In 2002, the Editorial that accompanied the launch of Nature Reviews Drug Discovery stated that “[d]espite all the excitement that accompanies each wave of technical innovation, from protein structure determination to proteomics, and from combinatorial chemistry to e-clinical trials, the fundamental truth is that pharmaceutical companies are not producing drugs any faster than they were before these innovations came along” (Anon, 2002).
A subsequent Editorial written on the occasion of the fifth anniversary of the journal asked whether the industry was in a better state than it had been 5 years previously, and concluded that the answer was a “resounding no” (Anon, 2007). In the intervening period, Leroy Hood and Roger Perlmutter had already predicted that the pharmaceutical industry would lose ∼US$80 billion in revenue by 2008, and stated that the current approaches to drug discovery and development could not keep up with demand (Hood & Perlmutter, 2004). This article now seems to have been prophetic: in a valedictory article published in the Harvard Business Review in 2008, Jean-Pierre Garnier, former chief executive officer of GlaxoSmithKline, pointed out that between December 2000 and February 2008, the industry lost ∼US$850 billion in shareholder value, with a concomitant decline in share prices from an average of 32 times earnings to 13 times earnings (Garnier, 2008).
During the past 10 years, the cost of bringing a single new drug to market has escalated hugely to somewhere in the region of US$1 billion; however, the success in doing so has approximately halved compared with the early 1990s, and the time taken has doubled to ∼12 years, although efforts are succeeding in reducing this to some extent. Industry statistics show an increased productivity in recent years in delivering candidate drugs from discovery into development; however, this has not been matched by success in the clinic, and there have been several well-publicized failures of drugs in the later stages of development (Anon, 2004, 2007; Frantz, 2007). This indicates that, although the output of projects from discovery into development has increased, the quality of the output has not. Responding to this situation by simply playing the ‘numbers game'—increasing the number of projects and speed of delivery through the pipeline in the hope of increasing the chances of hitting a successful outcome—would be similar to continually pumping more fuel into an inefficient engine just to maintain its output. Alternative approaches need to be considered to tackle this problem.
The key weakness of the current reductionist practice is that it cannot predict how the wider physiological system will behave in a quantitative way...
Despite the wealth of detailed information generated by the technical advances made during the past 20 years, the delivery of innovative medicines that target complex diseases remains a major challenge for the pharmaceutical industry. The failure of projects at late stages in their development owing to lack of efficacy, undesirable side effects or toxicity remains a major problem. The question is, why? Were the hopes and expectations generated in the post-genome era unrealistically high or is there something fundamentally wrong with the way in which we approach the problem? The answer, probably, is a little bit of both.
...[system biology's] potential impact on interpreting the complexity of physiological systems that underpin the development of new medicines still needs to be tested
An increasingly popular view is that, although we now have a lot of detailed information—anatomical, physiological, genetic, metabolic and so on—about the human organism, we still suffer from an incomplete understanding of the complex cellular and physiological interactions that determine how patients respond to their treatments. This might seem odd given the progress made in biological and medical science in recent years; however, pharmaceutical research and development generally has been an empirically data-driven and qualitatively oriented activity. Typically in this process, each drug and target combination tends to be considered in isolation, and removed from its physiological context. This can often be misleading, as the mechanisms contributing to the development of complex diseases are not just the result of a single gene and its protein product. Targets, and their response to putative therapies, need to be studied in their physiological context if new medicines are to be effective. The key weakness of the current reductionist practice is that it cannot predict how the wider physiological system will behave in a quantitative way: the dynamic ‘context'—pathway, cell, patient or population of patients—is missing. Approaches that combine mathematics, engineering, statistics and experimental science to create models that can be used to simulate complex networks, and to generate testable hypotheses, offer the hope that we can now begin to look at the dynamics of physiological network responses rather than static, isolated entities. Such approaches are encompassed in the term ‘systems biology'.
The impression that systems biology is something new—‘the next big thing' in science—has led almost unavoidably to a measure of hype in the lay, financial and industry media, which in turn has elicited responses ranging from deep scepticism to blind enthusiasm. However, the concepts of systems biology are not new; many agree that the foundations were laid by the work of Alan Hodgkin and Andrew Huxley in the 1950s on ion channels and nerve impulses (Hodgkin & Huxley, 1952). Current awareness of what is systems biology has increased largely through its application to tackle biological complexity. Many would agree that a dictionary could be compiled to cover the diversity of definitions that have been devised to describe systems biology; however, one definition from Leroy Hood lays the foundations for the others: “systems biology represents an analytical approach to the relationships among elements of a system with the goal of understanding its [the system's] emergent properties” (Hood & Perlmutter, 2004).
