorylation patterns of exponentially growing wild-type cells treated or mock-treated with cycloheximide, a translation elongation inhibitor that, by unknown mechanisms, hyperactivates TORC1 (Urban et al. 2007). In a second screen, protein phosphorylation patterns of wild-type cells and SCH93E cells (SCH93E is functionally identical to SCH9DE) were compared both with and without rapamycin treatment. For the third screen, protein phosphorylation patterns of wild-type cells and tap42-11 cells were similarly compared both with and without rapamycin treatment.
In total, 30 phosphorylation patterns were generated, containing 2256 distinct phosphopeptides mapping to 751 phosphoproteins (Supplemental Table S3; Supplemental File F1). The numbers of significantly up-regulated and down-regulated phosphopeptides in each screen are summarized in Supplemental Table S4. The summarized list of regulated phosphoproteins identified in both screens 2 and 3 and of proteins that were tested in independent phosphorylation assays is shown as Table 1. The comprehensive lists of regulated phosphopeptides are published as Supplemental Material (Supplemental File F2).
Table 1.
Summary of the phosphoproteomic screens
List of phosphoproteins predicted to be regulated by rapamycin in both screens and/or tested to be rapamycin-sensitive by independent assays. Proteins are sorted according to their reported functions.
a,b(↗) Up-regulated phosphopeptides; (↘) down-regulated phosphopeptides; (—) no regulated phosphopeptides. In parentheses, regulation observed in confirmation experiments [(WB) migration shift assay in Western blot; (PS) Phosphostaining; (↗) up-regulation of phosphorylation; (↘) down-regulation of phosphorylation; (—) no change in phosphorylation].
cPrevious reports showing that the phosphorylation of the indicated proteins is regulated by rapamycin in a similar manner as in the phosphoproteomic screens.
dRps6a and Rps6b have identical protein sequences.
eThe peptides identified for Hrb1 and Gbp2 mapped to regions of the proteins that are identical in sequence and therefore could not be attributed to one or the other.
Quality assessment
To assess the quality of our data we first asked whether our screens were consistent with one another, and compared the phosphorylation patterns generated from independent experiments performed as part of our different screens. The overlap between the rapamycin-sensitive phosphopeptides of screens 2 and 3 was statistically significant (Fig. 1C).
Importantly, rapamycin-sensitive phosphorylation was found in proteins known to function downstream from TORC1, including Sch9, Gln3, Gat1, Atg13, Maf1, Mks1, and Rtg3 (Table 1). Thus, from our repeated independent experiments we observed consistent regulatory events, including those described in the literature. These results show the versatility of label-free quantitation, and confirm that our experimental and biological pipeline is highly reproducible.
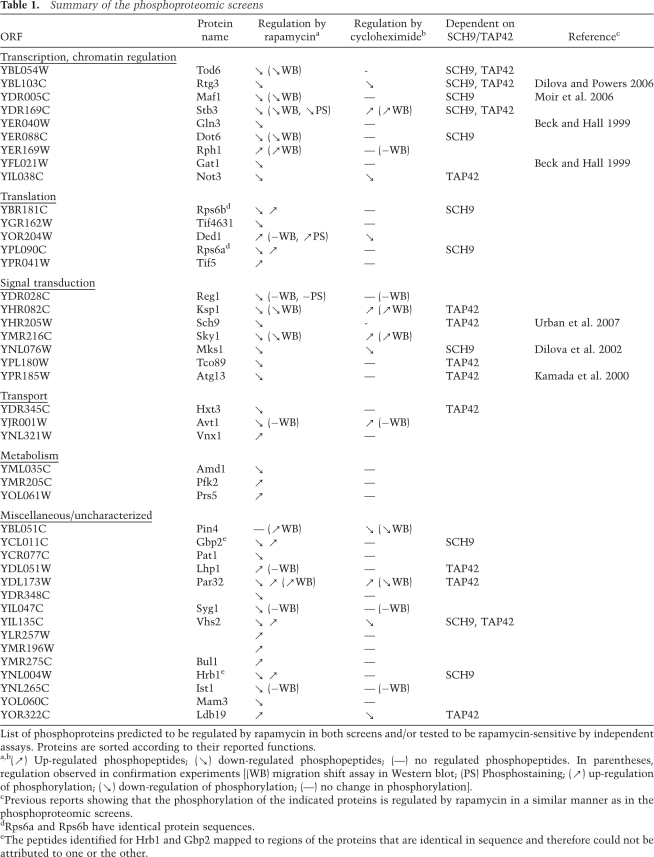
In order to illustrate that our experimental and computational pipeline generates data of high quality, we chose to further validate the regulated phosphorylation sites (Table 1). We selected proteins from all three screens, with preference for those found regulated in two independent screens, showing opposite regulation by cycloheximide and rapamycin and/or displaying a high number of regulated phosphopeptides. HA-epitope-tagged versions of these selected proteins were expressed in yeast, and their migration in SDS-PAGE was monitored by Western blotting in the hope that the regulated phosphorylation sites would generate a migration “shift.”
Dot6, Tod6, Ksp1, Sky1, and Stb3 were predicted to become hypophosphorylated after rapamycin treatment in the phosphoproteomic screens. Consistently, rapamycin treatment resulted in a faster migration of these proteins in SDS-PAGE (Fig. 2A,B), while cycloheximide had the inverse effect. Mutagenesis of some of the predicted phosphoserines in Dot6 and Tod6 to alanines resulted in variants whose migration was no longer altered by rapamycin treatment, indicating our protocol can accurately assign phosphorylation sites (Supplemental Fig. S1A). Ydl173w (renamed Par32 for phosphorylated after rapamycin, 32 kDa) and Rph1 were predicted to become hyperphosphorylated after rapamycin treatment (although some additional sites in Par32 were predicted to be hypophosphorylated after rapamycin treatment) and Pin4 was predicted to be dephosphorylated upon cycloheximide addition. Rapamycin treatment resulted in a slower migration of these proteins, while cycloheximide had the inverse effect. Syg1 and Avt1 were predicted to become hypophosphorylated after rapamycin treatment; however, no migration shift was observed after rapamycin or cycloheximide treatment for these two proteins.
Figure 2.
New TORC1 effectors. (A) Migration shift assays of proteins identified in the phosphoproteomic screens. Yeast cells expressing HA-tagged Ksp1, Sky1, Stb3, Par32, Pin4, Rph1, Syg1, and Avt1 were grown in YPD and treated with rapamycin or cycloheximide (CHX). Proteins were extracted under denaturing conditions and their SDS-PAGE migration was assayed by Western blotting. (B) Migration shift assays of phosphoproteins found to be regulated by SCH9 or TAP42. Reporter plasmids expressing HA-tagged Par32, Tod6, Dot6, or Maf1 were transformed into cells of the indicated genotype. Cells were grown in YPD treated as indicated with rapamycin and assayed as in A. (C) Wild-type cells expressing HA-tagged Ksp1, Sky1, Stb3, Dot6, Tod6, Par32, Pin4, or Rph1 were grown in YPD and treated 15 min with rapamycin or cycloheximide where indicated. Proteins were extracted under native conditions and HA immunoprecipitates were incubated with λ phosphatase in the presence or absence of phosphatase inhibitors.
