Abstract
A central question in Hedgehog (Hh) signaling is how evolutionarily conserved components of the pathway might use the primary cilium in mammals but not fly. We focus on Suppressor of fused (Sufu), a major Hh regulator in mammals, and reveal that Sufu controls protein levels of full-length Gli transcription factors, thus affecting the production of Gli activators and repressors essential for graded Hh responses. Surprisingly, despite ciliary localization of most Hh pathway components, regulation of Gli protein levels by Sufu is cilium-independent. We propose that Sufu-dependent processes in Hh signaling are evolutionarily conserved. Consistent with this, Sufu regulates Gli protein levels by antagonizing the activity of Spop, a conserved Gli-degrading factor. Furthermore, addition of zebrafish or fly Sufu restores Gli protein function in Sufu-deficient mammalian cells. In contrast, fly Smo is unable to translocate to the primary cilium and activate the mammalian Hh pathway. We also uncover a novel positive role of Sufu in regulating Hh signaling, resulting from its control of both Gli activator and repressor function. Taken together, these studies delineate important aspects of cilium-dependent and cilium-independent Hh signal transduction and provide significant mechanistic insight into Hh signaling in diverse species.
Keywords: Hedgehog, signal transduction, evolution, primary cilium, Sufu, Gli
Elucidating the molecular mechanism of Hedgehog (Hh) signal transduction is critical for understanding normal embryonic patterning and pathological conditions such as human congenital anomalies and cancers arising from misregulated Hh signaling. The main components of the Hh signaling pathway appear to be conserved between invertebrates and vertebrates; however, recent studies indicate that several aspects of Hh signaling have diverged (Lum and Beachy 2004; Ogden et al. 2004; Hooper and Scott 2005; Nieuwenhuis and Hui 2005; Huangfu and Anderson 2006; Ingham and Placzek 2006; Jia and Jiang 2006; Aikin et al. 2008; Dessaud et al. 2008; Varjosalo and Taipale 2008). In particular, the primary cilium, an ancient and evolutionarily conserved organelle, is essential for mammalian Hh signal transduction but dispensable for Hh signaling in Drosophila. The extent of Hh pathway divergence in different organisms is a major unresolved issue. Delineating cilium-dependent and cilium-independent processes of Hh signal transduction is crucial to understanding how the mammalian Hh pathway has evolved. Insight into this question will not only advance our mechanistic understanding of Hh signaling but also serve as a paradigm for investigating the evolution of signal transduction pathways.
Most vertebrate cells possess a nonmotile primary cilium (Huangfu and Anderson 2005). Primary cilia contain a long microtubular axoneme that extends from the basal body and is surrounded by an external membrane that is continuous with the plasma membrane (Rosenbaum and Witman 2002). Assembly and maintenance of the primary cilium are mediated by a process called intraflagellar transport (IFT), which involves bidirectional movement of IFT particles powered by anterograde kinesin (Kif3a, Kif3b, and Kif3c) and retrograde dynein motors (Rosenbaum and Witman 2002). Mouse ethylnitrosourea (ENU) mutants in genes encoding IFT proteins, or the respective motors, have defective Hh signaling (Huangfu et al. 2003), providing strong evidence that the primary cilium plays a key role in mammalian Hh signaling. Moreover, most core mammalian Hh pathway components including Smoothened (Smo), Patched1 (Ptch1), Suppressor of fused (Sufu), and Gli1, Gli2, and Gli3 localize to the primary cilium (Michaud and Yoder 2006; Eggenschwiler and Anderson 2007). Furthermore, production of both Gli protein activators and repressors appears to be affected in IFT mutant mice (Huangfu and Anderson 2005; Liu et al. 2005). These results establish a strong connection between primary cilia and mammalian Hh signaling. Interestingly, primary cilia are only present in sensory neurons in Drosophila, and mutations in either the anterograde kinesin motor or components of the IFT particles cause neuronal phenotypes in fly but do not seem to disrupt Hh signaling (Ray et al. 1999; Han et al. 2003), highlighting a unique role of the cilium in mammalian Hh signaling.
Binding of Hh to its 12-pass transmembrane receptor Ptch1 triggers ciliary localization of Smo, a seven-pass transmembrane protein (Corbit et al. 2005), with concomitant loss of Ptch1 on the cilium (Rohatgi et al. 2007), leading to Smo activation. Otherwise, Ptch1 inhibits Smo activity via unknown mechanisms. In the absence of the Hh ligand, Gli2 and Gli3 transcription factors, homologs of Drosophila Cubitus interruptus (Ci), undergo limited proteolysis in which the C-terminal activator domains are cleaved, thus generating transcriptional repressors (Aza-Blanc et al. 1997; B Wang et al. 2000; Pan et al. 2006). The Hh signal transduction cascade represses Ci/Gli2/3 proteolysis, promoting the generation of transcriptional activators that presumably are derived from full-length Ci/Gli proteins. In contrast, the Gli1 protein lacks an N-terminal repressor domain and functions solely as a transcriptional activator. The delicate balance between Gli activators and repressors is believed to be the major determinant of graded Hh responses. It is widely speculated that many key events of cytoplasmic Hh signal transduction downstream from Smo occur on the cilium, the absence of which is known to perturb the ratio of full-length Gli (the putative activators) and Gli repressors (Liu et al. 2005). However, functional studies to demonstrate how the primary cilium controls Hh signaling are largely lacking. Whether the production of Gli activators and repressors occurs on the primary cilium has never been demonstrated. In fact, even whether Smo is activated on the cilium, and how this occurs, is unclear. It is also unknown if Gli protein levels or activities can be regulated by processes independent of the primary cilium.
Studies on Hh signal transduction downstream from Smo in a number of model organisms suggest a modified pathway design (Huangfu and Anderson 2006). In particular, efforts to understand the roles of Fused (Fu), a putative serine-threonine kinase, and Sufu, a novel PEST domain protein, in Hh signaling shed light on pathway divergence. Sufu is dispensable for viability in fly and was identified as an extragenic suppressor of fu mutations (Preat 1992; Preat et al. 1993). In fly, Fu functions in concert with the atypical kinesin Costal-2 (Cos2) (Robbins et al. 1997; Sisson et al. 1997), Ci (Orenic et al. 1990), and Sufu in transducing the Hh signal. In the absence of extracellular Hh ligand, Cos2 functions as a scaffold in a multimolecular protein complex comprised of Fu, Ci, Cos2, and a small amount of Sufu. Cos2 recruits the kinases PKA, GSK3, and CK1 to promote Ci phosphorylation (Zhang et al. 2005), targeting Ci for limited proteolysis via the Slimb/β-TrCP-Cul1 E3 ubiquitin ligase. This cleavage event removes its C-terminal activation domain from Ci and produces a transcriptional repressor capable of inhibiting Hh target gene expression. Hh signal transduction leads to dissociation of the kinases from the Cos2-scaffolded complex and subsequent inhibition of Ci proteolysis. Instead, Cos2, Fu, and possibly Ci are recruited to Smo at the plasma membrane through direct associations between Cos-2 and the Smo C terminus (Stegman et al. 2000, 2004; Jia et al. 2003; Lum et al. 2003; Ogden et al. 2003; Ruel et al. 2003). In this process, Ci is converted into an activator through unknown mechanisms in order to activate Hh target genes (Ohlmeyer and Kalderon 1998). Fu and Sufu were proposed to exert opposite effects in controlling the processing, activity, and shuttling of Ci between the nucleus and the cytoplasm (Methot and Basler 2000; Wang and Holmgren 2000; G Wang et al. 2000; Lefers et al. 2001). Sufu is believed to tether Ci in the cytoplasm and repress Hh signaling, a function that could be antagonized by Fu.
Sufu is a major regulator of mammalian Hh signaling (Cooper et al. 2005; Svard et al. 2006), but the molecular mechanisms by which Sufu controls vertebrate Hh signaling are unknown. Sufu-deficient mice die ∼9.5 d post-coitus (dpc) with a ventralized neural tube due to global up-regulation of Hh signaling. This is in stark contrast to the lack of overt phenotypes in sufu mutant flies (Preat 1992). Interestingly, fly fused (fu) is essential for Hh signaling, and fu defects are suppressed by sufu inactivation (Preat 1992). In contrast, Fu-deficient mice are viable and display no embryonic phenotypes (Chen et al. 2005; Merchant et al. 2005). Hh signaling in zebrafish appears to represent a transitional state since morpholino-mediated knockdown of fu-produced phenotypes consistent with loss of Hh activity, whereas loss of sufu resulted in mild elevation of Hh signaling (Wolff et al. 2003; Koudijs et al. 2005). These results indicate that diverse species use a modified regulatory circuitry and/or distinct cellular microenvironments for Hh signaling. Given the vital role of Sufu in controlling mammalian Hh signaling, as well as the fact that it is physically present on the cilium, studies of Sufu provide a unique tool to clarify mechanisms of Hh signal transduction and their possible divergence during evolution.
Results
Hh pathway components localize to the primary cilium in a dynamic manner
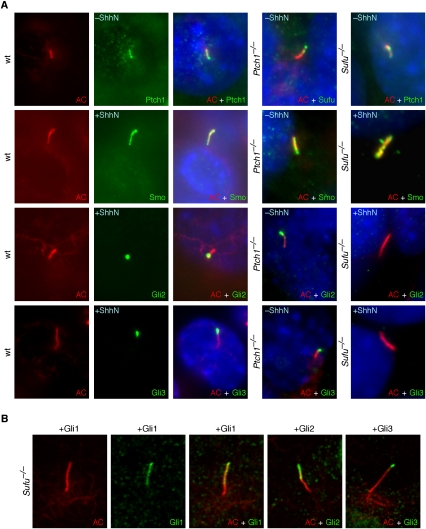
Most mammalian Hh pathway components, including Smo, Ptch1, Sufu, and Gli1, Gli2, and Gli3, localize to the primary cilium (Fig. 1A; Michaud and Yoder 2006; Eggenschwiler and Anderson 2007). Smo translocates to the primary cilium upon Hh pathway activation, with concomitant loss of Ptch1 from the cilium. While ciliary localization of endogenous Smo and Ptch1 has been extensively documented (Corbit et al. 2005; Rohatgi et al. 2007), endogenous Gli1, Gli2, and Gli3 and Sufu proteins on the cilium have not been examined thoroughly (Haycraft et al. 2005). We generated antibodies against Gli2 and Gli3 that are capable of detecting endogenous Gli proteins (Supplemental Fig. S1). Gli2 and Gli3 are present at the primary cilium at low levels in the absence of Hh ligand, and their levels on the cilium dramatically increase with the addition of exogenous Shh ligand (Fig. 1A; Supplemental Fig. S2A). This dynamic change of endogenous Gli2 and Gli3 levels on the cilium in response to Hh signaling differs from results using overexpressed Gli2 and Gli3 proteins, which can be detected on the cilium regardless of the state of Hh pathway activation (Haycraft et al. 2005; data not shown). In contrast, ciliary localization and intensity of Sufu are unaffected upon Hh pathway activation (Supplemental Fig. S2A). In some cells, Gli2, Gli3, and Sufu immunofluorescence is detected primarily at the tip of the primary cilium, while in other cells, it is distributed along the entire cilium, perhaps reflecting trafficking of these proteins on the cilium (Fig. 1A; Supplemental Fig. S3; data not shown). Gli2 and Gli3 localize to the primary cilium even in the absence of Hh stimulation in Ptch1−/− mouse embryonic fibroblasts (MEFs) (Fig. 1A), in which the Hh pathway is maximally activated in a ligand-independent manner. Together, these localization studies suggest that the primary cilium could constitute a critical site for Hh signaling, but functional studies are required for clarifying how the cilium regulates Hh signaling.
Figure 1.
Endogenous Hh pathway components display dynamic patterns of ciliary localization in response to Hh signaling, while overexpressed Gli proteins localize to the primary cilium in the absence of Sufu. (A) Immunofluorescence of wild-type (wt), Sufu−/−, and Ptch1−/− MEFs using antibodies against acetylated tubulin (AC) (labeling primary cilia, red) and various endogenous Hh pathway components including Smo, Ptch1, Gli2, Gli3 and Sufu (green). Smo translocates to the primary cilium upon Hh pathway activation, which is associated with concomitant loss of Ptch1 from the cilium. Low levels of Gli2 and Gli3 can be detected on the primary cilium by immunofluorescence without Hh ligand stimulation (data not shown), and their levels on the cilium significantly increase upon exposure to exogenous Shh ligand. In contrast, ciliary localization and intensity of Sufu are unchanged upon Hh pathway activation (data not shown). Gli2, Gli3, and Sufu immunofluorescence is detected primarily at the end of the primary cilium in some cells, while in others it decorates the entire cilium, perhaps due to dynamic ciliary trafficking of these proteins. Gli2 and Gli3 localize to the primary cilium in the absence of Hh stimulation in Ptch1−/− MEFs, in which the Hh pathway is maximally activated. Ciliary localization of Gli2 and Gli3 is completely abolished in Sufu−/− MEFs. (B) Immunofluorescence of Sufu−/− MEFs expressing mouse Flag-tagged Gli1, Gli2, or Gli3 using antibodies against acetylated tubulin (red) and Flag antibodies against Gli1, Gli2, or Gli3 (green). Overexpressed Gli proteins localize to the primary cilium in the absence of Sufu, and ciliary localization is unaffected by Hh stimulation. We speculate that the amount of overexpressed Gli proteins exceeds the capacity of Gli-degradation machinery in the absence of Sufu (see Fig. 2A).