In other words, systems biology is the quantitative analysis of how the components of complex biological systems interact dynamically to enable the systems to function. Such an analysis can be applied at the level of molecules, cells, organs or entire organisms. One possible approach to address the pharmaceutical challenges described earlier is to apply these principles to integrate and interpret the wealth of complex biological network information that we have accumulated and to apply it to drug development. In contrast to reductionist scientific methods, in which individual parts of a complex system are studied in isolation and in detail, this approach uses a combination of experimentally derived data—that is, parameters—and computational models of the behaviour of a system to analyse the complexity of multi-parameter interactions that underpin biological functions.
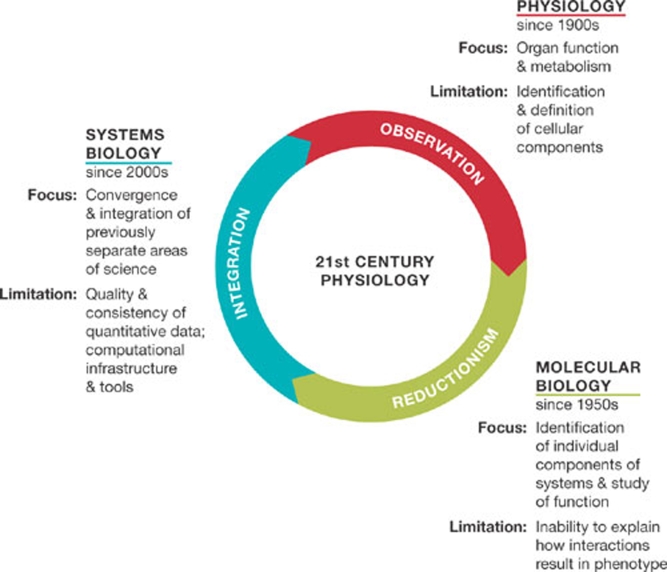
Systems biology is widely regarded as the natural successor to the Human Genome Project, as it provides the means to integrate complex data sets and the tools to undertake physiological studies (Fig 1). Mathematical modelling and computer simulation have been used to provide an integrated, dynamic view of how biological systems might respond to various interventions. Large amounts of time, effort and money have been devoted to such studies, and many examples of academic success now demonstrate that they can work. The potential value of models and simulations to industrial projects in general is recognized, and their application, particularly to help reduce the failure of drug projects, is gathering favour (Bangs & Paterson, 2003; Uehling, 2004; van der Greef & McBurney, 2005; McGee, 2005; Bangs, 2005; Chiswell et al, 2007; Jefferys et al, 2008; Dollery & Kitney, 2007).
Figure 1.
Systems biology brings physiology full circle: from the focus on organ function and metabolism at the start of the twentieth century, through the acquisition of detailed components and individual function, to the point where an ‘integrator' of complex, dynamic and quantitative data was needed to provide the toolbox for the twenty-first century.
These reports outline the opportunities for the application of systems biology in drug discovery, and broadly advocate a move towards a ‘predict and test' strategy rather than the current reductionist, high-throughput approaches, which are based on an element of ‘guess and pray'. The Economist, reporting on an analysis done in 1999 by PricewaterhouseCoopers, pointed out that the application of modelling and simulation to drug discovery could save US$200 million and 2–3 years in development time for each drug candidate (Anon, 2005). However, industry remains largely sceptical, as there are few success stories with tangible commercial effects, and concerns remain over the ability to reduce these approaches to routine practice. Although systems biology might be widely accepted as having proven itself in an academic sense, its potential impact on interpreting the complexity of physiological systems that underpin the development of new medicines still needs to be tested. At a time of significant economic and regulatory constraint for the industry, when the appetite for exploring unproven technologies is likely to be low, the challenge is to find ways to prototype and exemplify ‘blue-sky thinking'—being able to risk failure intelligently in order to improve our understanding. Therefore, an opportunity exists to explore how the academic advances in systems biology can be exploited to help overcome some of the hurdles and challenges of drug development.