As shifts in migration could result from other post-translational modifications we wished to confirm that the shifts that we had observed were indeed the result of changes in phosphorylation. To this end we treated selected immunoprecipitated phosphoproteins with λ-phosphatase in the absence or presence of phosphatase inhibitors. In each case, phosphatase treatment converted the protein to its fastest migrating species, confirming that differential phosphorylation was responsible for the observed migration shifts (Fig. 2C). In summary, eight of the 14 proteins we tested in migration shift assays (Fig. 2A,B; data not shown) gave the expected rapamycin-induced mobility shift as predicted in our phosphoproteomic screens. Immunoprecipitation experiments and staining for phosphorylated residues showed that Ded1, which did not show any migration shift, becomes hyperphosphorylated after rapamycin treatment as predicted (Supplemental Fig. S1B). This suggests that at least some of the identified proteins that did not present observable migration shifts are nonetheless rapamycin-sensitive phosphoproteins.
The other objective of our phosphoproteome screens was to partition rapamycin-sensitive phosphorylation events amongst the two known TORC1 substrates, Tap42 and Sch9 (Supplemental Fig. S2; Supplemental Files F1, F2). Again, we used SDS-PAGE migration shift assays to determine if this objective had been met. For this analysis we chose Par32 (rapamycin-induced hyperphosphorylation predicted to be mediated by Tap42), Tod6 (rapamycin-induced hypophosphorylation predicted to be mediated by Sch9 and Tap42), Dot6 (rapamycin-induced hypophosphorylation predicted to be mediated by Sch9), and Maf1 (rapamycin-induced hypophosphorylation predicted to be mediated by Sch9). As predicted, the rapamycin-induced hyperphosphorylation of Par32 was largely blocked in tap42-11 cells (Fig. 2B). Rapamycin-induced dephosphorylation of Tod6 and Dot6 was blocked in SCH9DE cells and was delayed in tap42-11 cells (Tap42 dependence was predicted for Tod6 but not Dot6, which fell just under the applied cutoff for Tap42 regulation) (Fig. 2B). Lastly, Maf1 dephosphorylation showed the predicted Sch9 dependency (Fig. 2B).
Altogether, these control experiments demonstrate that our integrated experimental and computational pipeline allowed us to identify and quantify novel rapamycin-sensitive phosphorylation sites on a system-wide scale and to accurately partition these signaling events downstream from Tap42 and Sch9.
Novel TORC1 targets
We identified >100 novel TORC1-dependent phosphorylation events (Supplemental Table S4; Supplemental File F2), and we wanted to assess whether these phosphoproteins are important for TORC1 to regulate cell growth. Preliminary results suggest they are: 38 of 102 corresponding deletion strains tested gave a moderate to strong rapamycin phenotype (Supplemental Fig. S3; Supplemental Table S5). Thus, from our data we can anticipate novel functions for TORC1 and we can begin to explain how TORC1 signals to previously established readouts:
Previous studies demonstrated a role for TORC1 in starvation-induced developmental transitions (Cutler et al. 2001). TORC1 may mediate these transitions via Ksp1, a protein kinase that is required for filamentous growth in yeast (Bharucha et al. 2008).
Pin4 and Rph1 were shown to become hyperphosphorylated upon DNA damage (EM Kim et al. 2002; Pike et al. 2004). Curiously, we found that rapamycin induced a similar hyperphosphorylation suggesting cross-talk between TORC1 and DNA damage response pathways.
Our data suggest many new links to ribosome biogenesis. We found rapamycin-sensitive Sch9-dependent phosphorylation sites in Stb3 and Dot6/Tod6 (two homologous myb-like HTH transcription factors), and these proteins have been shown recently to function as transcriptional regulators of ribi genes (Liko et al. 2007; Badis et al. 2008; Zhu et al. 2009). Sky1 is a conserved Ser/Thr kinase that phosphorylates pre-mRNA splicing factors of the SR family (Siebel et al. 1999). Given the prevalence of introns in RP genes, this could suggest that TORC1 also plays a more direct role in ribosome biogenesis.
We chose to focus our attention on yet another protein implicated in ribosome biogenesis regulation. Specifically, we were intrigued that Maf1, a conserved repressor of RNA Pol III, was predicted to be regulated by TORC1 via Sch9 (Fig. 2B), as this regulation was thought to occur via PP2A phosphatases (Oficjalska-Pham et al. 2006).
Maf1 is a direct target of Sch9
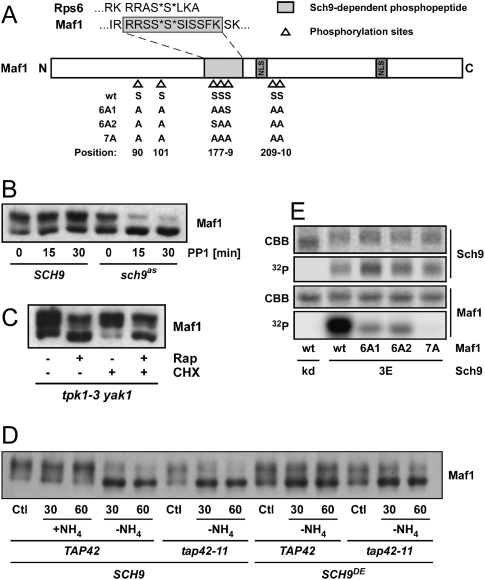
Previously, we demonstrated that Sch9 phosphorylated two serines near the C terminus of Rps6 (Urban et al. 2007). Notably, the sequence surrounding the phosphoserines now identified in Maf1 bears a striking similarity to this S6 sequence (Fig. 3A). Maf1 was reported to be dephosphorylated after rapamycin treatment, but the mechanism by which TORC1 regulates Maf1 phosphorylation was not determined (Oficjalska-Pham et al. 2006; Roberts et al. 2006). Although PP2A family protein phosphatases have been implicated in Maf1 dephosphorylation, we confirmed earlier reports (Willis et al. 2004; Willis and Moir 2007) that Tap42 does not play a role in rapamycin-induced Maf1 dephosphorylation (Fig. 2B).
Figure 3.
TORC1 regulates Maf1 phosphorylation via SCH9, independently of PKA. (A) Maf1 schematic. Maf1 features including phosphorylation sites and NLSs are pictured. Serines predicted to be phosphorylated in the MS data are followed by asterisks as are the Sch9 target residues in the C terminus of Rps6. The various alanine-substituted versions of Maf1 used in the kinase assays shown in D are summarized below the scheme. (B) Sch9 inhibition leads to Maf1 dephosphorylation. (C) TORC1 regulates Maf1 phosphorylation independently of PKA. (B,C) Protein extracts were prepared from cells of the indicated genotype following treatment (15 min in C) with the indicated drugs (PP1: 1NM-PP1). Phosphorylation of Maf1-3HA was determined by SDS-PAGE and Western blotting. (D) Sch9 couples nitrogen-dependent signals to Maf1. Prototroph cells of the indicated genotype were grown to exponential phase in SD, filtered, and resuspended in control (+NH4) or in ammonium-deprived medium (−NH4). Samples were taken at the indicated time points and analyzed for Maf1 phosphorylation by Western blotting. (E) Sch9 phosphorylates 7 serines in Maf1 in vitro. Maf1 mutants, purified from Escherichia coli, were tested as substrates for GST-Sch93E purified from yeast. GST-Sch9kd is a point mutant lacking catalytic activity and was used as a negative control. Reactions were resolved by SDS-PAGE, proteins were stained with Coomassie (CBB) and the dried gel was analyzed for 32P incorporation.