Gli2 and Gli3 protein levels are greatly reduced in the absence of Sufu
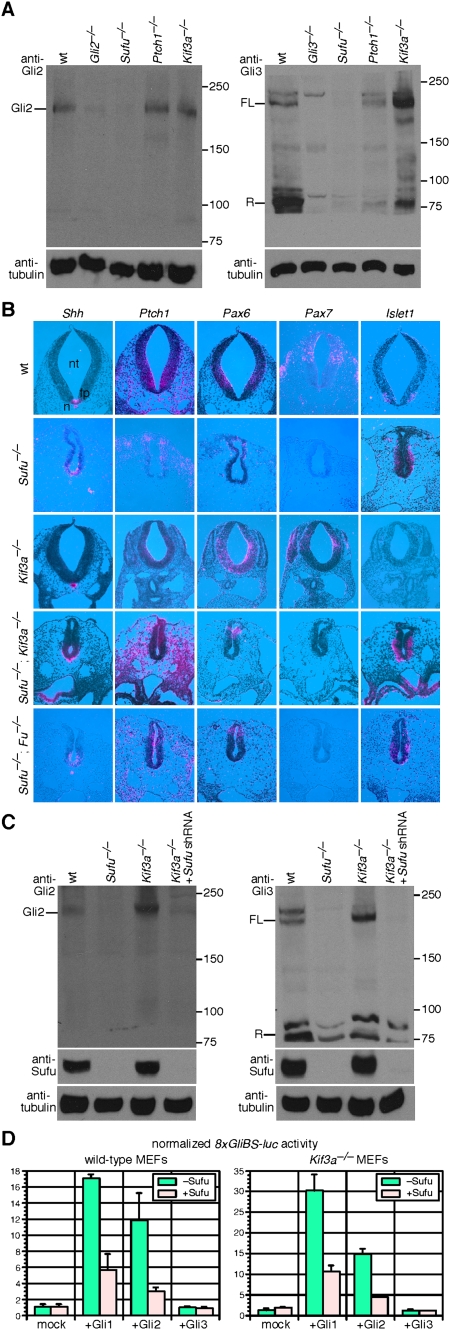
To investigate how Sufu regulates Hh signaling, we examined Gli protein levels in Sufu−/− embryos and MEFs. We found that endogenous Gli2 and Gli3 proteins (both full-length and repressor) are barely detectable on Western blots in the absence of Sufu (Fig. 2A; Supplemental Fig, S4; Supplemental Table S1). Consistent with this finding, ciliary localization of Gli2 and Gli3 is completely abolished in Sufu−/− MEFs (Fig. 1A). This is not due to reduced mRNA levels of Gli2 and Gli3, which are comparable between wild-type and Sufu−/− MEFs (Supplemental Fig. S5). These results reveal a major unappreciated role of mammalian Sufu in controlling full-length Gli protein levels and consequently the production of Gli activators and repressors.
Figure 2.
Mouse Sufu regulates Gli protein levels independent of the primary cilium. (A) Western blots of lysates derived from wild-type (wt), Gli2−/−, Gli3−/−, Sufu−/−; Ptch1−/− and Kif3a−/− MEFs probed with anti-Gli2 and anti-Gli3 antibodies. Endogenous Gli2 and Gli3 protein levels (including full-length Gli3 and Gli3 repressor) are greatly reduced in the absence of Sufu. Both full-length Gli3 and Gli3 repressor can be detected in Ptch1−/− albeit the Gli3 repressor level is reduced. The ratio of full-length Gli3 to Gli3 repressor is altered in Kif3a−/− MEFs as reported previously (Liu et al. 2005). Gli2 processing is known to be extremely inefficient, and the Gli2 repressor form cannot be readily detected without additional enrichment steps using specific Gli-binding oligonucleotides (Pan et al. 2006). We also cannot accurately assess the full-length to repressor ratios for Gli2 and Gli3 in Sufu mutants. Tubulin was used as the loading control, and numbers on the right indicate locations of protein size standards. (FL) Full-length; (R) repressor. (B) Isotopic in situ hybridization using 33P-UTP-labeled ribo-probes (pink) on paraffin sections of wild-type (wt), Sufu−/−, Kif3a−/−, Sufu−/−; Kif3a−/−, and Sufu−/−; Fu−/− mouse embryos at 9.5 dpc. Loss of Sufu resulted in global up-regulation of Hh signaling and ventralization of the neural tube. Shh, whose expression is restricted to the notochord and floor plate in wild type, is extended dorsally in the absence of Sufu. Similarly, Hh target genes such as Patched 1 (Ptch1) are expanded dorsally, suggesting ventralization of the neural tube. The expression domains of neuronal progenitor markers (Class I genes, including Pax7 and Pax6, repressed by Hh signaling and Class II genes, including Nkx6.1, Nkx2.2, and Foxa2, activated by Hh signaling) are shifted. For instance, Pax7, the dorsal-most marker, is not expressed, and Pax6 expression is confined to the dorsal neural tube of Sufu−/− embryos. Dorsal expansion of Nkx6.1, Nkx2.2, and Foxa2 was also observed in the absence of Sufu (data not shown). Similarly, the expression domains of markers for differentiated interneurons and motoneurons are changed. For instance, Sufu−/− neural tube displayed dorsal expansion of Islet1 and Oligo2 (data not shown), which label motoneurons. By comparison, marker analysis revealed a partially dorsalized Kif3a−/− neural tube. The neural tube defects in Sufu−/−; Kif3a−/− or Sufu−/−; Fu−/− embryos resemble those in Sufu−/− mutants. (n) Notochord; (fp) floor plate; (nt) neural tube. (C) Western blots of lysates derived from wild-type, Sufu−/−, Kif3a−/− MEFs and Kif3a−/− MEFs expressing Sufu shRNA probed with anti-Gli2 and anti-Gli3 antibodies. Efficient Sufu knockdown in Kif3a−/− MEFs was verified by anti-Sufu antibodies. Gli2 and Gli3 protein levels are reduced in Kif3a−/− MEFs expressing Sufu shRNA to the same extent as in Sufu−/− MEFs. (D) Hh reporter assays using the 8xGliBS-luc reporter in wild-type and Kif3a−/− MEFs. Expression of Gli1 or Gli2 (but not Gli3) activates the Hh reporter, and Hh pathway activation is repressed when Sufu is coexpressed. Gli2 is known to activate Hh reporters less efficiently than Gli1 (Gerber et al. 2007). Loss of the primary cilium in Kif3a−/− MEFs does not impair Sufu's ability in repressing Gli-mediated Hh pathway activation. Error bars are standard deviation (s.d.).
Sufu functions independently of Fu and the primary cilium in controlling Gli protein levels
Fu-deficient mouse embryos display no Hh phenotype (Chen et al. 2005; Merchant et al. 2005). To investigate whether the antagonistic genetic interaction of Fu and Sufu is conserved in mammals, we asked whether loss of Fu can rescue Hh defects in Sufu mutants. We observed that Sufu phenotypes cannot be rescued by loss of Fu in mice and Sufu−/−; Fu−/− mutants phenocopy Sufu−/− embryos (Fig. 2B). This suggests a modified regulatory circuitry in transducing the mammalian Hh signal downstream from Smo, and Fu is dispensable in this process.
The observation that Sufu, Gli1, Gli2, and Gli3 localize to the primary cilium, coupled with biochemical data showing that Sufu physically interacts with all three Gli proteins, raised the interesting possibility that Sufu regulates Gli protein function on the primary cilium. To directly test whether Sufu's function is mediated by the cilium, we generated mice deficient in Sufu and Kif3a (in which primary cilia fail to form) (Supplemental Fig. S6; Marszalek et al. 1999). Unlike Sufu-deficient mice, Kif3a−/− embryos display reduced Hh signaling in a dorsalized neural tube (Fig. 2B; Huangfu et al. 2003), and Kif3a−/− MEFs are unresponsive to Hh agonists (Supplemental Figs. S7, S8). If the primary cilium is required for Sufu's function in controlling Gli protein levels, we expected to observe a blockade of elevated Hh signaling in Sufu mutants once primary cilia are eliminated. Surprisingly, we found that the neural tube defects in Sufu−/−; Kif3a−/− mutants are identical to Sufu mutants (Fig. 2B). Marker analysis described below revealed increased Hh signaling as shown by dorsal expansion of Hh target genes (e.g., Ptch1) and ventral neural tube markers in both Sufu−/−; Kif3a−/− and Sufu−/− mutants (Fig. 2B; data not shown), indicating that Sufu's function does not require an intact primary cilium.
Loss of Sufu resulted in global up-regulation of Hh signaling and ventralization of the neural tube. Shh expression, which localizes to the notochord and floor plate in the ventral midline, is extended dorsally. This is accompanied by similar dorsal expansion of Hh target genes such as Ptch1, suggesting ventralization of the neural tube. Consistent with this, the expression domains of neuronal progenitor markers (Class I genes, including Pax7 and Pax6, repressed by Hh signaling and Class II genes, including Nkx6.1, Nkx2.2, and Foxa2, activated by Hh signaling) are shifted. For instance, the dorsal-most marker Pax7 is not expressed, and Pax6 expression is restricted to the dorsal neural tube of Sufu−/− embryos. Expression of Nkx6.1, Nkx2.2, and Foxa2 is expanded dorsally in the absence of Sufu (data not shown). Similarly, the expression domains of markers for differentiated interneurons and motoneurons are altered. For example, expression of Islet1 and Oligo2 (data not shown), which label motoneurons, is expanded dorsally in Sufu−/− neural tube. By comparison, marker analysis revealed a partially dorsalized Kif3a−/− neural tube. The neural tube defects in Sufu−/−; Kif3a−/− embryos resemble those in Sufu−/− mutants (Fig. 2B).
To confirm that regulation of Gli protein levels by Sufu can still occur in the absence of cilia, we knocked down Sufu in wild-type and Kif3a−/− MEFs via lentiviral delivery of shRNA (Hannon 2003) and demonstrated that Gli2 and Gli3 protein levels are greatly reduced to the same extent in both cell lines (Fig. 2C). Taken together, these results indicate that Sufu functions independently of the primary cilium in controlling Gli protein function, highlighting the importance of functional studies in illuminating the mechanisms by which the primary cilium regulates Hh signaling, as opposed to relying solely on protein localization.
Hh signaling is up-regulated in Ptch1 and Sufu mutants via distinct mechanisms
To further define cilium-dependent and cilium-independent Hh signaling events, we examined molecular defects in Ptch1−/− and Sufu−/− MEFs. Ptch1 and Sufu are two major regulators of mammalian Hh signaling, and both Ptch1- and Sufu-deficient mouse embryos display a severely ventralized neural tube due to elevated Hh signaling (Fig. 2B; Goodrich et al. 1997; Cooper et al. 2005; Svard et al. 2006). Gli2 and Gli3 protein are barely detectable in Sufu−/− MEFs (Fig. 2A). Instead, in Ptch1−/− MEFs, which display ligand-independent maximal activation of Hh signaling, Gli2 and Gli3 localize to the primary cilium in the absence of Hh stimulation (Fig. 1A), and Gli2 and Gli3 protein levels are similar to those in wild-type MEFs with a reduction in Gli3 repressor levels (Fig. 2A).
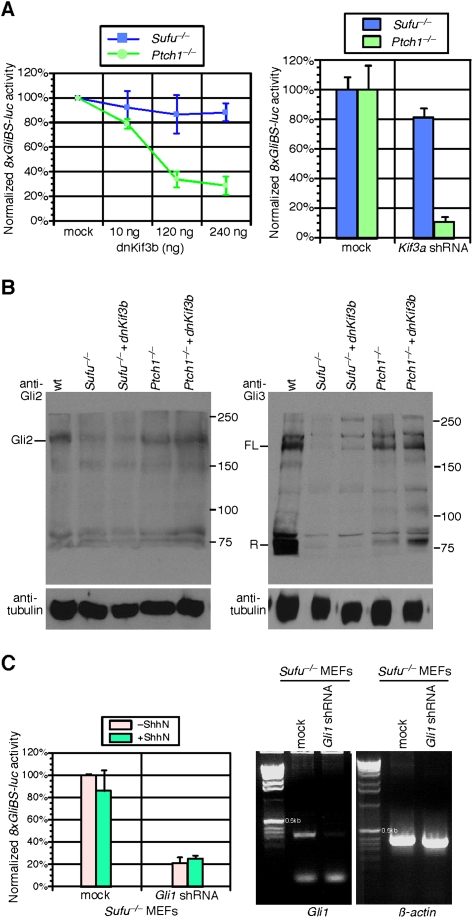
To assess the requirement of the primary cilium in mediating Ptch1 or Sufu function, we inhibited primary cilium function in either Ptch1−/− or Sufu−/− MEFs by expressing a dominant-negative form of Kif3b (dnKif3b), a subunit of the kinesin-II motor that participates in IFT (Fan and Beck 2004), or Kif3a shRNA. Inhibition of ciliary function in Sufu−/− MEFs has no effect on Gli protein levels or Hh pathway activity (Fig. 3A,B). Furthermore, overexpressed Gli2 and Gli3 localize to the primary cilium in Sufu−/− MEFs, suggesting that Sufu is not essential for ciliary localization of Gli proteins (Fig. 1B; Supplemental Fig. S9A). Moreover, Sufu is able to suppress Gli-mediated Hh activation in Kif3a−/− MEFs (Fig. 2D), indicating an essential negative role of Sufu in regulating Gli function independent of the primary cilium. In contrast, the constitutive Hh signaling in Ptch1−/− MEFs is greatly reduced compared with Sufu−/− MEFs when dnKif3b or Kif3a shRNA is expressed (Fig. 3A). Defective ciliary function in Ptch1−/− MEFs also changed the ratio of full-length Gli3 to Gli3 repressor, resembling the ratio observed in Kif3a−/− mutants or MEFs (Fig. 3B; Supplemental Fig. S10). Thus, despite ciliary localization of Ptch1 and Sufu, and Hh pathway up-regulation in Ptch1−/− and Sufu−/− mutants resulting in similar phenotypes, the molecular mechanisms of Ptch and Sufu are different. Ptch1 function, like Smo, is dependent on the primary cilium, while Sufu functions independently of the cilium.