The opportunity to exploit academic advances in systems biology is undoubtedly real, but the question remains as to who will do it and how
Here, I present three examples of the application of modelling and simulation to drug-discovery projects. First, working at the cellular level, a model of the epidermal growth factor receptor pathway was used to explore the mechanisms that might be responsible for increased sensitivity to Iressa™ (AstraZeneca, London, UK) in patients carrying a receptor mutation (Lynch et al, 2004; Paez et al, 2004). Computational biologists were able to run a range of simulations and generate a series of hypotheses that could explain the published data, and which were evaluated experimentally in laboratory studies (Hendricks et al, 2006a, b).
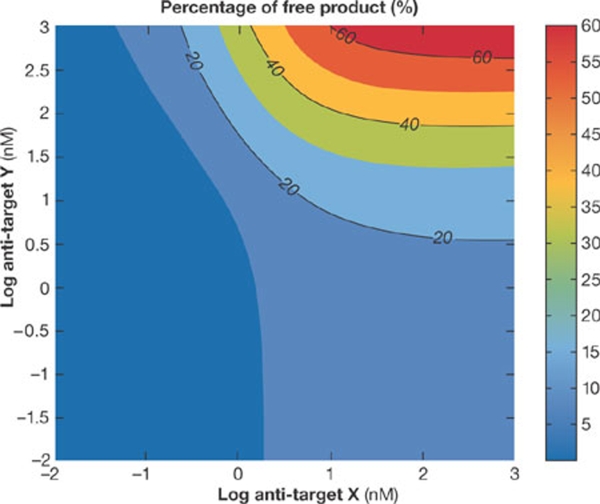
Second, another area of increasing interest is the potential to investigate combination therapies. Fig 2 shows an example of such an application. In this case, the objective was to find the optimal strategy to increase the level of ‘free product' in the relevant biological compartment. To address this question, a model of the network that regulates levels of the target product was designed, validated and used to run simulations of various drug combinations. The model predicted that tenfold lower doses of a combination would deliver significantly greater levels of the target product than either of the individual therapeutic options on their own. Using a model to explore and test a wide range of options and to generate some specific, testable hypotheses cuts down the amount of expensive laboratory experiments needed to achieve the same endpoint, and allows teams to focus attention on specific, prioritized options.
Figure 2.
Evaluation of combination therapy. The objective is to provide a therapeutic that can maximize the amount of ‘free product' in the relevant biological effect compartment. A tailored mathematical model of the biological network involved in regulating the target ‘product' was developed, validated and used to explore different combinations of therapeutic intervention to select those that might be optimal. Here, it can be seen that a 1-μM dose of either anti-X or anti-Y on their own would deliver <20% or <5% free product, whereas in combination, at a tenfold lower dose of 100 nM, they delivered >40% free product. These are hypotheses generated by a validated model that can be used to focus expensive and time-consuming experimental effort more effectively.
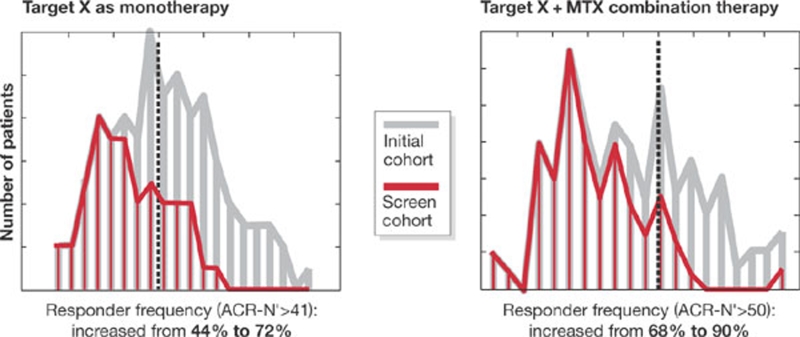
Third, at the physiological level, the biotechnology company Entelos (Foster City, CA, USA) has collaborated with major pharmaceutical companies to apply computer simulations in clinical trials. Using large-scale physiological models of human diseases, they have applied simulations to support target validation, biomarker selection, and the design and optimization of clinical trials. For example, their diabetes platform has helped to deliver a 40% reduction in time in a phase I trial, using 66% fewer patients (Kansal & Trimmer, 2005). Working with an exploratory group in AstraZeneca (London, UK) on a rheumatoid arthritis project, their models were used to identify and propose markers of patient response to a novel therapeutic used alone or in combination with standard therapy. The model identified five analytes that might be useable as patient segmentation markers. The hypothesis was that these markers could be used to identify responders and thereby segment patient populations to improve the response to therapy (Fig 3).