We wished to determine if Sch9 inhibition alone could cause dephosphorylation of Maf1. In our hands, SCH9 deletion mutants grow very slowly but rapidly accumulate suppressive mutations potentially confounding conclusions derived from these strains. We therefore took advantage of a previously described analog-sensitive allele of SCH9 (sch9as) encoding a protein that can be specifically inhibited by the bulky ATP analog 1NM-PP1 (Jorgensen et al. 2004). Addition of 1NM-PP1 to sch9as, but not wild-type cells, resulted in a rapid dephosphorylation of Maf1 (Fig. 3B).
As Maf1 was reported to be regulated by the RAS–PKA pathway and to be phosphorylated in vitro by PKA (Moir et al. 2006), we wished to further explore the relative contributions of Sch9 and PKA to Maf1 phosphorylation. PKA is encoded by three genes in yeast (TPK1, TPK2, and TPK3) and is regulated by glucose in parallel to the TOR pathway (Dechant and Peter 2008). Deletion of the three TPK genes is lethal but can be rescued by the deletion of YAK1 (Garrett and Broach 1989). We found that Maf1 is still phosphorylated in tpk1 tpk2 tpk3 yak1 cells, and this phosphorylation is still sensitive to rapamycin treatment (Fig. 3C). In contrast to the results obtained with sch9as cells, Maf1 phosphorylation was only slightly affected by addition of 1NM-PP1 to tpk1as tpk2as tpk3as cells (Supplemental Fig. S4A,B), suggesting a minor role of PKA under these conditions. Maf1 dephosphorylation during transit through diauxic shift did not show any significant differences in tpk1 tpk2 tpk3 yak1 versus wild-type cells (Supplemental Fig. S4C). In addition, Maf1 dephosphorylation following nitrogen starvation (Fig. 3D) was partially blocked in cells expressing the SCH9DE allele. Altogether, these data suggest that Sch9 is the major kinase upstream of Maf1 in exponentially growing cells.
While this manuscript was in preparation, Lee et al. (2009) reported that Sch9 phosphorylates Maf1; but, in contrast to our results, their work suggested that Sch9 and PKA perform equally important roles in Maf1 regulation. We believe that this discrepancy is due to protocol differences in the handling of yeast cells prior to protein extract. In our studies, whenever possible, TCA was added to growing cultures to quench all enzymatic activity prior to further manipulations. In contrast, Lee et al. (2009) cooled the cells on ice prior to lysis. We found that this cooling step elicits a PKA-dependent rephosphorylation of Maf1 (Supplemental Fig. S8B). This observation suggests a more prominent role of PKA in Maf1 phosphorylation at lower temperatures and explains the overestimated importance in Maf1 regulation at 30°C assigned to PKA by Lee et al. (2009).
Next, we asked if Sch9 could directly phosphorylate Maf1 in vitro. We found that Sch93E, but not a kinase-dead mutant, was able to phosphorylate purified recombinant Maf1 (Fig. 3E). Maf1 contains six motifs fitting the R[R/K]xS consensus, which is often attributed to AGC family kinases (S90, S101, S177, S178, S209, and S210). The phosphopeptide identified in our phosphoproteomic screens (Fig. 3A) contains two overlapping copies of this motif. Unfortunately, neither the Sequest algorithm used to annotate the tandem mass spectra nor manual inspection allowed us to determine with certainty which of the four consecutive serines in this peptide were phosphorylated. To resolve this issue we generated various alanine substituted versions of Maf1 (Fig. 3A) and assayed the ability of Sch9 to phosphorylate these proteins in vitro. First we replaced all six serines fitting the R[R/K]xS consensus with alanine. This version (Maf16A1) was still phosphorylated in vitro, albeit very poorly, by Sch9 (Fig. 3E). Based on the phosphopeptide that we identified in our screen we chose to substitute an additional serine residue (S179) generating a version of Maf1 we refer to as Maf17A. Maf17A was not a substrate for Sch9 in vitro, suggesting that we mapped all of the Maf1 residues phosphorylated by Sch9.
TORC1 regulates RNA Pol III through Sch9
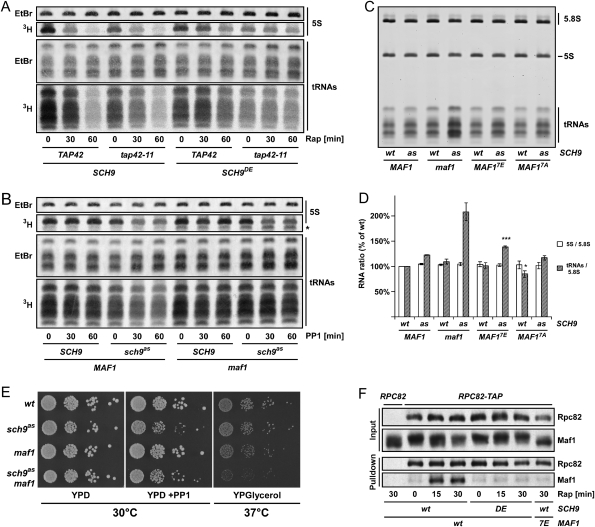
Having established that TORC1, via Sch9, regulates Maf1 phosphorylation we next asked whether this cascade is physiologically important for the regulation of RNA Pol III function. To begin, we confirmed that rapamycin treatment causes a dramatic reduction in tRNA and 5S rRNA synthesis as determined by 3H-uracil pulse labeling (Fig. 4A). This drop in RNA Pol III activity was largely blocked in cells expressing SCH9DE, whereas tap42-11 alone seemed to play little if any role in this process (Fig. 4A). These observations demonstrate that Sch9 influences RNA Pol III activity.
Figure 4.
TORC1 regulates RNA Pol III via Sch9 and Maf1. (A) Rapamycin inhibits 5S rRNA and tRNA synthesis via SCH9. (B) Sch9 inhibition leads to a Maf1-dependent inhibition of tRNA synthesis. (A,B) RNA synthesis in cells of the indicated genotype following the indicated drug treatment was determined by metabolic labeling with 3H-uracil. Total RNA loaded was determined by staining with ethidium bromide (EtBr). Asterisk (*) indicates an unstable RNA species that accumulates in maf1 cells. (C) MAF1 phosphorylation regulates tRNA levels. Cells of the indicated genotype were grown in SC −URA −LEU −TRP −HIS 0.2% Gln 300 nM 1NM-PP1 to log phase (OD600 < 0.8) and total RNA was extracted. Total levels of the 5S and 5.8S rRNA and tRNA were assayed by PAGE and EtBr staining. (D) Quantification of C and two other independent experiments. 5S:5.8S and tRNA:5.8S ratios were calculated and plotted relative to untreated wild-type control. (*) P < 0.05; (***) P < 0.001 versus wild-type control. (E) Genetic interactions between SCH9 and MAF1. Ten-fold dilutions of the indicated strains were spotted and grown on YPD ± 1NM-PP1 (2 d, 30°C) or on YPGlycerol (4 d, 37°C). (F) Sch9 regulates Maf1 association with RNA Pol III. Interaction of Maf1-3HA with RNA Pol III was assessed by Rpc82-TAP pull-downs followed by SDS-PAGE and Western blotting. Relevant genotypes and rapamycin treatments are indicated.