Figure 3.
Loss of the primary cilium impairs ligand-independent Hh pathway activation in Ptch1−/− MEFs but has no effect on Sufu−/− MEFs. (A) Hh reporter assays using the 8xGliBS-luc reporter in Sufu−/− and Ptch1−/− MEFs expressing increasing amounts of a dominant-negative (dn) Kif3b construct (shown in the left panel) or Kif3a shRNA, both of which inhibits the function of the primary cilium. While dnKif3b or Kif3a shRNA have no effect on Hh pathway activation in Sufu−/− MEFs, increasing quantities of dnKif3b or Kif3a shRNA reduce Hh pathway activation in Ptch1−/− MEFs. (B) Western blots of lysates derived from wild-type (wt), Sufu−/−, and Ptch1−/− MEFs and Sufu−/− and Ptch1−/− MEFs expressing dnKif3b probed with anti-Gli2 and anti-Gli3 antibodies. Inhibition of ciliary function by dnKif3b has no effect on endogenous Gli2 and Gli3 protein levels in Sufu−/− MEFs, which are greatly reduced in the absence of Sufu. In contrast, defective ciliary function in Ptch1−/− MEFs changed the ratio of full-length Gli3 to Gli3 repressor. Tubulin was used as the loading control, and numbers on the right indicate locations of protein size standards. (FL) Full-length; (R) repressor. (C, left panel) Hh reporter assays using the 8xGliBS-luc reporter in Sufu−/− MEFs and Sufu−/− MEFs expressing Gli shRNA in the presence or absence of exogenous Shh. Hh pathway activation is significantly compromised in Sufu−/− MEFs in which Gli1 is efficiently knocked down, consistent with the notion that Gli1 contributes to Hh pathway activation in the absence of Sufu. Similar results were obtained using three pairs of Gli1 shRNA directed against different regions of Gli1. In contrast, Sufu−/− MEFs and Sufu−/− MEFs expressing Gli1 shRNA display normal responsiveness to the canonical Wnt ligand Wnt3a assayed by the SuperTOPflash reporter (data not shown). (Right panel) Semiquantitative RT–PCR demonstrates that Gli1 is efficiently knocked down via Gli1 shRNA; β-actin serves as the control.
Drosophila and zebrafish Sufu restore Gli protein levels in Sufu-deficient MEFs, while Drosophila Smo fails to rescue Smo-deficient MEFs
We reasoned that if involvement of the primary cilium in mammalian Hh signaling represents a major shift from fly Hh signaling, events mediated by Sufu and thus independent of the cilium would likely be evolutionarily conserved. In contrast, cilium-dependent processes of Hh signal transduction including ciliary localization of Smo and Ptch and their interactions on the cilium would be divergent among different model organisms.
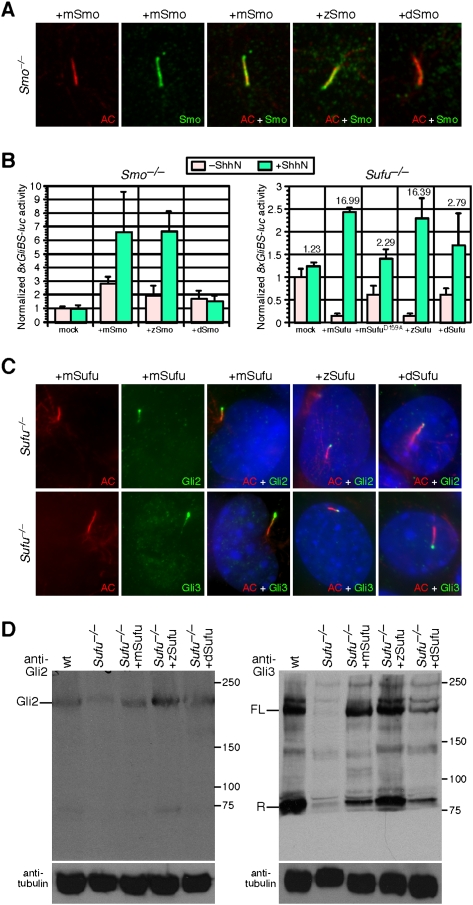
To test this idea, we introduced Smo cDNAs from mouse, zebrafish, or fly into Smo−/− MEFs via transient transfection (Supplemental Fig. S11) and assayed both ciliary localization of Smo by immunofluorescence and Hh pathway activation by transfecting MEFs with a Hh reporter construct (8xGliBS-luc) that has eight Gli-binding sites driving firefly luciferase expression (Sasaki et al. 1999). As expected, expression of mouse Smo in Smo−/− MEFs resulted in Smo ciliary localization (Fig. 4A; Supplemental Fig. S11) and conferred Hh responses (Fig. 4B, left panel). Interestingly, zebrafish Smo was detected with our antibody directed against mouse Smo, and both localized to the cilium (Fig. 4A; Supplemental Fig. S11) and activated the Hh pathway (Fig. 4B, left panel) upon Hh ligand stimulation in Smo−/− MEFs. Although the role primary cilia play in mediating Hh signaling in zebrafish has not been fully defined, our finding suggests that both cilium-dependent and cilium-independent processes exist. In contrast, Drosophila Smo expressed in Smo−/− MEFs failed to reach the shaft or tip of the cilium (Fig. 4A; Supplemental Fig. S11), and no Hh responses were observed (Fig. 4B, left panel). These results imply distinct requirements for Smo activation in different species and underscore the unique role of the primary cilium in vertebrate Hh signaling.
Figure 4.
Zebrafish and fly Sufu restore Gli protein levels in mouse Sufu−/− MEFs, while Drosophila Smo fails to rescue Hh defects in mouse Smo−/− MEFs. (A) Immunofluorescence of Smo−/− MEFs expressing Smo from different species including mouse (m), zebrafish (z), and Drosophila (d) using antibodies against acetylated tubulin (AC) (labeling the primary cilium, red) and Smo (green). While mSmo and zSmo introduced into Smo−/− MEFs via transient transfection led to ciliary localization of Smo, dSmo mainly resides in the cytoplasm and is not found on the cilium. (B, left panel) Hh activity assays using the 8xGliBS-luc reporter in Smo−/− MEFs expressing Smo from different species via transient transfection. Both mouse (m) and zebrafish (z) Smo restored Hh responsiveness in Smo−/− MEFs, while expression of Drosophila Smo (dSmo) has no effect on Hh activation. (Right panel) Sufu−/− MEFs were transfected with mouse Sufu (mSufu), mouse Sufu with the D159A mutation (mSufuD159A), zebrafish Sufu (zSufu), or Drosophila Sufu (dSufu). Both mSufu and zSufu repressed basal 8xGliBS-luc activity in the absence of ShhN and promoted an increase in ShhN-mediated response. In contrast, the mSufuD159A and dSufu constructs had a less pronounced effect, which may partially be attributed to their weaker Gli-binding capacity (Supplemental Fig. S9). The numbers indicate the ratios of Hh responsiveness in the presence and absence of exogenous Shh (e.g., the ratio is 1.23 when no Sufu is added and is 16.99 when mSufu is added). Error bars are s.d. (C) Immunofluorescence of Sufu−/− MEFs expressing Sufu from different species via retroviral infection using antibodies against acetylated tubulin (red) and Gli2 or Gli3 (green). Mouse, zebrafish, or Drosophila Sufu was capable of restoring ciliary localization of endogenous Gli2 and Gli3 to the cilium when expressed in Sufu−/− MEFs, suggesting an evolutionarily conserved function of Sufu. The percentage of cilia that exhibit Gli2 and Gli3 immunoreactivity is lower in Sufu−/− MEFs expressing dSufu compared with mSufu or zSufu, consistent with a partial rescue of Hh defects in Sufu−/− MEFs by dSufu. (D) Western blots of lysates derived from wild-type (wt), Sufu−/− MEFs, and Sufu−/− MEFs expressing mouse, zebrafish, and Drosophila Sufu via retroviral infection probed with anti-Gli2 and anti-Gli3 antibodies. Endogenous Gli2 and Gli3 protein levels are restored when Sufu from different species is expressed, suggesting an evolutionarily conserved biochemical function of Sufu. Tubulin was used as the loading control, and numbers on the right indicate locations of protein size standards. (FL) Full-length; (R) repressor.
We then assessed the effects of Sufu from different species on Hh pathway activation in Sufu−/− MEFs. Sufu from mouse, zebrafish, or fly was introduced into Sufu−/− MEFs via retroviral infection, and Gli2 and Gli3 protein levels and localization were assayed by immunofluorescence and Western blotting. We first showed that expression of mouse Sufu in Sufu−/− MEFs restored both Gli2 and Gli3 protein levels (Fig. 4D) and their ciliary localization (Fig. 4C; Supplemental Fig. S2B). Similarly, zebrafish Sufu was also capable of restoring Gli protein levels (Fig. 4D; Supplemental Fig. S12) and their ciliary localization (Fig. 4C; Supplemental Fig. S2B) in Sufu−/− MEFs, again consistent with the presence of both cilium-dependent and cilium-independent processes in this organism. Remarkably, expression of Drosophila Sufu in Sufu−/− MEFs led to a partial rescue of Gli protein levels (Fig. 4D; Supplemental Fig. S12) and ciliary localization (Fig. 4C; Supplemental Fig. S2B). These results are consistent with a conserved biochemical function of Sufu.
Sufu antagonizes Spop in regulating Gli2 and Gli3 protein levels
To further test the idea that Sufu-mediated regulation of Gli protein levels is conserved among different organisms, we asked whether Spop (speckle-type POZ protein) (Supplemental Fig. S13), a homolog of the Drosophila MATH and BTB domain-containing protein Hib, antagonizes Sufu in regulating Gli protein levels. Hib forms a complex with Ci and Cullin3 (Cul3) and promotes Ci ubiquitination by the Cul3-based E3 ubiquitin ligase, resulting in complete degradation of Ci (Zhang et al. 2006). Sufu appears to protect Ci from Hib-mediated degradation through competitive binding with Hib for Ci (Zhang et al. 2006).
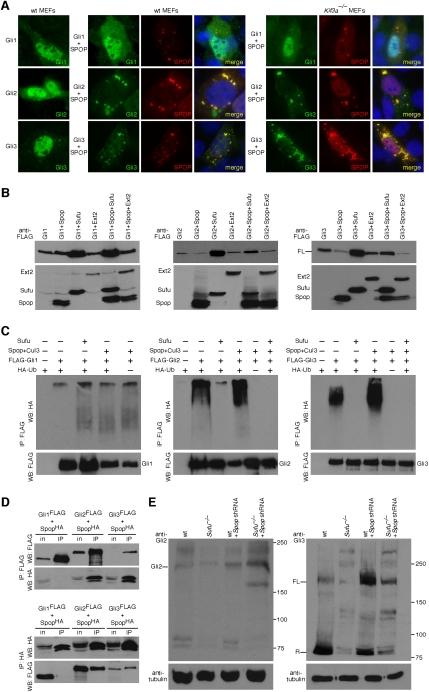
We first investigated the subcellular distribution of overexpressed Spop and Gli1, Gli2, and Gli3 in MEFs and established cell lines. While Gli proteins are distributed relatively evenly in the cytoplasm and nucleus (Fig. 5A), a prominent feature of Spop protein distribution is the presence of focal densely stained nuclear speckles (Fig. 5A; Supplemental Fig. S14; Hernandez-Munoz et al. 2005). We also observed foci of Spop staining in the cytoplasm in a subset of cell types examined, indicating that cell line-specific factors may determine the proportion of nuclear and cytosolic Spop. Interestingly, coexpression of Gli2 and Gli3 with Spop recruited Gli2 and Gli3 into Spop-positive speckles (Fig. 5A; Supplemental Figs. S14, S9B). In contrast, the distribution of Gli1 remains unchanged in the presence of Spop (Fig. 5A; Supplemental Fig. S9B). These results suggested that Spop might physically interact with Gli2 and Gli3. To test this, we transfected HEK 293T cells with constructs encoding Spop-HA and Gli1-Flag, Gli2-Flag, or Gli3-Flag. Spop physically associates with Gli2 and Gli3, but not Gli1 (Fig. 5D). Furthermore, coexpression of Spop with Gli proteins in HEK 293T cells led to a significant reduction in Gli2 and Gli3 protein levels, while Gli1 is unaffected (Fig. 5B; Supplemental Fig. S15; data not shown). Finally, we reasoned that if Spop preferentially interacts with Gli2 or Gli3 and reduces the protein levels, transcriptional activation could be compromised. Coexpression of Spop with Gli1 or Gli2 resulted in a substantial, dose-dependent reduction in Gli2-mediated but not Gli1-mediated Hh pathway activation (Supplemental Fig. S16). Thus, we conclude that Spop binds Gli2 and Gli3 and causes a reduction in global Gli2 and Gli3 levels, resulting in a decrease in Gli2-dependent transcriptional activity. Gli3 cannot be assessed in this assay owing to its weak transactivation ability (Gerber et al. 2007; data not shown).