To extend these observations we wished to determine if Sch9 signals to RNA Pol III via Maf1. 1NM-PP1 addition to sch9as cells resulted in a rapid inhibition in tRNA and 5S rRNA synthesis (Fig. 4B). MAF1 deletion abrogated the 1NM-PP1-induced reduction of tRNA synthesis but seemingly did not abrogate the reduction of 5S rRNA synthesis. An explanation for this result is explored below. Pre-tRNAs are rapidly processed in exponentially growing cells, and their abundance can thus be used to infer RNA Pol III activity. Quantitative RT–PCR analyses of the pre-tRNAPro levels were consistent with the 3H-uracil pulse labeling experiments and confirmed the epistasis between SCH9 and MAF1 (Supplemental Fig. S5).
These observations prompted us to determine whether the deregulation of tRNA synthesis observed in our sch9as maf1 mutant could lead to an altered abundance of tRNA relative to other RNA species. To this end we compared 5.8S, 5S, and tRNA levels in exponentially growing cells in the presence or absence of Sch9 and/or Maf1 function. Although total 5S rRNA levels relative to RNA Pol I-derived 5.8S rRNA were unchanged in any of the strains examined, the tRNA:5.8S rRNA ratio was approximately twofold higher in 1NM-PP1-treated sch9as maf1 cells compared with wild-type cells (Fig. 4C,D). An explanation for this result is explored below. To evaluate the importance of Maf1 phosphorylation in RNA Pol III regulation we employed Maf1 mutants where all seven Sch9 phosphorylated residues were replaced with either glutamate (Maf17E) or alanine residues (Maf17A). Inhibition of Sch9 in cells expressing Maf17E leads to tRNA accumulation intermediate to that observed in MAF1 and maf1 cells; cells expressing the Maf17A mutant had reduced basal levels of tRNA (Fig. 4C,D). 3H-uracil pulse labeling experiments following Sch9 inhibition and rapamycin treatment in MAF1, maf1, MAF17A, and MAF17E strains similarly demonstrate that Maf1 phosphosite mutants are partially functional (Supplemental Fig. S6A,B). These results are consistent with Sch9 regulating RNA Pol III via Maf1, but suggest that Maf1 phosphorylation plays only a partial role in this regulation, implying that Sch9 targets additional factors to regulate RNA Pol III albeit in a Maf1-dependent manner.
Additional genetic observations further support our observations. In the presence of 1NM-PP1, sch9as cells grew slowly, dividing every 150 min ± 8 min (compared with 103 min ± 4 min for wild-type cells). This slow growth rate was slightly, but significantly, improved to 135 min ± 5 min by deletion of MAF1, suggesting that reduced RNA pol III activity is one of multiple growth-limiting consequences resulting from loss of Sch9 function. Conversely, we observed a synthetic growth defect when sch9as maf1 cells were grown at 37°C on the nonfermentable carbon source glycerol (Fig. 4E). This result fits with the previously proposed model that accumulation of tRNA in maf1 cells is detrimental to mitochondrial function (Boguta et al. 1997).
Sch9 regulates Maf1 localization and association with the RNA Pol III machinery
How does phosphorylation by Sch9 alter the ability of Maf1 to inhibit RNA Pol III activity? We explored two potential mechanisms by which Sch9 might regulate Maf1: Maf1 localization and the capacity of Maf1 to bind to RNA Pol III. Maf1 was both nuclear and cytoplasmic in our strain background and promptly accumulated in the nucleus upon rapamycin treatment (Supplemental Fig. S7A,B). Sch9DE did not block the rapamycin-induced nuclear accumulation; but, probably due to the hypomorphic nature of these alleles (Jorgensen et al. 2004; Urban et al. 2007), SCH9DE and sch9as cells showed increased basal nuclear accumulation of Maf1 that could be further enhanced in sch9as cells by 1NM-PP1 treatment (Supplemental Fig. S7A–D). From these results, we propose that Maf1 phosphorylation by Sch9 contributes but is not sufficient to prevent Maf1 nuclear localization. This hypothesis is consistent with previous models (Moir et al. 2006) suggesting that TORC1 regulates the two redundant nuclear localization sequences (NLSs) found in Maf1 (Fig. 3A) independently; activity of the N-terminal NLS was proposed to be regulated via phosphorylation (i.e., Sch9-dependent) while the C-terminal NLS is regulated independently of phosphorylation (i.e., Sch9-independent).
In contrast to localization, the ability of Maf1 to directly bind to RNA Pol III appears to be a more important mechanism by which Maf1 phosphorylation regulates RNA Pol III activity (Oficjalska-Pham et al. 2006; Roberts et al. 2006). Therefore we wished to test if phosphorylation by Sch9 alters the ability of Maf1 to interact with the RNA Pol III subunit Rpc82. Using a coprecipitation assay we confirmed that rapamycin treatment strongly increases the association of Maf1 with Rpc82. Importantly, we found that Sch9DE blocked this rapamycin-induced interaction (Fig. 4F). Consistent with this observation, the Maf17E mutant did not associate with Rpc82 upon rapamycin treatment (Fig. 4F), while the Maf17A mutant showed constitutive interaction in untreated wild-type cells (Supplemental Fig. S8A). We were not, however, able to detect an induction of the interaction upon Sch9 inhibition in the TB50 genetic background (Supplemental Fig. S8A), which is likely due to Maf1 rephosphorylation that occurs during cooling prior to nondenaturing protein extraction (Supplemental Fig. S8B). We observed a small induction of the interaction upon Sch9 inhibition in the W303 background that could be further enhanced by concomitant inhibition of PKA (Supplemental Fig. S8C). Taken together, these results demonstrate that Sch9 (and PKA in some conditions) regulates the capacity of Maf1 to bind RNA Pol III.
Sch9 mediates the TORC1 signal to RNA Pol I
As noted above (Fig. 4C,D), we observed that the ratio of tRNA:5.8S rRNA is increased in 1NM-PP1-treated sch9as maf1 cells relative to untreated sch9as maf1 cells. It was not clear why tRNA should be accumulated relative to 5.8S rRNA in these exponentially growing cells; i.e., when rRNA expression should be fully derepressed. We rationalized that this result could be explained if both RNA Pol I and III activities are reduced upon Sch9 inhibition, with MAF1 deletion suppressing only the latter. Thus, tRNA synthesis was not up-regulated per se, but rather RNA Pol I-dependent rRNA synthesis was decreased in these cells. This hypothesis could also explain the apparent failure of MAF1 deletion to rescue the decrease in RNA Pol III-dependent 5S rRNA synthesis resulting from Sch9 inhibition (Fig. 4B): 5S rRNA transcription, we speculate, is rescued by MAF1 deletion. However, because RNA Pol I activity is reduced, 5S rRNA is produced in excess relative to other rRNAs and, consequently, is unstable and rapidly degraded. The shorter unstable RNA species whose levels increased when Sch9 activity was inhibited in maf1 cells (asterisk in Fig. 4B) fits well with this notion. These deductions prompted us to test if TORC1 regulates RNA Pol I via Sch9.
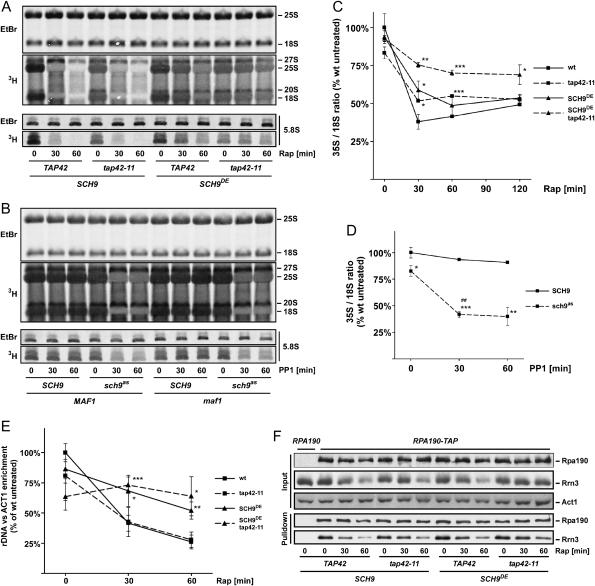
We first confirmed that rapamycin treatment results in a rapid decline in the synthesis of 25S, 18S, and 5.8S rRNAs, as judged by 3H-uracil pulse labeling assays (Fig. 5A). This effect was dramatically blocked in SCH9DE cells but unaltered in tap42-11 cells. Interestingly, 3H-uracil incorporation into these rRNAs was virtually insensitive to rapamycin in SCH9DE tap42-11 cells. Consistently, addition of 1NM-PP1 to sch9as cells resulted in a rapid, Maf1-independent, decrease in the synthesis of 25S, 18S and 5.8S rRNAs (Fig. 5B).
Figure 5.
TORC1 regulates RNA Pol I via Sch9. (A) Rapamycin treatment decreases the processing/expression of RNA pol I-derived rRNA species. (B) Sch9 inhibition decreases the processing/expression of RNA pol I-derived rRNA species. (A,B) Synthesis/processing of rRNA was assayed by metabolic labeling with 3H-uracil. Total RNA loaded was determined by staining with ethidium bromide (EtBr). (C) Rapamycin treatment decreases 35S pre-rRNA synthesis. (D) Sch9 inhibition decreases 35S pre-rRNA synthesis. (C,D) 35S and 18S rRNA levels were determined by primer extension—gels are shown in Figure S9—and their ratios were plotted. Values are means of three independent experiments ± SD. (E) RNA Pol I recruitment at the rDNA locus depends on Sch9. Association of RPA190-13myc with the rDNA locus was determined by ChIP. Values are means of four independent experiments ± SD. Statistical confidences for C–E: (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 versus wild-type control; (##) P < 0.01 versus untreated isogenic control. (F) Sch9 does not regulate Rrn3–RNA Pol I interaction. Association of Rrn3-5HA with Rpa190-TAP was assayed using TAP pull-downs and SDS-PAGE/Western blotting. (A–F) Relevant genotypes and rapamycin/1NM-PP1 treatment times are indicated.
These results clearly demonstrated that Sch9 is indeed important in regulating the synthesis of RNA pol I-derived rRNA species and beg the question: How? Sch9 could regulate RNA pol I transcription or rRNA processing or a combination thereof. To begin to discriminate between these possibilities we first determined the relative amounts of unstable rRNA species to try to gauge the flux from pre-rRNAs to mature end products. Rapamycin treatment and Sch9 inhibition led to decreased rRNA processing as judged by dramatically increased 27S:25S and 20S:18S ratios (Fig. 5A,B). Rapamycin-induced processing defects were largely blocked by the SCH9DE allele. Thus, Sch9 activity promotes rRNA processing.
We also explored if Sch9 might regulate RNA Pol I activity by several approaches. First, we used primer extension assays to determine the relative abundance of the short-lived 35S pre-rRNA. In wild-type cells, 35S levels dropped approximately threefold when assayed after 30 min of rapamycin treatment (Fig. 5C). SCH9DE and tap42-11 cells showed slight, but significant resistance, while SCH9DE tap42-11 cells showed pronounced resistance to this rapamycin-induced drop in 35S pre-rRNA levels. Consistent with these observations, addition of 1NM-PP1 to sch9as cells resulted in an approximately twofold drop in 35S pre-rRNA levels (Fig. 5D). Although it is difficult to separate processing effects from RNA pol I activity, these observations suggest that Sch9 and Tap42 each play a role in regulating RNA Pol I transcription.
Next, we used chromatin immunoprecipitations (ChIPs) to more directly evaluate a role for Sch9 and/or Tap42 in RNA Pol I activity. ChIPs with Rpa190, the catalytic subunit of RNA Pol I, showed that RNA Pol I occupancy at the rDNA locus decreased more than twofold following 30 min of rapamycin treatment (Fig. 5E). Mirroring the 3H-uracil labeling assays, this rapamycin-induced eviction of RNA Pol I from the rDNA locus was strongly blocked in SCH9DE cells and completely blocked in SCH9DE tap42-11 cells. Thus, TORC1 promotes the recruitment of RNA Pol I to the rDNA locus primarily via Sch9.
Previously, Claypool et al. (2004) had proposed that TORC1 influenced RNA Pol I recruitment to the rDNA locus by promoting an interaction between RNA Pol I and Rrn3, an essential initiation factor. We therefore asked if Sch9 and/or Tap42 impinge on this Rrn3–RNA Pol I interaction by pull-down of TAP-tagged Rpa190. This appears not to be the case: The rapamycin-induced dissociation of Rrn3 and RNA Pol I in SCH9DE tap42-11 cells was essentially indistinguishable from the dissociation observed in wild-type cells (Fig. 5F). These data suggest that the dissociation of Rrn3 from RNA Pol I is not the primary mechanism by which TORC1 inhibition causes a reduction in rDNA transcription. We do note, however, that Rrn3 levels drop during rapamycin treatment, and that this is blocked in SCH9DE tap42-11 cells.
Discussion
Sch9 is a central coordinator of protein synthesis
Previous work (Upadhya et al. 2002) had shown that phosphorylation of the RNA Pol III inhibitor Maf1 is regulated downstream from TORC1 and, consistently, a Maf1 phosphopeptide was found to be down-regulated after rapamycin treatment in our phosphoproteomic screens. However, our screens further predicted that Maf1 phosphorylation is regulated by TORC1 via Sch9. Subsequent biochemical studies demonstrated that Maf1 is likely a direct substrate of Sch9 and genetic experiments demonstrated that RNA Pol III down-regulation upon Sch9 inhibition is Maf1-dependent. At the molecular level, we found that Maf1 phosphorylation by Sch9 was important for Maf1 association with RNA Pol III. However, RNA Pol III activity is still sensitive to rapamycin in cells expressing a “phosphomimetic” version of Maf1 (Maf17E). Thus, we believe that Sch9 must target a factor(s) in addition to Maf1 to regulate RNA Pol III activity.