Figure 5.
Mouse Spop colocalizes with Gli proteins and antagonizes Sufu in controlling Gli protein levels. (A) Double immunostaining of MEFs transfected singly with Flag-tagged Gli1, Gli2, or Gli3 or cotransfected with Myc-tagged Spop using Flag and Myc antibodies against Flag-tagged Gli1, Gli2, or Gli3 (green) and Myc-tagged Spop (red). Both cytoplasmic and nuclear staining of Gli1, Gli2, and Gli3 was detected. Punctate Spop immunoreactivity in the nucleus and cytoplasm was evident, consistent with previous reports (Hernandez-Munoz et al. 2005). Immunoreactivity of Gli2 and Gli3 (and not Gli1) was reduced when coexpressed with Spop, and Gli2/3 distribution extensively overlaps with Spop, particularly in the cytoplasm. Loss of the primary cilium in Kif3a−/− MEFs has no effect on the subcellular distributions and interactions of Gli1, Gli2, Gli3, and Spop. (B) Western blots of lysates derived from HEK 293T cells expressing Flag-tagged Gli3 singly or in combination with Flag-tagged Spop, Sufu, or Ext2 probed with anti-Flag antibodies. Lack of apparent Gli3 processing in cultured cells has been previously reported (B Wang et al. 2000). Coexpression of Gli2 or Gli3 with Sufu notably enhanced Gli2 and Gli3 protein levels. In contrast, coexpression of Gli2 or Gli3 with Spop (but not the control protein Ext2) significantly reduces Gli2 and Gli3 protein levels, which can then be restored when Sufu is coexpressed. We noticed that reduction in Gli2 protein levels is not as dramatic as Gli3 when Spop is coexpressed. Gli1 or Ext2 protein levels are unaffected when Spop is overexpressed (data not shown). α-Tubulin serves as the loading control (data not shown). (C) Western blot of immunoprecipitated Gli1, Gli2, and Gli3 (epitope-tagged with one copy of Flag) to detect polyubiquitinated Gli proteins. Spop promotes ubiquitination of Gli2 and Gli3 but not Gli1; Gli2 and Gli3 ubiquitination is abolished when Sufu is coexpressed. (WB) Western blot. (D) Western blot of immunoprecipitated Gli1, Gli2, and Gli3 (epitope-tagged with one copy of Flag) to detect physical interaction with Spop (epitope-tagged with one copies of HA) from HEK 293T lysates. Spop physically associates with Gli2 and Gli3 but not Gli1. (in) Input; (IP) immunoprecipitation. (E) Western blots of lysates derived from wild-type and Sufu−/− MEFs and wild-type and Sufu−/− MEFs expressing Spop shRNA probed with anti-Gli2 and anti-Gli3 antibodies. Efficient knockdown of Spop was verified by semiquantitative RT–PCR (data not shown). Gli2 and Gli3 levels are partially restored in Sufu−/− MEFs when Spop is knocked down, consistent with a model in which Sufu and Spop antagonize each other in regulating Gli2 and 3 (but not Gli1) protein levels. Lack of complete rescue of Gli protein levels could be attributed to the presence of additional mammalian Spop homologs (e.g., Spop-like and Tdpoz proteins) (Huang et al. 2004). (FL) Full-length; (R) repressor.
We then asked whether Spop binding to Gli2 and Gli3 promotes their ubiquitination and subsequent degradation by the 26S proteasome. We carried out an in vivo ubiquitination assay in HEK 293T cells and showed that Spop promotes ubiquitination of Gli2 and Gli3 but not Gli1 (Fig. 5C; Supplemental Fig. S17). The Spop-dependent reduction in Gli2 and Gli3 protein levels was rescued with the proteasome inhibitor MG132 (Supplemental Fig. S18), suggesting that Spop directs Gli2 and Gli3 degradation in a proteasome-dependent manner. This is further supported by the observation that coexpression of Spop with Gli2/3 and Cul3 recruited Gli2/3 into Spop-positive foci, which also contain Cul3 (Supplemental Fig. S19). In contrast, Cul3 and Gli2/3 do not colocalize in the absence of Spop, supporting the hypothesis that Spop targets Gli2 and Gli3 for Cul3-mediated proteasomal degradation.
Since Sufu is required for maintaining Gli2 and Gli3 protein levels, we tested whether Sufu antagonizes Spop and thus preserves Gli2 and Gli3 protein stability. Expression of Sufu was able to block Spop-mediated Gli2 and Gli3 protein reduction (Fig. 5B) and ubiquitination (Fig. 5C), suggesting that Spop and Sufu antagonize each other in regulating Gli protein levels. Consistent with this notion, shRNA-mediated knockdown of Spop in Sufu−/− MEFs partially restored Gli protein levels (Fig. 5E) and enhanced Hh pathway activity (Supplemental Fig. S20). In addition, recruitment of Gli2 and Gli3 to Spop-positive foci can occur in Kif3a−/− MEFs (Fig. 5A; Supplemental Fig. S9B), suggesting that the Sufu–Spop–Gli circuit is evolutionarily conserved and is independent of the primary cilium. This is further supported by the observation that Spop does not localize to the primary cilium when overexpressed (Supplemental Fig. S21), and mammalian Gli2 and Gli3 are stabilized when expressed in Drosophila deficient in Hib activity (Zhang et al. 2006). We speculate that Sufu sequesters Gli2/3 protein in the cytoplasm and protects them from Spop-mediated protein degradation, providing a Gli protein pool for the production of Gli2/3 activators and repressors (Fig. 7, below). Despite a conserved mechanism of Spop and Sufu in regulating Gli protein levels, it is interesting to note that Gli1 appears to be refractory to Spop regulation, but its transactivation potential is still inhibited by Sufu. This selective regulation could allow the production of a wide range of Hh responses.
Figure 7.
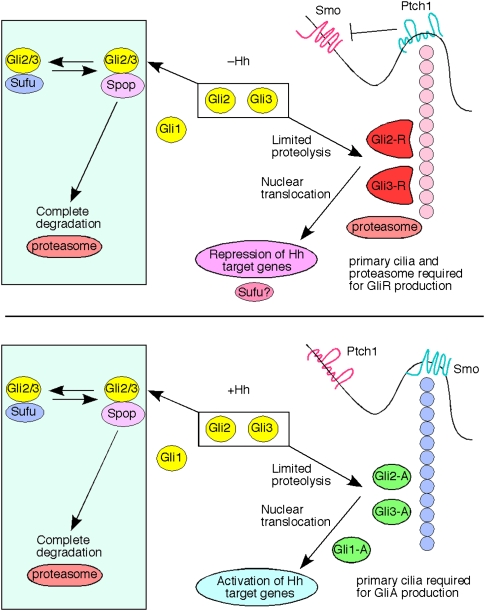
A model of mammalian Hh signaling. Sufu plays a pivotal role in controlling Gli protein levels. Sufu protects full-length Gli2 and Gli3 proteins from Spop-mediated ubiquitination and complete degradation by the proteasome. In this way, Sufu functions as an adaptor to preserve a pool of Gli2 and Gli3 that can be readily converted into Gli activators and repressors. This aspect of Hh signaling is evolutionarily conserved and independent of the primary cilium. In contrast, the primary cilium is required for generating Gli repressors via limited proteolysis in the absence of Hh signaling and converting full-length Gli proteins into activators through unknown mechanisms upon Hh pathway activation. These events occurs downstream from Smo, which translocates to the primary cilium when Hh ligand binds to Ptch1 and removes it from the cilium. How other Hh pathway components or ciliary proteins particulate in cilium-dependent and cilium-independent activity needs to be investigated further.
Sufu has an unexpected positive role in controlling mammalian Hh signaling
Elevated Hh signaling in the Sufu-deficient neural tube indicates that Sufu is a negative regulator of mammalian Hh signaling (Fig. 2B; Cooper et al. 2005; Svard et al. 2006). With the new insight that Sufu regulates full-length Gli protein levels, we surmised that the function of Gli activators (derived from full-length Gli proteins) could be compromised in the absence of Sufu, resulting in submaximal Hh pathway activation. To test this hypothesis, we investigated the mechanisms by which Sufu controls Hh signaling in MEFs. We first examined Hh responses in wild-type and Sufu-deficient MEFs. We showed that wild-type MEFs are Hh-responsive by transfecting wild-type MEFs with the 8xGliBS-luc reporter construct both in the absence and presence of Shh-conditioned media (Supplemental Fig. S8). Hh responsiveness in Sufu−/− MEFs is mildly elevated in the absence of Hh stimulation (Supplemental Fig. S8), consistent with the demonstrated negative role of Sufu. Interestingly, exogenous Shh fails to support Hh responses in Sufu−/− MEFs to the same extent as in wild-type MEFs (Supplemental Fig. S8), consistent with the requirement of Sufu for maximal Hh pathway activation.
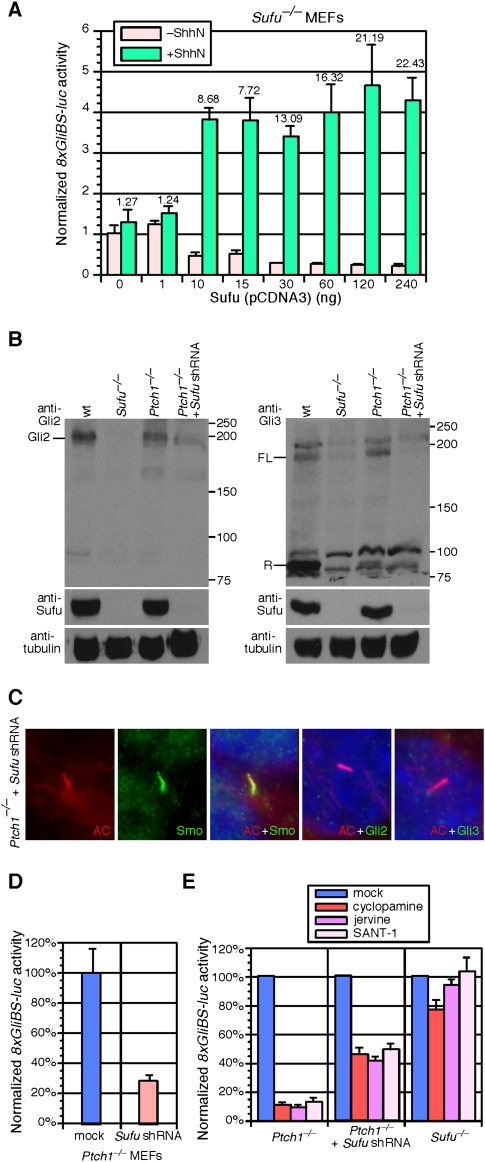
To further test this hypothesis, we transfected increasing amounts of Sufu cDNA in Sufu−/− MEFs in the absence or presence of exogenous Shh and assessed its effects on Hh pathway activation, as measured by luciferase activity from the transfected 8xGliBS-luc reporter. We found that in the absence of exogenous Shh, increasing amounts of Sufu transfected in Sufu−/− MEFs gradually decreased basal reporter levels (Fig. 6A). This is consistent with negative regulation of Hh signaling by Sufu. Interestingly, in the presence of exogenous Shh, increasing the amount of Sufu cDNA in Sufu−/− MEFs instead promoted Hh responsiveness (Fig. 6A). This suggests that Sufu is also required for maximal Hh signaling and that Sufu is required for generation of a broader dynamic range of Hh responses. In contrast, Ptch1 does not display a positive role in Hh signaling using a similar assay (Supplemental Fig. S22).
Figure 6.
Mouse Sufu has positive and negative roles in regulating Hh signaling. (A) Hh activity assays using the 8xGliBS-luc reporter in Sufu−/− MEFs transfected with varying quantities of Sufu. Addition of increasing amounts of Sufu to Sufu−/− MEFs reduces Hh responsiveness in the absence of exogenous Shh, while promoting Hh activation in the presence of Shh. The numbers indicate the ratios of Hh responsiveness in the presence and absence of exogenous Shh (e.g., the ratio is 1.27 when no Sufu is added and is 22.43 when 240 ng of Sufu is added). Similar results were seen in two additional Sufu−/− cell lines and with the Smo agonist purmorphamine instead of ShhN (data not shown). Error bars are s.d. (B) Western blots of MEF lysates derived from wild type (wt), Sufu−/−, Ptch1−/−, or Ptch1−/− expressing Sufu shRNA probed with anti-Gli2 and anti-Gli3 antibodies. Gli2 and Gli3 protein levels are greatly reduced in Ptch1−/− MEFs when Sufu is knocked down. (C) Immunofluorescence of Ptch1−/− MEFs stably expressing Sufu shRNA using antibodies against acetylated tubulin (labeling the primary cilium) (red) and various Hh pathway components including Smo, Gli2, and Gli3 (green). Smo, Gli2, and Gli3 localize to the primary cilium in Ptch1−/− MEFs in the absence of exogenous Hh stimulation, consistent with maximal Hh pathway activation. While Smo localization to the primary cilium is unaffected in Ptch1−/− MEFs when Sufu is knocked down, ciliary localization of Gli2 and Gli3 in Ptch1−/− MEFs is abolished when Sufu is eliminated, suggesting compromised Hh pathway activation. (D) Hh reporter assays using the 8xGliBS-luc reporter in Ptch1−/− MEFs and Ptch1−/− MEFs expressing Sufu shRNA. Sufu knockdown leads to reduced Hh pathway activity in Ptch1−/− MEFs. (E) Hh reporter assays using the 8xGliBS-luc reporter in Sufu−/− and Ptch1−/− MEFs and Ptch1−/− MEFs expressing Sufu shRNA in the presence of various Hh antagonists that inhibit Smo function (Chen et al. 2002a,b). Hh pathway activation in Ptch1−/− MEFs is efficiently knocked down in the presence of Hh antagonists, but these Smo inhibitors have no effect on Hh pathway activity in Sufu−/− MEFs. When Sufu is knocked down in Ptch1−/− MEFs, these cells become partially insensitive to Hh antagonists.
When introduced into Sufu−/− MEFs via transient transfection, zebrafish and fly Sufu were also able to promote Hh responsiveness in the presence of exogenous Shh (Fig. 4B, right panel), again supporting a conserved role of Sufu. The effect of fly Sufu is less pronounced in this assay than either mouse or zebrafish Sufu. We speculate that this is due to reduced affinity of fly Sufu for Gli proteins (Supplemental Fig. S23). Supporting this idea, the mouse SufuD159A mutant (Merchant et al. 2004), which exhibits reduced binding to Gli proteins (Supplemental Fig. S23), also displays reduced efficacy in restoring Hh responsiveness in the presence of exogenous Shh (Fig. 4B, right panel).
To further validate a dual function of Sufu in controlling Hh signaling, we reasoned that if Sufu is required for maximal Hh pathway activation, knockdown of Sufu in Ptch1−/− MEFs would compromise pathway activation. Indeed, when Sufu is efficiently knocked down via shRNA in Ptch1−/− MEFs, ciliary localization of Gli2 and Gli3 is largely eliminated (Fig. 6C; Supplemental Fig. S2C), Gli2 and Gli3 protein levels are greatly reduced (Fig. 6B), and Hh pathway activity is compromised (Fig. 6D). Consistent with these findings, neural tube defects in Sufu−/− embryos (Fig. 2B; Cooper et al. 2005; Svard et al. 2006) are slightly less severe than Ptch1−/− (Goodrich et al. 1997; data not shown), which can be attributed to a requirement of Sufu in Hh pathway activation. We also assessed how Sufu knockdown affects the efficacy of Hh antagonists in Ptch1−/− MEFs. Hh pathway activation in Ptch1−/− MEFs is efficiently inhibited by Hh antagonists cyclopamine, jervine, and SANT-1 (Chen et al. 2002b), but these Smo inhibitors have no effect on Hh pathway activity in Sufu−/− MEFs (Fig. 6E). When Sufu is knocked down in Ptch1−/− MEFs, these cells become partially insensitive to Smo inhibitors (Fig. 6E), consistent with the observation that activation in Sufu−/− MEFs is independent of Smo function. These studies also argue that Sufu functions downstream from Ptch1 and Smo. By controlling full-length Gli protein levels, Sufu could shift the contribution of Gli activators and repressors to Hh signaling outputs in various tissues. Finally, we anticipate that conditional inactivation of Sufu in different Hh-responsive tissues such as the limb will lead to increased Hh signaling, as determined by expression of Hh target genes such as Ptch1, Hip1, and Gli1, consistent with increased cell-autonomous activation of the Hh pathway in the absence of Sufu. We also found that the expression levels of Ptch1 are reduced in Hh-responsive cells adjacent to the Hh source (data not shown). This would suggest that maximal Hh pathway activation fails to occur in the absence of Sufu, consistent with the in vitro data and our hypothesis of a dual role of Sufu in controlling Hh signaling.
Discussion
Our studies delineate important aspects of cilium-dependent and cilium-independent Hh signal transduction (Fig. 7; Supplemental Fig. S24). Hh binding to Ptch relieves its repression of Smo and induces a conformational change in Smo that results in dimerization of Smo cytoplasmic tails and is essential for pathway activation (Zhao et al. 2007). Although these events are common across different species, the execution of these steps appears to be accomplished in distinct microenvironments in insects and mammals. The requirement of the primary cilium in mammalian but not fly Hh pathway activation suggests that important modifications in Hh pathway design, including ciliary localization of Smo and its activation, have occurred during evolution (Huangfu and Anderson 2006). In contrast with nonconserved, cilium-dependent processes, Sufu antagonizes the action of the conserved Gli-degrading protein Spop downstream from Smo, and thus preserves a pool of full-length Gli proteins. Consequently, the production of Gli activators and repressors is dependent on the presence of Sufu, but does not require an intact primary cilium. Duplication of the Gli genes coupled with selective regulation of Gli2 and Gli3 by Spop allows the production of a more complex and robust Hh response in mammals.
Mammalian Sufu regulates Gli protein function independent of the primary cilium
Despite ciliary localization of Sufu and Gli proteins, our genetic studies and cell-based assays unambiguously demonstrated that Sufu controls Gli protein levels independent of the primary cilium. Furthermore, Sufu is not essential for Gli trafficking to the cilium. This highlighted the importance of functional studies to assess the physiological relevance of the presence of Hh pathway components, or unrelated nonstructural proteins, on the primary cilium. In contrast to our results, a prior report showed that knockdown of Sufu in Ift172wim or Dync2h1ttn MEFs (in which primary cilium function is disrupted) caused no detectable activation of Hh reporters (Ocbina and Anderson 2008). This was interpreted as Sufu acting within cilia to keep the Hh pathway off in the absence of ligand (Ocbina and Anderson 2008). We suspect that the discrepancy could be due to incomplete knockdown of Sufu using RNAi-based approaches that could potentially complicate interpretations of genetic epistasis. We cannot rule out the possibility that Sufu has additional roles in Hh signaling that are regulated or mediated by the primary cilium. However, our data suggest that these processes most likely either have minor effects on Hh signaling or are redundant with other events.
Distinct combinations of Gli activators and repressors and Hh outputs
A major obstacle in understanding Hh pathway activation by Gli proteins is our inability to understand how a ratio of Gli activators and repressors is mechanistically converted into defined transcriptional events. In vertebrates, this is complicated by the fact that Gli3 and Gli1 are negative and positive targets, respectively, of Hh signaling (Marigo et al. 1996; Lee et al. 1997; Bai et al. 2002). This issue is underlined by the observation that the Hh pathway is activated in both Ptch1 and Sufu mutants, yet Gli protein levels are affected only in the absence of Sufu. This suggests that different combinations of Gli activators and repressors can lead to Hh pathway activation. Our knockdown of Gli1 in Sufu−/− MEFs suggests that Gli1 contributes to transcriptional activation in Sufu mutants (Fig. 3C). We hypothesize that reductions in full-length Gli2 and Gli3 proteins, and consequently a reduction in Gli repressor levels, leads to up-regulation of Gli1 and thus ligand-independent activation of the Hh pathway. Supporting this, Gli1 was shown to be a target of the Gli3 repressor by chromatin immunoprecipitation (Hu et al. 2006; Vokes et al. 2008). Previous studies on Gli-deficient neural tubes suggest that Gli activator function predominates over Gli repressors and that Gli1 is able to compensate for the loss of Gli2 activator (Bai and Joyner 2001; Bai et al. 2002; Huangfu and Anderson 2006). Thus, we predict that Gli1, whose expression is expanded dorsally in Sufu−/− neural tubes (Svard et al. 2006), is responsible for the up-regulated Hh signaling in this tissue. Reduction in Gli repressor levels could also directly contribute to Hh target gene expression. However, since full-length Gli2 and Gli3 protein levels are reduced, maximal Hh signaling fails to occur in Sufu-deficient neural tubes.
A conserved Sufu/Spop/Gli circuit
Our studies on Sufu provide important mechanistic insight into how Sufu regulates Hh signaling. Largely based on physical interactions between Sufu and Gli proteins, the traditional model proposed that Sufu tethers Gli protein in the cytoplasm, preventing nuclear translocation and subsequent activation of target genes (Kogerman et al. 1999). In this study, we showed that Sufu antagonizes Spop, preventing degradation of full-length Gli2 and Gli3. The process of Sufu–Spop antagonism is evolutionarily conserved since Drosophila Sufu protects Ci from Hib-mediated degradation through competitive binding to Ci (Zhang et al. 2006). As a result, loss of Sufu affects production of Gli2/Gli3 activator and repressor forms, which are both derived from full-length proteins. This is achieved by Hib/Spop forming a complex with Ci/Gli2/Gli3 and Cul3, thus promoting Ci/Gli ubiquitination through the Cul3-based E3 ubiquitin ligase and resulting in complete degradation by the 26S proteasome (Zhang et al. 2006).
Drosophila Sufu is able to partially restore the defects in Gli2/Gli3 protein levels, ciliary localization, and Hh pathway activation in Sufu−/− MEFs, supporting conservation of this process. Interestingly, overexpression of Drosophila Sufu in imaginal discs inhibits Hh target gene expression in anterior cells that receive the Hh signal, but activates Hh target gene expression in the most anterior region that does not receive the Hh signal (Dussillol-Godar et al. 2006). This is consistent with a dual role of fly Sufu and whether a conserved mechanism underlies these effects needs to be further investigated.
Nevertheless, important differences in the Sufu–Spop–Gli circuit exist between flies and mammals. Gli1, unlike Gli2 and Gli3, does not appear to be subject to Spop regulation. Furthermore, while sufu mutant flies are viable (Preat 1992), Sufu−/− mice die during early embryogenesis (Cooper et al. 2005; Svard et al. 2006). Therefore, the gain-of-function phenotype in Sufu-null mice may result from increased levels of Gli1, triggered by Spop-mediated degradation of full-length Gli2/Gli3. Gli1 may have lost a requisite Spop-interacting domain, allowing it to escape regulation by Spop. Identification of domains in Gli2 and Gli3 that interact with Spop will further clarify this issue. Notably, full-length Ci and Ci repressor levels appear to be proportionately reduced in sufu mutant flies, implying that sufu affects Ci protein stability (Ohlmeyer and Kalderon 1998). Duplication of the ancestral Ci/Gli gene, coupled with subfunctionalization (including the distribution of activator and repressor function) and evolution of negative and positive transcriptional regulatory loops, may account for the vastly different effects of loss of Sufu in insects and vertebrates.
Multiple degradation and processing signals in the Gli proteins
Regulation of Gli protein stability is a key step in controlling Hh pathway activity, and multiple, distinct degradation signals have been identified in the three Gli proteins. For instance, two degradation signals are present in Gli1, one of which contains recognition sequences for the β-TrCP adapter protein, and two β-TrCP-binding motifs also exist in Gli2 (Bhatia et al. 2006; Huntzicker et al. 2006; Pan et al. 2006). This allows utilization of the β-TrCP adapter protein for Gli1/2 proteolysis via the Cul1-based E3 ligase, distinct from Spop-mediated Gli2/3 degradation through the Cul3-based E3 ligase. β-TrCP is also required for limited proteolysis of Gli3 into a truncated repressor form (Wang and Li 2006). A critical unresolved issue is to understand how multiple degradation signals in Gli proteins are used to regulate full-length protein stability as well as generation of repressor forms. Further investigation is required to determine if the role of Sufu is specific in antagonizing Spop-mediated degradation, or if it is capable of opposing additional degradative pathways (Di Marcotullio et al. 2006, 2007). It is also formally possible that Sufu has a direct effect on Gli repressor stability.
Where is the site of action for Sufu?