Expectedly, MAF1 deletion, which mostly affects tRNA levels, did not suppress the sch9 growth phenotype. However, it would be interesting in the future to study the impact of Maf1 on other Sch9 phenotypes such as cell size regulation and longevity (Jorgensen et al. 2004; Kaeberlein et al. 2005). In particular, regulation of tRNA and especially initiator tRNAMet levels by Maf1 could affect translation and, via Gcn4, longevity as recently observed (Steffen et al. 2008).
During the course of these studies we found that, in addition to RNA Pol III, Sch9 also regulates the synthesis of RNA Pol I transcripts. Specifically, Sch9 promotes both processing of rRNA species and recruitment of RNA Pol I to the rDNA locus. rRNA processing could be an indirect function of Sch9 as Sch9 controls the expression of many rRNA processing factors in the ribi regulon (Jorgensen et al. 2004). We do not know the mechanism by which Sch9 promotes recruitment of RNA Pol I to the rDNA locus but it does not appear to involve the association of RNA Pol I with its initiation factor Rrn3. Interestingly, RNA Pol I activity was proposed to be determinant for the expression of other ribosomal components (RPs and 5s rRNA), which could suggest that Sch9 regulates RP genes expression indirectly via RNA Pol I activity (Laferte et al. 2006).
Sch9 was thought previously to be the ortholog of mammalian Akt. However, we proposed recently that Sch9 functions more similarly to mammalian ribosomal S6 kinase (S6K1) (Urban et al. 2007). This present study strengthens the functional similarities between Sch9 and S6K1: Both Sch9 and S6K1 have now been shown to regulate the activities of the three nuclear RNA polymerases (Zhang et al. 2005; Woiwode et al. 2008). Recent studies have shown that elevated RNA Pol III transcription is necessary, or in some cases sufficient, for cellular transformation (Johnson et al. 2008; Marshall et al. 2008). Extrapolating from our results, it will be of interest to determine if dysregulation of S6K1 and/or Maf1 likewise contribute to cellular transformation.
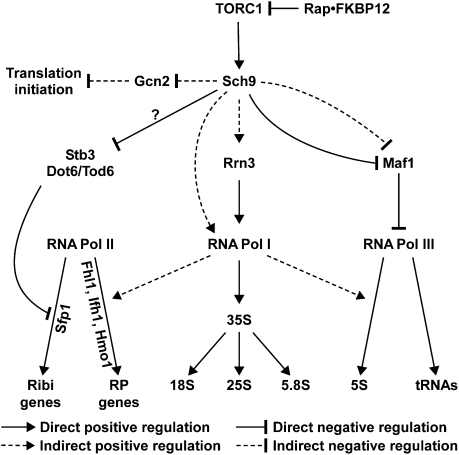
In summary, the present study, together with previous work, demonstrates that Sch9 regulates the activities of all three nuclear RNA polymerases. In addition, we showed previously that Sch9 also regulates translation initiation (Urban et al. 2007). Thus, Sch9 appears to play a central role in coupling environmental cues to the coordinated expression, assembly, and activity of the protein synthesis machinery (Fig. 6). In addition to Maf1, our characterization of the rapamycin-sensitive phosphoproteome also uncovered other TORC1/Sch9 targets implicated in ribosome biogenesis; namely, Stb3 and Dot6/Tod6, which have been shown previously to respectively bind RRPE and PAC elements in ribi gene promoters (Liko et al. 2007; Badis et al. 2008; Zhu et al. 2009). We found Sch9-dependent rapamycin-sensitive phosphorylation sites in Stb3 and Dot6/Tod6, many of which fit the R[R/K]xS consensus motif, suggesting that these proteins could be directly phosphorylated by Sch9. Western blot analyses confirmed that Dot6 and Tod6 are indeed phosphorylated downstream from Sch9 (Fig. 2B), and preliminary experiments suggest that Sch9 signals antagonize the ability of Stb3, Dot6, and Tod6 to inhibit ribi gene expression (Fig. 6; Supplemental Figs. S1A, S3; data not shown).
Figure 6.
Model of TORC1 signaling highlighting the central role that Sch9 plays in coordinating the expression, assembly and activity of the protein synthesis machinery. See the Discussion for details.
Label-free quantitative phosphoproteomic screens
The integrated experimental and computational framework that we present in this work enables relative quantification of phosphorylation patterns. The procedure is technically robust and sensitive, and the data acquired is accurate and, as we illustrate in this manuscript, highly reproducible. The main advantage of our method is that it allows us to quantify thousands of phosphorylation sites between, in principle, an unlimited number of samples or biological states. In addition, no a priori knowledge of phosphopeptide ions is required for quantitation as targeted LC-MS/MS methods can be employed to identify regulated ions of interest (Schmidt et al. 2008). This improves the sensitivity and especially the achievable throughput compared with quantification based on isotope labeling.
Although our data identified many of the rapamycin-sensitive phosphorylation events described in the literature, many were missed. Indeed, high coverage of a given phosphoproteome remains a major challenge in current phosphoproteomics (Bodenmiller et al. 2007b). Nevertheless, this study demonstrates that even with an incomplete coverage of a phosphoproteome our experimental and computational pipeline can elucidate novel and important biological phenomena as demonstrated with the characterization of Maf1 phosphorylation.
Materials and methods
Yeast cultures and assays
Saccharomyces cerevisiae strains and plasmids are described in Supplemental Tables S1 and S2, respectively. Strains were constructed according to standard protocols. SCH9 and TAP42 were deleted in diploid strains and complemented with plasmids encoding wild-type alleles before sporulation. Wild-type alleles were subsequently replaced with mutant alleles in haploids by plasmid shuffling.
Unless specified otherwise, rapamycin was used at 200 nM (from a 1 mM stock solution in 90% ethanol, 10% Tween-20), 1NM-PP1 at 200 nM (from a 1 mM stock in DMSO), and cycloheximide at 25 μg/mL (from a 10 mg/mL stock solution in H2O).
Label-free phosphoproteomics
Cells were grown in SC −LEU 0.2% Gln at 23°C to OD600 0.8 and subjected to the indicated treatments for 30 min. All biochemical activities were then quenched by the addition of trichloroacetic acid and proteins were extracted under denaturing conditions. Three 400-μL aliquots for each condition were processed separately for disulfide bond reduction, cysteine alkylation, trypsin digestion, and phosphopeptide enrichment as described in more detail in the Supplemental Material.
The phosphopeptides were separated by reverse phase chromatography on an Eksigent nano-LC system and were analyzed on a hybrid LTQ-Orbitrap mass spectrometer (Thermo Electron Corporation) interfaced with a nano-electrospray ion source as detailed in the Supplemental Material. The LC-MS/MS data was searched against the SGD yeast protein database as described in the Supplemental Material.
For the detection of the regulated features (peak picking and integration of the area from the LC-MS data, alignment of features over multiple runs) the SuperHirn version 2.0 algorithm (Mueller et al. 2007; Rinner et al. 2007) was used. Of note, as we used a label-free approach, the peptide sequence information from all LC-MS/MS runs was usable to assign the LC-MS features present in the SuperHirn output file, called MasterMap (the relevant parameters used are published as Supplemental Material). The MasterMap was post-processed as follows: Of all of the phosphopeptide features the deconvoluted masses were computed and the peak areas of phosphopeptides present in different charge states were merged. Based on these areas, the statistical significance was computed using a t-test as implemented in the Corra software environment (Brusniak et al. 2008).