Sufu was postulated to function in both the nucleus and the cytoplasm, as overexpressed Sufu protein in cultured cells could be detected in both compartments (Ding et al. 1999; Kogerman et al. 1999). Furthermore, Sufu can be coimmunoprecipitated with all three Gli proteins and was shown to cooperate with SAP18–Sin3 corepressor complex in repressing transcription from a multimerized Gli-binding site luciferase reporter (Ding et al. 1999; Kogerman et al. 1999; Cheng and Bishop 2002; Paces-Fessy et al. 2004). Thus, it was proposed that Sufu may have a direct role in repressing Gli-mediated transcription in the nucleus in addition to sequestering Gli proteins in the cytoplasm. Recent work has challenged Sufu's cytoplasmic function by demonstrating that an overexpressed Gli1-eGFP fusion protein has a similar cytoplasmic distribution in wild-type or Sufu-deficient MEFs; in both cell types, Gli1 is largely cytoplasmic and becomes predominantly nuclear when nuclear export is blocked (Svard et al. 2006). However, the distributions of overexpressed Gli proteins may fail to reflect those of endogenous Gli proteins. Importantly, conclusions based on Gli1 studies may not be applicable to Gli2 and Gli3 given their distinct properties. We observed that knockdown of Spop in Sufu−/− MEFs partially restored levels of cytoplasmic Gli2 and Gli3, resembling the wild-type nuclear–cytoplasmic distribution (data not shown). While the data suggest that Sufu may have minimal effect on shuttling Gli1, Gli2, and Gli3, we cannot at this time rule out potential alternations in kinetics of Gli trafficking or possible post-transcription degradation events. Contrary to previous reports (Cheng and Bishop 2002; Paces-Fessy et al. 2004), we failed to observe any discernable effects of SAP18 either singly or in conjunction with other Hh pathway components on Hh pathway activity in MEFs (Supplemental Fig. S25). Nevertheless, although our studies highlight a major function of Sufu in regulating cytoplasmic Gli protein levels, we cannot conclusively exclude potential minor roles in the nucleus.
We would like to emphasize that previous work that involved manipulating other mammalian Hh pathway components in the absence of Sufu should be considered in light of the fact that Sufu regulates Gli2/Gli3 stability. For instance, protein kinase A (PKA) phosphorylates Gli2 and Gli3, promoting β-TrCP binding and limited proteolysis to generate repressor forms (Pan et al. 2006; Wang and Li 2006). Treatment of multiple Sufu−/− MEF clones with either forskolin or IBMX, known PKA agonists, has no convincing effect on elevated Hh pathway levels (Supplemental Fig. S26). Since Gli2/Gli3 protein levels are drastically reduced in Sufu−/− MEFs, the effects of PKA are expected to be minimized.
The primary cilium and changes in Hh pathway design during evolution
Zebrafish or Drosophila Sufu can restore Gli protein levels in Sufu−/− MEFs, yet Smo from Drosophila fails to rescue Hh signaling defects in Smo−/− MEFs. This supports our proposal that cilium-independent steps in Hh signaling are evolutionarily conserved. The primary cilium has been shown to be essential for proper Hh pathway activation and the production of a proper ratio of Gli activator and repressor forms in mammals. Thus, the primary cilium likely provides an environment in which Hh pathway components dynamically interact with each other in response to varying extracellular Hh ligand concentrations. Such obligatory intracellular interactions may have coevolved to an extent where Drosophila Hh pathway components cannot functionally substitute for their mammalian counterparts. Supporting this, prior studies heterologously expressing human SMOH (De Rivoyre et al. 2006) or FU (Daoud and Blanchet-Tournier 2005) in the developing fly wing disc showed that the mammalian proteins could not rescue loss of the cognate fly gene. Hh signaling occurs in fly in the absence of the primary cilium, raising the fundamental question of whether utilization of the primary cilium in mammalian Hh signaling simply adds to the complexity of Hh signaling or represents a major redesign of the pathway. Identifying ciliary components that contribute to mammalian Hh signaling and elucidating their function will help resolve this critical issue. Further investigation of a possible role of the primary cilium in zebrafish Hh signaling, which may represent a transitional state between fly and mouse, as well as in other primitive vertebrate species will provide additional insight into how various vertebrate species have adopted the primary cilium in Hh transduction.
Outstanding questions in Hh signaling
A major unresolved issue in mammalian Hh signaling is understanding the molecular mechanisms of signal transduction from Smo to the Gli proteins. Smo encodes a seven-pass transmembrane protein that resembles a G-protein-coupled receptor (GPCR). Gαi has been implicated as a mediator of Smo activity in Drosophila (Ogden et al. 2008), yet manipulation of vertebrate Gαi has little effect on Gli3 processing and chick neural tube patterning (Low et al. 2008). These findings not only stress the necessity of additional genetic analysis, particularly loss-of-function studies, and biochemical characterization to settle the role of G proteins in transducing the Hh signal downstream from Smo, but again point to potential divergence in pathway design between species. We consider three unresolved areas of future investigation based on our studies of the relationship between Sufu and Gli proteins.
First, it is unclear if Hh signaling modulates Sufu activity (Supplemental Fig. S27), and if Sufu interacts with other Hh pathway components to control Gli function. This is complicated by the involvement of the primary cilium in mammalian Hh transduction, as well as the current lack of evidence for a Cos2-scaffolded counterpart to the Drosophila Hh signaling complex (HSC) that contains Cos2, Fu, Ci, and possibly Sufu (Stegman et al. 2000; Varjosalo et al. 2006). Furthermore, the Fused kinase, which opposes Sufu activity in the fly, is dispensable for mammalian Hh signaling and shows no epistatic relationship with Sufu (Chen et al. 2005; Merchant et al. 2005). Sufu likely functions downstream from Ptch1 and Smo, since expression of a constitutively active form of Smo (SmoM2) in Sufu−/− MEFs failed to further activate the Hh pathway (Supplemental Fig. S28), and Sufu knockdown in Ptch1−/− MEFs led to reduced Gli protein levels (Fig. 6B,C). It has been hypothesized that Smo signals to Sufu (Svard et al. 2006), but no biochemical evidence for this has yet been presented. Definitive in vivo loss-of-function studies on the mouse Cos2 orthologs Kif7 and Kif27 as well as further biochemical characterization of the mammalian Smo–Kif3a–β-arrestin complex (Kovacs et al. 2008) will address whether a conserved cytoplasmic HSC exists in mammals, or if Smo–Kif3a complexes on the primary cilium have replaced the scaffolding function of the HSC. Examination of a potential relationship of Sufu to a mammalian HSC will permit a greater understanding of Hh signal transduction.
Second, the mechanisms of Smo and Gli trafficking to and on the primary cilium are poorly understood, but the cilium is critical for proper pathway activation and formation of Gli activators and repressors (Corbit et al. 2005; Huangfu and Anderson 2005; Liu et al. 2005). Our data indicate that Sufu is not essential for Gli trafficking to the primary cilium. Thus, identifying signals that confer ciliary localization and trafficking of Hh pathway components and their interacting partners is crucial to understand the role of the cilium in controlling their function. Interestingly, genetic studies of the vesicle transport protein Rab23, a GTPase that is a cell-autonomous negative regulator of vertebrate Hh signaling, showed that Rab23 controls Gli2 and Gli3 activity (Eggenschwiler et al. 2006). This raises the possibility that Rab23 regulates trafficking of Hh pathway components that inhibit Gli activator function. In contrast, the GTPase Arl13b appears to be required for generation of the Gli2 activator (Caspary et al. 2007). These, and other Rab proteins involved in biogenesis of the primary cilium (Oro 2007; Yoshimura et al. 2007), are likely to be useful targets for investigating the dynamics of Smo and Gli movement on the primary cilium and their relationships to states of pathway activation. In addition, the steps required for conversion of full-length Gli to Gli activators are poorly understood (Methot and Basler 1999; Smelkinson et al. 2007), and biochemical identification of these events and their relationship to the primary cilium is essential.
Finally, our comprehension of Gli activator and repressor function at endogenous target promoters or enhancers is lacking, but significant progress has recently been made in identifying bona fide Gli-binding sites via chromatin immunoprecipitation (Vokes et al. 2007, 2008). Many Gli-binding sites are occupied by both Gli1 activator and an artificial Gli3 repressor (Vokes et al. 2007, 2008), yet the dynamics of activator and repressor binding of all three Gli proteins in the absence or presence of Hh ligand and the transcriptional outputs of Gli protein binding remain to be investigated. Further experiments of this nature, focusing on Gli partner proteins such as Sufu and Hoxd12 (Y Chen et al. 2004), and potential coactivators (e.g., CBP) (Akimaru et al. 1997) and corepressors (e.g., Sin3a, SAP18, and Ski) (Cheng and Bishop 2002; Paces-Fessy et al. 2004), will illuminate the mechanism of Gli transcription factor action, and possible Sufu function, at endogenous binding sites.
Materials and methods
Animal husbandry
A conditional allele of Sufu was generated by flanking exons 4–8 with loxP sites using gene targeting (Joyner 2000). A null allele of Sufu was subsequently produced by Cre-mediated excision of sequences between the two loxP sites. Kif3a mice were obtained from MMRRC (Mutant Mouse Regional Resource Centers); Ptch1 mice were provided by Dr. Matt Scott (Stanford University); and Smo mice were provided by Dr. Andy McMahon (Harvard University). Fu mutant mice were genotyped and maintained as described (Chen et al. 2005). Gli2zfd and Gli3xt mice have been described previously (Hui and Joyner 1993; Mo et al. 1997).
Histology and in situ hybridization
Embryos collected at various developmental stages were fixed in 4% paraformaldehyde overnight at 4°C and processed and embedded in paraffin (Nagy et al. 2003). All the embryos collected were sectioned at 6-μm thickness for histological analysis and in situ hybridization (Nagy et al. 2003). Whole-mount in situ hybridization using digoxigenin-labeled probes and section in situ hybridization using 33P-labeled riboprobes were performed as described (MH Chen et al. 2004).
Molecular biology and constructs
Standard molecular biology techniques, including molecular cloning, genomic DNA preparation, RNA isolation, PCR, RT–PCR, and Southern analysis, were performed as described (Sambrook and Russell 2001; Ausubel et al. 2003; Nagy et al. 2003).
Flag-mGli1, Flag-mGli2, and Flag-hGli3 were described previously (Gerber et al. 2007). Mouse Smo was obtained from Haruhiko Akiyama. The zebrafish Smo cDNA was a gift from Monte Westerfield, and the fly Smo cDNA was a gift from Jin Jiang. Smo cDNAs were C-terminally tagged with Flag or Myc and were cloned into pcDNA3, pEF-V5-His-TOPO, or pCS2+ for transient overexpression. Mouse Sufu cDNA was N-terminally tagged with Flag or Myc. The fly Sufu cDNA was a gift from Jin Jiang. cDNAs encoding zebrafish Sufu and mouse Spop and Ext2 were obtained from Open BioSystems and N- or C-terminally tagged with Flag or Myc. Gli, Sufu, and Spop cDNAs were cloned into pcDNA3 (Invitrogen) for transient overexpression or pBABE-puro for retroviral overexpression. Detailed cloning strategies and maps are available upon request. A dominant-negative eGFP-Kif3b construct was a gift from Andy Peterson, and the Cul3-Myc construct was from P. Renee Yew. The Flag-FoxC2 and Flag-MyoD constructs were gifts from Brian Black.
Derivation of MEFs
MEFs were derived from wild-type, Sufu−/−, Kif3a−/−, Ptch1−/−, Smo−/−, Gli2−/−, and Gli3−/− embryos at 9.5 dpc and cultured in DMEM, supplemented with 15% fetal bovine serum (FBS), L-glutamine (Invitrogen), nonessential amino acids (Invitrogen), 1 mM sodium pyruvate (Invitrogen), and penicillin/streptomycin (Invitrogen). These cells were subsequently immortalized with recombinant retroviruses encoding the simian virus (SV) 40 large T antigen (Brown et al. 1986). To immortalize MEFs, 2 mL of viral conditioned-medium mixed with polybrene (8 μg/mL) were added to MEFs at ∼50% confluence on a 6-cm plate. After 1 h of incubation, viral conditioned medium was removed and replaced with fresh medium. Viral infection was repeated several times in 48 h to achieve maximum infection efficiency. MEFs were subsequently selected by adding G418 (500 μg/mL) to the medium 24 h after viral infection was completed. G418-resistant clones that appeared after 2 wk of G418 selection were picked and expanded. Immortalized MEFs were maintained in culture medium supplemented with 100–200 μg/mL G418.
Antibody production
Partial mouse cDNAs encoding Smoothened (Smo, amino acids 550–793), Patched 1 (Ptch1, amino acids 1235–1414), Gli2 (amino acids 327–442), and Gli3 (amino acids 395–500) were cloned into pRSET (Invitrogen), pVCH6, or pGEX (Amersham) expression vectors. 6xHis or GST fusion proteins were expressed in BL21 (DE3) pLys bacteria and purified on Ni-NTA (Qiagen) or glutathione-Sepharose resin (Amersham) according to the manufacturer's instructions. Purified antigen was injected into rabbits (Animal Pharm) for generation of polyclonal antibodies. Antibodies were affinity-purified from crude serum using Affigel-10 beads (Bio-Rad) conjugated with Ni-NTA-purified 6xHis-antigen fusion proteins. Sufu antibodies (sc-28847) were purchased from Santa Cruz Biotechnologies.
Cell culture, transient transfections, Western blotting, and immunoprecipitation
HEK 293T cells and transformed MEF lines were maintained in DMEM supplemented with 10% fetal bovine serum (Invitrogen), penicillin/streptomycin (Invitrogen), and L-glutamate (Invitrogen). For protein expression, cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Forty-eight hours later, cells were harvested and lysed in RIPA buffer (50 mM Tris-Cl at pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% SDS, 1% Na deoxycholate, protease inhibitors). Lysates were sheared with a 20-gauge 0.5-in needle, and 6× Laemmli loading buffer was added. For detecting Gli2 and Gli3 proteins, samples were resolved on 5% SDS-PAGE gels and transferred to PVDF membranes following standard procedures (Harlow and Lane 1999). After transfer, membranes were blocked for 1 h in TBST (Tris-buffered saline with 0.1% Tween 20)/5% nonfat dry milk at room temperature and incubated with rabbit anti-Gli2 (1:3000) or rabbit anti-Gli3 (1:1000) antibodies in TBST/3% bovine serum albumin (BSA) overnight at 4°C. The membranes were then washed extensively with TBST and incubated with donkey anti-rabbit HRP (1:3000) for 1 h at room temperature. For detecting mouse Sufu, Flag-Spop, Myc-Spop, Flag-Ext2, Flag-FoxC2, Flag-MyoD, and tubulin, the following primary and secondary antibodies were used: rabbit anti-Sufu (1:500; Santa Cruz Biotechnologies), rabbit anti-Flag (1:1000; Sigma), mouse anti-Flag (1:1000; Sigma), rabbit anti-Myc (1:1000; Sigma), mouse anti-α-tubulin (1:2000; Sigma); donkey anti-rabbit HRP (1:2000; Jackson Laboratories), donkey anti-mouse HRP (1:2000; Jackson Laboratories). Chemiluminescent detection was performed using ECL Plus detection reagents (Amersham).
For immunoprecipitation, cells were harvested 48 h post-transfection and lysed in lysis buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris-Cl at pH 7.5, 1 mM EDTA, 0.5 mM PMSF, 2 μg/mL pepstatin A, 10 μg/mL leupeptin, 5 μg/mL aprotinin). Lysates were sheared with a 20-gauge needle and remained on ice for 30 min. Lysates were then clarified by centrifugation at 20,817g for 20 min at 4°C. The supernatant was removed and bound to 50 μL of anti-Flag M2 or anti-HA agarose beads (Sigma) for 4 h at 4°C with constant nutation. Beads were washed three times with lysis buffer prior to addition of sample buffer. Immunoprecipitated proteins were analyzed by 7.5% SDS-PAGE and transferred to PVDF for immunoblotting. Antibodies used were rabbit anti-Flag (1:2000; Sigma) and rabbit anti-HA (1:1000; Sigma).
Retroviral generation and transduction of stable cell lines
HEK 293T cells were transfected with pCL-ECO (Naviaux et al. 1996) and pBABE-puro containing Smo or Sufu cDNAs derived from mouse, zebrafish, or fly. Supernatant was collected 72 h post-transfection, filtered through a 0.45-μm syringe filter (Nalgene) and added to 50% confluent MEFs with 8 μg/mL polybrene (Sigma). Two days after the addition of retroviral supernatant, MEFs were split 1:10 and selected with puromycin (2.5 μg/mL). Presence of stable expression was verified by Western blotting and/or immunofluorescence.
shRNA design, lentiviral design, production, and infection
shRNAs were designed using the pSicOligomaker application (Reynolds et al. 2004). To select shRNA sequences with minimal homology with other mouse transcripts, sequences were compared against the mouse non-Refseq RNA database using BLAST. Oligonucleotides encoding shRNAs were annealed and inserted into the pLentiLox3.7 vector. To create lentiviral supernatants, HEK 293T cells were transfected with the appropriate pLentiLox3.7 vector and the packaging vectors pLP1, pLP2, and pLP/VSV-G using Lipofectamine (Invitrogen). Seventy-two hours post-transfection, supernatants were harvested and filtered through a 0.45-μm cellulose acetate filter (Nalgene). Lentivirus was concentrated either 10-fold using a Centriprep Ultracel YM-10 device (Millipore) or 100-fold by ultracentrifugation. MEFs at 50% confluence were infected with concentrated lentivirus supplemented with 8 μg/mL polybrene. Knockdown was verified by Western blotting if appropriate antibodies were available or, in other cases, knockdown was assessed by extracting RNA using an RNA Midi kit (Qiagen) followed by RT–PCR following standard procedures. The following 19-mer sequences were used for shRNA-mediated knockdown: mouse Sufu (NM_015752): GAGTTGACGTTTCGTCTGA (nucleotides 540–558), GTAGTGACTTTCTTCCAGA (nucleotides 765–783), GGCGGGGACTGGAGATTAA (nucleotides 1126–1144), GGAGGACTTAGAAGATCTA (nucleotides 1520–1538); mouse Kif3a (NM_008443): GAACTATCACCGTCCATAA (nucleotides 278–296), GGAGAGAGACCCATTTGAA (nucleotides 2070–2088), GACCGTAATTGATTCTTTA (nt 2226-2244); mouse Gli1 (NM_010296): TCGGAGTTCAGTCAAATTA (nucleotides 383–391), ACATGCTCCGTGCCAGATA (nucleotides 1927–1945), AAGCTCAGCTGGTGTGTAA (nucleotides 2848–66); mouse Spop (NM_025287): GACTCAGTTTAACCTTCAA (nucleotides 163–181), GAAAGGGCTAGATGAAGAA (nucleotides 407–425), GTACAAGACTCTGTCAATA (nucleotides 670–696), GAAGCGGTAGGATTTATTT (nucleotides 2615–2633)
Immunofluorescence
Cells were fixed in 4% paraformaldehyde for 15 min at room temperature. Standard procedures for immunostaining were subsequently followed. The primary antibodies used were mouse anti-acetylated tubulin (1:2000; Sigma), mouse anti-Flag (1:1000; Sigma), mouse anti-Myc (1:1000; Santa Cruz Biotechnologies), rabbit anti-Myc (1:1000; Sigma), rabbit anti-Smo (1:500), rabbit anti-Gli2 (1:500), rabbit anti-Gli3 (1:500), and rabbit anti-Sufu (1:100; Santa Cruz Biotechnologies). Secondary antibodies and conjugates used were donkey anti-mouse AlexaFluor 594 (1:2000; Molecular Probes), donkey anti-rabbit AlexaFluor 488 (1:2000; Molecular Probes), and DAPI (1:10,000; Sigma). Fluorescent images were acquired using a SPOT 2.3 camera connected to a Nikon E1000 epifluorescence microscope. Adjustment of RGB histograms and channel merges were performed using Advanced SPOT and NIH Image J. Fluorescent confocal images were acquired using a Nikon TE2000U inverted microscope with a Yogokawa CSU22 spinning disk confocal (Solamere Technology Group), a Photometrics Cascade II Camera, and MicroManager software (Vale laboratory, University of California at San Francisco). Images were acquired with a 100× oil-immersion lens and a 1.5× zoom adapter (Nikon) using two laser lines (488 nm and 568 nm). Confocal stacks were collected using a 0.25-μm step size along the Z-axis. Stacks were analyzed and xy projections generated using NIH ImageJ. Deconvolution was performed with the Iterative Deconvolve 3D plugin (Robert Dougherty, OptiNav, Inc.).
Hh reporter activity assays
Prior to the day of transfection, MEFs were seeded at a density of 5 × 104 to 1 × 105 cells per milliliter (dependent on cell line) in 24-well plates. The next day, cells were transfected with a 4:5:1 ratio of pcDNA3/expression construct 8xGliBS-luc:pRL-TK using Fugene 6 (Roche) according to the manufacturer's instructions. Forty-eight hours after transfection, cells were changed to low-serum medium (DMEM supplemented with 0.5% newborn calf serum and penicillin/streptomycin) for a further 30–48 h. Cells were harvested, and firefly and Renilla luciferase activities were determined using the Dual Luciferase Reporter Assay System (Promega) and an LmaxII 384 luminometer (Molecular Devices). Multiple (three or more) assays were performed; each sample was assayed in triplicate.
Ubiquitination assays
HEK 293T cells were transfected with HA-tagged Ub, Flag-tagged Glis (Gli1, Gli2, or Gli3), Sufu, Spop, and Cullin3 using Lipofectamine 2000 (Invitrogen). Forty hours post-transfection, cells were treated with 50 μM MG132 (Sigma). Six hours later, cells were harvested and lysed in 1% SDS lysis buffer (50 mM Tris-HCl at pH 7.5, 0.5 mM EDTA, 1% SDS) and boiled for 10 min. Lysates were cleared by centrifugation at 14,000g for 10 min. The supernantant was diluted 10 times with NETN buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris-HCl at pH 7.5, 0.1% NP-40), and then immunoprecipitated with mouse anti-Flag agarose beads (Sigma) overnight at 4°C. The beads were washed with ice-cold NETN buffer three times and then subjected to immunoblot analysis with mouse anti-HA antibody.
Acknowledgments
We thank Phil Beachy, Brian Black, Jin Jiang, Tom Kornberg, Michael McManus, Geeta Narlikar, Andy Peterson, Danny Reinberg, Thea Tlsty, and P. Renee Yew for constructs and reagents; Erica Yao for technical assistance; and Jau-Nian Chen and Ross Metzger for critical reading of the manuscript. Some data for this study were acquired at the Nikon Imaging Center at UCSF/QB3. This work was supported by a post-doctoral fellowship from the American Heart Association to M.-H.C., grants from the National Cancer Institute of Canada and Canadian Institutes of Health Research to C.-c.H., and grants from the National Institutes of Health and a Career Investigator Award from the American Lung Association to P.-T.C.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1794109.
Supplemental material is available at http://www.genesdev.org.
References
- Aikin RA, Ayers KL, Therond PP. The role of kinases in the Hedgehog signalling pathway. EMBO Rep. 2008;9:330–336. doi: 10.1038/embor.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimaru H, Chen Y, Dai P, Hou DX, Nonaka M, Smolik SM, Armstrong S, Goodman RH, Ishii S. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature. 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. Wiley; New York: 2003. [Google Scholar]
- Aza-Blanc P, Ramirez-Weber FA, Laget MP, Schwartz C, Kornberg TB. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- Bai CB, Joyner AL. Gli1 can rescue the in vivo function of Gli2. Development. 2001;128:5161–5172. doi: 10.1242/dev.128.24.5161. [DOI] [PubMed] [Google Scholar]
- Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- Bhatia N, Thiyagarajan S, Elcheva I, Saleem M, Dlugosz A, Mukhtar H, Spiegelman VS. Gli2 is targeted for ubiquitination and degradation by β-TrCP ubiquitin ligase. J Biol Chem. 2006;281:19320–19326. doi: 10.1074/jbc.M513203200. [DOI] [PubMed] [Google Scholar]
- Brown M, McCormack M, Zinn KG, Farrell MP, Bikel I, Livingston DM. A recombinant murine retrovirus for simian virus 40 large T cDNA transforms mouse fibroblasts to anchorage-independent growth. J Virol. 1986;60:290–293. doi: 10.1128/jvi.60.1.290-293.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes & Dev. 2002a;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci. 2002b;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Li Y-J, Kawakami T, Xu SM, Chuang P-T. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes & Dev. 2004;18:641–659. doi: 10.1101/gad.1185804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Knezevic V, Ervin V, Hutson R, Ward Y, Mackem S. Direct interaction with Hoxd proteins reverses Gli3-repressor function to promote digit formation downstream of Shh. Development. 2004;131:2339–2347. doi: 10.1242/dev.01115. [DOI] [PubMed] [Google Scholar]
- Chen MH, Gao N, Kawakami T, Chuang P-T. Mice deficient in the fused homolog do not exhibit phenotypes indicative of perturbed hedgehog signaling during embryonic development. Mol Cell Biol. 2005;25:7042–7053. doi: 10.1128/MCB.25.16.7042-7053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SY, Bishop JM. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18–mSin3 corepressor complex. Proc Natl Acad Sci. 2002;99:5442–5447. doi: 10.1073/pnas.082096999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AF, Yu KP, Brueckner M, Brailey LL, Johnson L, McGrath JM, Bale AE. Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development. 2005;132:4407–4417. doi: 10.1242/dev.02021. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Daoud F, Blanchet-Tournier MF. Expression of the human FUSED protein in Drosophila. Dev Genes Evol. 2005;215:230–237. doi: 10.1007/s00427-005-0469-x. [DOI] [PubMed] [Google Scholar]
- De Rivoyre M, Ruel L, Varjosalo M, Loubat A, Bidet M, Therond P, Mus-Veteau I. Human receptors patched and smoothened partially transduce hedgehog signal when expressed in Drosophila cells. J Biol Chem. 2006;281:28584–28595. doi: 10.1074/jbc.M512986200. [DOI] [PubMed] [Google Scholar]
- Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: A sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- Di Marcotullio L, Ferretti E, Greco A, De Smaele E, Po A, Sico MA, Alimandi M, Giannini G, Maroder M, Screpanti I, et al. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol. 2006;8:1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- Di Marcotullio L, Ferretti E, Greco A, De Smaele E, Screpanti I, Gulino A. Multiple ubiquitin-dependent processing pathways regulate hedgehog/gli signaling: Implications for cell development and tumorigenesis. Cell Cycle. 2007;6:390–393. doi: 10.4161/cc.6.4.3809. [DOI] [PubMed] [Google Scholar]
- Ding Q, Fukami S, Meng X, Nishizaki Y, Zhang X, Sasaki H, Dlugosz A, Nakafuku M, Hui C. Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr Biol. 1999;9:1119–1122. doi: 10.1016/s0960-9822(99)80482-5. [DOI] [PubMed] [Google Scholar]
- Dussillol-Godar F, Brissard-Zahraoui J, Limbourg-Bouchon B, Boucher D, Fouix S, Lamour-Isnard C, Plessis A, Busson D. Modulation of the Suppressor of fused protein regulates the Hedgehog signaling pathway in Drosophila embryo and imaginal discs. Dev Biol. 2006;291:53–66. doi: 10.1016/j.ydbio.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenschwiler JT, Bulgakov OV, Qin J, Li T, Anderson KV. Mouse Rab23 regulates hedgehog signaling from smoothened to Gli proteins. Dev Biol. 2006;290:1–12. doi: 10.1016/j.ydbio.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Fan J, Beck KA. A role for the spectrin superfamily member Syne-1 and kinesin II in cytokinesis. J Cell Sci. 2004;117:619–629. doi: 10.1242/jcs.00892. [DOI] [PubMed] [Google Scholar]