Phosphopeptides were considered to be cycloheximide- or rapamycin-sensitive if their abundance relative to untreated cells was altered twofold or more (P-value, <0.05) by cycloheximide or rapamycin treatment, respectively. Rapamycin-sensitive phosphopeptides were considered to be Sch9-dependent after fulfilling two selection criteria. First, rapamycin-induced changes in wild-type cells had to be blunted twofold or more in SCH93E cells. Second, the abundance of a phosphopeptide reduced (or induced) by rapamycin treatment in wild-type cells had to be ≥1.5-fold higher (or lower) in both untreated and rapamycin-treated SCH93E cells compared with rapamycin-treated wild-type cells. The same criteria were used to evaluate Tap42 dependence. All regulated phosphopeptide ions corresponding to the phosphorylation sites of interest were validated.
Data availability
All MS2 information will be made available via the Phosphopep database (http://www.phosphopep.org) (Bodenmiller et al. 2007a). The raw data in the mzXML format can be downloaded from the Peptide Atlas Web page at http://www.peptideatlas.org/repository (Desiere et al. 2006).
Copurification assays
Precultures grown in plasmid-selective synthetic medium were diluted in YPD and grown to OD600 0.7–1.0. One-hundred-milliliter aliquots were treated as described in the text and processed for native protein extraction as described in the Supplemental Material. Protein concentrations were normalized and aliquots were removed to control for input. TAP pull-downs were performed for 2 h at 4°C with 107 magnetic beads (Epoxy M270 Dynabeads, Invitrogen) covalently coated with purified rabbit IgG. Beads were washed three times with lysis buffer before resuspension in SDS-PAGE sample buffer. Samples were analyzed by SDS-PAGE and Western blotting.
3H-uracil labeling
Cells were grown at 25°C in SC −URA to OD600 0.6–0.9 and treated as described in the text. Five-milliliter aliquots were removed, supplemented with 50 μCi 5,6-3H-uracil, and incubated for 20 min. Chase was performed for 20 min with a 100-fold excess cold uracil before total RNA was extracted and analyzed as described in the Supplemental Material.
Primer extension assays
Primer extension assays were performed as described previously (Claypool et al. 2004) with slight modifications. The protocol is detailed in the Supplemental Material.
Recombinant protein expression and purification–kinase assays
Purification of GST-Sch9(3E/kd) from yeast cells was performed as described previously (Urban et al. 2007), except that SCH93E was used as the active SCH9 allele. Maf1 proteins were expressed in bacteria, affinity-purified using the pGEX6P1 system (Invitrogen), and assayed as substrates for GST-SCH93E/kd as described previously (Urban et al. 2007).
Acknowledgments
We thank Tom Sturgill, Claudio DeVirgilio, David Shore, Howard Riezman, and R.A. and R.L. laboratory members for comments on draft manuscripts. We also thank the Functional Genomics Center Zurich for the generous support with MS resources. This project was funded in part by ETH Zurich, the Swiss National Science Foundation under grant no. 31000-10767, with federal (U.S.) funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, under contract no. N01-HV-28179, and by SystemsX.ch, the Swiss initiative for systems biology. Work at the FGCZ was supported by the University Research Priority Program Systems Biology and Functional Genomics of the University of Zurich. R.A. was supported in part by a grant from F. Hoffmann-La Roche Ltd. (Basel, Switzerland) provided to the Competence Center for Systems Physiology and Metabolic Disease. B.B. was the recipient of a fellowship by the Boehringer Ingelheim Fonds. A.H. and A.U. were recipients of fellowships from the Novartis Foundation for Biology and Medicine and the Roche Research Foundation, respectively. R.L. is the recipient of a professorship from the Swiss National Science Foundation (PP00P3-110770/3100A0-108114) and receives generous support from the canton of Genève, the Leenaards Foundation, and the European Research Council (ERC-2007-StG 206173-TOR signaling).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.532109.
Supplemental material is available at http://www.genesdev.org.
References
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Beugnet A, Tee AR, Taylor PM, Proud CG. Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem J. 2003;372:555–566. doi: 10.1042/BJ20021266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharucha N, Ma J, Dobry CJ, Lawson SK, Yang Z, Kumar A. Analysis of the yeast kinome reveals a network of regulated protein localization during filamentous growth. Mol Biol Cell. 2008;19:2708–2717. doi: 10.1091/mbc.E07-11-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Malmstrom J, Gerrits B, Campbell D, Lam H, Schmidt A, Rinner O, Mueller LN, Shannon PT, Pedrioli PG, et al. PhosphoPep–a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol Syst Biol. 2007a;3:139. doi: 10.1038/msb4100182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods. 2007b;4:231–237. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
- Boguta M, Czerska K, Zoladek T. Mutation in a new gene MAF1 affects tRNA suppressor efficiency in Saccharomyces cerevisiae. Gene. 1997;185:291–296. doi: 10.1016/s0378-1119(96)00669-5. [DOI] [PubMed] [Google Scholar]
- Brusniak MY, Bodenmiller B, Campbell D, Cooke K, Eddes J, Garbutt A, Lau H, Letarte S, Mueller LN, Sharma V, et al. Corra: Computational framework and tools for LC-MS discovery and targeted mass spectrometry-based proteomics. BMC Bioinformatics. 2008;9:542. doi: 10.1186/1471-2105-9-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova VA, Hinnebusch AG. Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes & Dev. 2003;17:859–872. doi: 10.1101/gad.1069003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool JA, French SL, Johzuka K, Eliason K, Vu L, Dodd JA, Beyer AL, Nomura M. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol Biol Cell. 2004;15:946–956. doi: 10.1091/mbc.E03-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler NS, Pan X, Heitman J, Cardenas ME. The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol Biol Cell. 2001;12:4103–4113. doi: 10.1091/mbc.12.12.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant R, Peter M. Nutrient signals driving cell growth. Curr Opin Cell Biol. 2008;20:678–687. doi: 10.1016/j.ceb.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Desai N, Lee J, Upadhya R, Chu Y, Moir RD, Willis IM. Two steps in Maf1-dependent repression of transcription by RNA polymerase III. J Biol Chem. 2005;280:6455–6462. doi: 10.1074/jbc.M412375200. [DOI] [PubMed] [Google Scholar]
- Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R. The PeptideAtlas project. Nucleic Acids Res. 2006;34:D655–D658. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C, Loewith R. Cell growth control: Little eukaryotes make big contributions. Oncogene. 2006;25:6392–6415. doi: 10.1038/sj.onc.1209884. [DOI] [PubMed] [Google Scholar]
- Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes & Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- Dilova I, Powers T. Accounting for strain-specific differences during RTG target gene regulation in Saccharomyces cerevisiae. FEMS Yeast Res. 2006;6:112–119. doi: 10.1111/j.1567-1364.2005.00008.x. [DOI] [PubMed] [Google Scholar]
- Dilova I, Chen CY, Powers T. Mks1 in concert with TOR signaling negatively regulates RTG target gene expression in S. cerevisiae. Curr Biol. 2002;12:389–395. doi: 10.1016/s0960-9822(02)00677-2. [DOI] [PubMed] [Google Scholar]
- Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- Duvel K, Santhanam A, Garrett S, Schneper L, Broach JR. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol Cell. 2003;11:1467–1478. doi: 10.1016/s1097-2765(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Garrett S, Broach J. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP-dependent protein kinase. Genes & Dev. 1989;3:1336–1348. doi: 10.1101/gad.3.9.1336. [DOI] [PubMed] [Google Scholar]
- Geiduschek EP, Kassavetis GA. The RNA polymerase III transcription apparatus. J Mol Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Broach JR. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Dubeau L, Johnson DL. Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J Biol Chem. 2008;283:19184–19191. doi: 10.1074/jbc.M802872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes & Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, III, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim EM, Jang YK, Park SD. Phosphorylation of Rph1, a damage-responsive repressor of PHR1 in Saccharomyces cerevisiae, is dependent upon Rad53 kinase. Nucleic Acids Res. 2002;30:643–648. doi: 10.1093/nar/30.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laferte A, Favry E, Sentenac A, Riva M, Carles C, Chedin S. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes & Dev. 2006;20:2030–2040. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Moir RD, Willis IM. Regulation of RNA polymerase III transcription involves SCH9-dependent and SCH9-independent branches of the target of rapamycin (TOR) pathway. J Biol Chem. 2009;284:12604–12608. doi: 10.1074/jbc.C900020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiainen H, Uotila A, Urban J, Dohnal I, Ammerer G, Loewith R, Shore D. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol Cell. 2009;33:704–716. doi: 10.1016/j.molcel.2009.01.034. [DOI] [PubMed] [Google Scholar]
- Liko D, Slattery MG, Heideman W. Stb3 binds to ribosomal RNA processing element motifs that control transcriptional responses to growth in Saccharomyces cerevisiae. J Biol Chem. 2007;282:26623–26628. doi: 10.1074/jbc.M704762200. [DOI] [PubMed] [Google Scholar]
- Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- Moir RD, Lee J, Haeusler RA, Desai N, Engelke DR, Willis IM. Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc Natl Acad Sci. 2006;103:15044–15049. doi: 10.1073/pnas.0607129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LN, Rinner O, Schmidt A, Letarte S, Bodenmiller B, Brusniak MY, Vitek O, Aebersold R, Muller M. SuperHirn—a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics. 2007;7:3470–3480. doi: 10.1002/pmic.200700057. [DOI] [PubMed] [Google Scholar]
- Mueller LN, Brusniak MY, Mani DR, Aebersold R. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J Proteome Res. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- Nanahoshi M, Nishiuma T, Tsujishita Y, Hara K, Inui S, Sakaguchi N, Yonezawa K. Regulation of protein phosphatase 2A catalytic activity by α4 protein and its yeast homolog Tap42. Biochem Biophys Res Commun. 1998;251:520–526. doi: 10.1006/bbrc.1998.9493. [DOI] [PubMed] [Google Scholar]
- Oficjalska-Pham D, Harismendy O, Smagowicz WJ, Gonzalez de Peredo A, Boguta M, Sentenac A, Lefebvre O. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol Cell. 2006;22:623–632. doi: 10.1016/j.molcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Pike BL, Yongkiettrakul S, Tsai MD, Heierhorst J. Mdt1, a novel Rad53 FHA1 domain-interacting protein, modulates DNA damage tolerance and G(2)/M cell cycle progression in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:2779–2788. doi: 10.1128/MCB.24.7.2779-2788.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkse MWH, Uitto PM, Hilhorst MJ, Ooms B, Heck AJR. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-nanoLC-ESI-MS/MS and titanium oxide precolumns. Anal Chem. 2004;76:3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- Planta RJ. Regulation of ribosome synthesis in yeast. Yeast. 1997;13:1505–1518. doi: 10.1002/(SICI)1097-0061(199712)13:16<1505::AID-YEA229>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Rinner O, Mueller LN, Hubalek M, Muller M, Gstaiger M, Aebersold R. An integrated mass spectrometric and computational framework for the analysis of protein interaction networks. Nat Biotechnol. 2007;25:345–352. doi: 10.1038/nbt1289. [DOI] [PubMed] [Google Scholar]
- Roberts DN, Wilson B, Huff JT, Stewart AJ, Cairns BR. Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol Cell. 2006;22:633–644. doi: 10.1016/j.molcel.2006.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanam A, Hartley A, Duvel K, Broach JR, Garrett S. PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryot Cell. 2004;3:1261–1271. doi: 10.1128/EC.3.5.1261-1271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Beck T, Koller A, Kunz J, Hall MN. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Gehlenborg N, Bodenmiller B, Mueller LN, Campbell D, Mueller M, Aebersold R, Domon B. An integrated, directed mass spectrometric approach for in-depth characterization of complex peptide mixtures. Mol Cell Proteomics. 2008;7:2138–2150. doi: 10.1074/mcp.M700498-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel CW, Feng L, Guthrie C, Fu XD. Conservation in budding yeast of a kinase specific for SR splicing factors. Proc Natl Acad Sci. 1999;96:5440–5445. doi: 10.1073/pnas.96.10.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya R, Lee J, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell. 2002;10:1489–1494. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- Wanke V, Cameroni E, Uotila A, Piccolis M, Urban J, Loewith R, De Virgilio C. Caffeine extends yeast lifespan by targeting TORC1. Mol Microbiol. 2008;69:277–285. doi: 10.1111/j.1365-2958.2008.06292.x. [DOI] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Wei M, Fabrizio P, Hu J, Ge H, Cheng C, Li L, Longo VD. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008;4:e13. doi: 10.1371/journal.pgen.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis IM, Moir RD. Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem Sci. 2007;32:51–53. doi: 10.1016/j.tibs.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Willis IM, Desai N, Upadhya R. Signaling repression of transcription by RNA polymerase III in yeast. Prog Nucleic Acid Res Mol Biol. 2004;77:323–353. doi: 10.1016/S0079-6603(04)77009-9. [DOI] [PubMed] [Google Scholar]
- Woiwode A, Johnson SA, Zhong S, Zhang C, Roeder RG, Teichmann M, Johnson DL. PTEN represses RNA polymerase III-dependent transcription by targeting the TFIIIB complex. Mol Cell Biol. 2008;28:4204–4214. doi: 10.1128/MCB.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Zaragoza D, Ghavidel A, Heitman J, Schultz MC. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol Cell Biol. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Comai L, Johnson DL. PTEN represses RNA polymerase I transcription by disrupting the SL1 complex. Mol Cell Biol. 2005;25:6899–6911. doi: 10.1128/MCB.25.16.6899-6911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HL, Watts JD, Aebersold R. A systematic approach to the analysis of protein phosphorylation. Nat Biotechnol. 2001;19:375–378. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- Zhu C, Byers K, McCord R, Shi Z, Berger M, Newburger D, Saulrieta K, Smith Z, Shah M, Radhakrishnan M, et al. High-resolution DNA binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All MS2 information will be made available via the Phosphopep database (http://www.phosphopep.org) (Bodenmiller et al. 2007a). The raw data in the mzXML format can be downloaded from the Peptide Atlas Web page at http://www.peptideatlas.org/repository (Desiere et al. 2006).