- Gerber AN, Wilson CW, Li YJ, Chuang PT. The hedgehog regulated oncogenes Gli1 and Gli2 block myoblast differentiation by inhibiting MyoD-mediated transcriptional activation. Oncogene. 2007;26:1122–1136. doi: 10.1038/sj.onc.1209891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- Han YG, Kwok BH, Kernan MJ. Intraflagellar transport is required in Drosophila to differentiate sensory cilia but not sperm. Curr Biol. 2003;13:1679–1686. doi: 10.1016/j.cub.2003.08.034. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, editor. RNAi: A guide to gene silencing. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. [Google Scholar]
- Harlow E, Lane D. Using antibodies: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Munoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y, van Lohuizen M. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci. 2005;102:7635–7640. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper JE, Scott MP. Communicating with Hedgehogs. Nat Rev Mol Cell Biol. 2005;6:306–317. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- Hu MC, Mo R, Bhella S, Wilson CW, Chuang P-T, Hui CC, Rosenblum ND. GLI3-dependent transcriptional repression of Gli1, Gli2 and kidney patterning genes disrupts renal morphogenesis. Development. 2006;133:569–578. doi: 10.1242/dev.02220. [DOI] [PubMed] [Google Scholar]
- Huang CJ, Chen CY, Chen HH, Tsai SF, Choo KB. TDPOZ, a family of bipartite animal and plant proteins that contain the TRAF (TD) and POZ/BTB domains. Gene. 2004;324:117–127. doi: 10.1016/j.gene.2003.09.022. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: Conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Hui CC, Joyner AL. A mouse model of Greig cephalopolysyndactyly syndrome: The extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat Genet. 1993;3:241–246. doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]
- Huntzicker EG, Estay IS, Zhen H, Lokteva LA, Jackson PK, Oro AE. Dual degradation signals control Gli protein stability and tumor formation. Genes & Dev. 2006;20:276–281. doi: 10.1101/gad.1380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, Placzek M. Orchestrating ontogenesis: Variations on a theme by sonic hedgehog. Nat Rev Genet. 2006;7:841–850. doi: 10.1038/nrg1969. [DOI] [PubMed] [Google Scholar]
- Jia J, Jiang J. Decoding the Hedgehog signal in animal development. Cell Mol Life Sci. 2006;63:1249–1265. doi: 10.1007/s00018-005-5519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Tong C, Jiang J. Smoothened transduces Hedgehog signal by physically interacting with Costal2/Fused complex through its C-terminal tail. Genes & Dev. 2003;17:2709–2720. doi: 10.1101/gad.1136603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner AL. Gene targeting: A practical approach. 2nd ed. Oxford University Press; New York: 2000. [Google Scholar]
- Kogerman P, Grimm T, Kogerman L, Krause D, Unden AB, Sandstedt B, Toftgard R, Zaphiropoulos PG. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat Cell Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- Koudijs MJ, den Broeder MJ, Keijser A, Wienholds E, Houwing S, van Rooijen EM, Geisler R, van Eeden FJ. The zebrafish mutants dre, uki, and lep encode negative regulators of the hedgehog signaling pathway. PLoS Genet. 2005;1:e19. doi: 10.1371/journal.pgen.0010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs JJ, Whalen EJ, Liu R, Xiao K, Kim J, Chen M, Wang J, Chen W, Lefkowitz RJ. β-Arrestin-mediated localization of smoothened to the primary cilium. Science. 2008;320:1777–1781. doi: 10.1126/science.1157983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Platt KA, Censullo P, Ruiz i Altaba A. Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development. 1997;124:2537–2552. doi: 10.1242/dev.124.13.2537. [DOI] [PubMed] [Google Scholar]
- Lefers MA, Wang QT, Holmgren RA. Genetic dissection of the Drosophila Cubitus interruptus signaling complex. Dev Biol. 2001;236:411–420. doi: 10.1006/dbio.2001.0345. [DOI] [PubMed] [Google Scholar]
- Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- Low WC, Wang C, Pan Y, Huang XY, Chen JK, Wang B. The decoupling of Smoothened from Gαi proteins has little effect on Gli3 protein processing and Hedgehog-regulated chick neural tube patterning. Dev Biol. 2008;321:188–196. doi: 10.1016/j.ydbio.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L, Beachy PA. The Hedgehog response network: Sensors, switches, and routers. Science. 2004;304:1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- Lum L, Zhang C, Oh S, Mann RK, von Kessler DP, Taipale J, Weis-Garcia F, Gong R, Wang B, Beachy PA. Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol Cell. 2003;12:1261–1274. doi: 10.1016/s1097-2765(03)00426-x. [DOI] [PubMed] [Google Scholar]
- Marigo V, Johnson RL, Vortkamp A, Tabin CJ. Sonic hedgehog differentially regulates expression of GLI and GLI3 during limb development. Dev Biol. 1996;180:273–283. doi: 10.1006/dbio.1996.0300. [DOI] [PubMed] [Google Scholar]
- Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc Natl Acad Sci. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant M, Vajdos FF, Ultsch M, Maun HR, Wendt U, Cannon J, Desmarais W, Lazarus RA, de Vos AM, de Sauvage FJ. Suppressor of fused regulates Gli activity through a dual binding mechanism. Mol Cell Biol. 2004;24:8627–8641. doi: 10.1128/MCB.24.19.8627-8641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant M, Evangelista M, Luoh SM, Frantz GD, Chalasani S, Carano RA, van Hoy M, Ramirez J, Ogasawara AK, McFarland LM, et al. Loss of the serine/threonine kinase fused results in postnatal growth defects and lethality due to progressive hydrocephalus. Mol Cell Biol. 2005;25:7054–7068. doi: 10.1128/MCB.25.16.7054-7068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot N, Basler K. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell. 1999;96:819–831. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- Methot N, Basler K. Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development. 2000;127:4001–4010. doi: 10.1242/dev.127.18.4001. [DOI] [PubMed] [Google Scholar]
- Michaud EJ, Yoder BK. The primary cilium in cell signaling and cancer. Cancer Res. 2006;66:6463–6467. doi: 10.1158/0008-5472.CAN-06-0462. [DOI] [PubMed] [Google Scholar]
- Mo R, Freer AM, Zinyk DL, Crackower MA, Michaud J, Heng HH, Chik KW, Shi XM, Tsui LC, Cheng SH, et al. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 1997;124:113–123. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the mouse embryo: A laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. [Google Scholar]
- Naviaux RK, Costanzi E, Haas M, Verma IM. The pCL vector system: Rapid production of helper-free, high-titer, recombinant retroviruses. J Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis E, Hui CC. Hedgehog signaling and congenital malformations. Clin Genet. 2005;67:193–208. doi: 10.1111/j.1399-0004.2004.00360.x. [DOI] [PubMed] [Google Scholar]
- Ocbina PJ, Anderson KV. Intraflagellar transport, cilia, and mammalian Hedgehog signaling: Analysis in mouse embryonic fibroblasts. Dev Dyn. 2008;237:2030–2038. doi: 10.1002/dvdy.21551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden SK, Ascano M, Jr, Stegman MA, Suber LM, Hooper JE, Robbins DJ. Identification of a functional interaction between the transmembrane protein Smoothened and the kinesin-related protein Costal2. Curr Biol. 2003;13:1998–2003. doi: 10.1016/j.cub.2003.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden SK, Ascano M, Jr, Stegman MA, Robbins DJ. Regulation of Hedgehog signaling: A complex story. Biochem Pharmacol. 2004;67:805–814. doi: 10.1016/j.bcp.2004.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden SK, Fei DL, Schilling NS, Ahmed YF, Hwa J, Robbins DJ. G protein Gαi functions immediately downstream of Smoothened in Hedgehog signalling. Nature. 2008;456:967–970. doi: 10.1038/nature07459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmeyer JT, Kalderon D. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature. 1998;396:749–753. doi: 10.1038/25533. [DOI] [PubMed] [Google Scholar]
- Orenic TV, Slusarski DC, Kroll KL, Holmgren RA. Cloning and characterization of the segment polarity gene cubitus interruptus dominant of Drosophila. Genes & Dev. 1990;4:1053–1067. doi: 10.1101/gad.4.6.1053. [DOI] [PubMed] [Google Scholar]
- Oro AE. The primary cilia, a ‘Rab-id’ transit system for hedgehog signaling. Curr Opin Cell Biol. 2007;19:691–696. doi: 10.1016/j.ceb.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paces-Fessy M, Boucher D, Petit E, Paute-Briand S, Blanchet-Tournier MF. The negative regulator of Gli, Suppressor of fused (Sufu), interacts with SAP18, Galectin3 and other nuclear proteins. Biochem J. 2004;378:353–362. doi: 10.1042/BJ20030786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preat T. Characterization of Suppressor of fused, a complete suppressor of the fused segment polarity gene of Drosophila melanogaster. Genetics. 1992;132:725–736. doi: 10.1093/genetics/132.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preat T, Therond P, Limbourg-Bouchon B, Pham A, Tricoire H, Busson D, Lamour-Isnard C. Segmental polarity in Drosophila melanogaster: Genetic dissection of fused in a Suppressor of fused background reveals interaction with costal-2. Genetics. 1993;135:1047–1062. doi: 10.1093/genetics/135.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Perez SE, Yang Z, Xu J, Ritchings BW, Steller H, Goldstein LS. Kinesin-II is required for axonal transport of choline acetyltransferase in Drosophila. J Cell Biol. 1999;147:507–518. doi: 10.1083/jcb.147.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, Therond PP. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Ruel L, Rodriguez R, Gallet A, Lavenant-Staccini L, Therond PP. Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat Cell Biol. 2003;5:907–913. doi: 10.1038/ncb1052. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: A laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: Implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- Sisson JC, Ho KS, Suyama K, Scott MP. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90:235–245. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- Smelkinson MG, Zhou Q, Kalderon D. Regulation of Ci-SCFSlimb binding, Ci proteolysis, and hedgehog pathway activity by Ci phosphorylation. Dev Cell. 2007;13:481–495. doi: 10.1016/j.devcel.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegman MA, Vallance JE, Elangovan G, Sosinski J, Cheng Y, Robbins DJ. Identification of a tetrameric hedgehog signaling complex. J Biol Chem. 2000;275:21809–21812. doi: 10.1074/jbc.C000043200. [DOI] [PubMed] [Google Scholar]
- Stegman MA, Goetz JA, Ascano M, Jr, Ogden SK, Nybakken KE, Robbins DJ. The Kinesin-related protein Costal2 associates with membranes in a Hedgehog-sensitive, Smoothened-independent manner. J Biol Chem. 2004;279:7064–7071. doi: 10.1074/jbc.M311794200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svard J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergstrom A, Ericson J, Toftgard R, Teglund S. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Taipale J. Hedgehog: Functions and mechanisms. Genes & Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Li SP, Taipale J. Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev Cell. 2006;10:177–186. doi: 10.1016/j.devcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Vokes SA, Ji H, McCuine S, Tenzen T, Giles S, Zhong S, Longabaugh WJ, Davidson EH, Wong WH, McMahon AP. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development. 2007;134:1977–1989. doi: 10.1242/dev.001966. [DOI] [PubMed] [Google Scholar]
- Vokes SA, Ji H, Wong WH, McMahon AP. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes & Dev. 2008;22:2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QT, Holmgren RA. Nuclear import of cubitus interruptus is regulated by hedgehog via a mechanism distinct from Ci stabilization and Ci activation. Development. 2000;127:3131–3139. doi: 10.1242/dev.127.14.3131. [DOI] [PubMed] [Google Scholar]
- Wang B, Li Y. Evidence for the direct involvement of βTrCP in Gli3 protein processing. Proc Natl Acad Sci. 2006;103:33–38. doi: 10.1073/pnas.0509927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Wang G, Amanai K, Wang B, Jiang J. Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes & Dev. 2000;14:2893–2905. doi: 10.1101/gad.843900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff C, Roy S, Ingham PW. Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr Biol. 2003;13:1169–1181. doi: 10.1016/s0960-9822(03)00461-5. [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhao Y, Tong C, Wang G, Wang B, Jia J, Jiang J. Hedgehog-regulated Costal2-kinase complexes control phosphorylation and proteolytic processing of Cubitus interruptus. Dev Cell. 2005;8:267–278. doi: 10.1016/j.devcel.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006;10:719–729. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tong C, Jiang J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature. 2007;450:252–258. doi: 10.1038/nature06225. [DOI] [PubMed] [Google Scholar]