Figure 3.
Simulation of the impact of stratification on patient response to therapy. In this example, a physiological model representing rheumatoid arthritis was used to evaluate potential markers of patient response to a novel therapy, X, alone and in combination with standard methotrexate (MTX) therapy. The model identified five factors that, if measureable in vivo, could be used to stratify patient populations to improve response to therapy from 44% to 72% for monotherapy, and from 68% to 90% for combination therapy. Although this hypothesis would need to be tested, once again it helps to focus early experimental work on specific areas.
A reasonable question might be why, given all of the available information and evidence of apparent success, is systems biology not being embraced more enthusiastically by industry? All new scientific and technological developments are met with a degree of scepticism and initial resistance to implementation; systems biology is no different. A major reason is the fact that it is not seen as having been sufficiently ‘reduced to practice' for consistent, routine application in drug discovery. It is important to remember that the criteria for validation are much more stringent and demanding than those for academic peer review. The next phase, therefore, has to focus on compiling examples of applications that are recognized and acknowledged by industry to be relevant to its needs, that demonstrate consistency and reproducibility, and that will help to build confidence that systems biology has a key role in modernizing the development of new medicines.
...we have to find ways to test and evaluate these technologies [...] to understand what impact they can have and [...] how they can be incorporated into routine practice
Yet, how is this going to be achieved? The need to take up the challenge was identified several years ago by Hood & Perlmutter (2004) who suggested that systems biology “offers powerful new approaches” to deal with the problems in drug development. They ended their article by asking who would take the lead in effecting the necessary change. The implication was that, logically, this should be the pharmaceutical industry itself; however, as has already been mentioned in this article, the economic pressures on industry make this a difficult time to explore anything that is not going to deliver to the bottom line in the near term.
The opportunity to exploit academic advances in systems biology is undoubtedly real, but a question remains as to who will do it and how. The constraints faced by the industry have already been described. The challenge for academic centres is to find ways of focusing their activities to generate relevant evidence and applying their science to commercial benefit, while simultaneously maintaining the pursuit of academic excellence. Although this is achievable, it will require much closer, better-directed activities that work across academia and industry, which focus on making a tangible impact on delivering novel therapies.
How we might go about doing this was the subject of a closed workshop that brought together a small group of experts representing a range of areas of expertise from academia, industry, regulators and funding agencies. The product was a series of recommendations, which were not only supported by the strength of consensus among the group, but also backed up and validated by the structured analysis of the factual, evidence-based information collected as part of the pre-meeting process.
Briefly, it was proposed that the sort of impact and evidence that might help to build confidence in systems biology and influence its broader adoption in industry would come from focusing concerted efforts on a few key areas—cancer, metabolic disease and inflammation, and/or infectious disease—in which the data and tools currently available offered the greatest chances of early success. In addition, it was agreed that much could also be achieved by applying systems approaches to predictive toxicology. Rather than a lack of funding, it was the coordination and focus of work to achieve the ends needed to influence industry that was seen as the problem. It was felt that the level of complexity and size of the tasks involved effectively demanded some form of coordinated effort along the lines of that used to support the Single Nucleotide Polymorphism Consortium or the Human Genome Project. A strong consensus favoured the creation of some form of consortium to coordinate and drive forward this agenda, including the exploration of setting standards where appropriate, to ensure consistency and reproducibility of data collection and model construction. The points that emerged from this ‘think tank' were communicated initially in the form of a Commentary in Nature (Henney & Superti-Furga, 2008), backed up by an online discussion forum (network.nature.com/groups/systbiohumanhealth/forum/topics). The intention was to use this workshop, and the resulting output, as a route to inform and stimulate further debate within the scientific community, with the ultimate purpose of influencing broader understanding of the topic in general, and increased adoption by industry in particular.
These fundamentally important issues are relevant to the development of innovative medicines to tackle complex disease. The discussion about how we improve our ability to deliver effective therapies, in my view, cannot ignore the contribution that systems approaches can make. It is not sufficient to collect anecdotal examples of success; we have to find ways to test and evaluate these technologies thoroughly and in the right context, to understand what impact they can have and, if appropriate, to understand how they can be incorporated into routine practice.
The jury is still out; however, the time is right for us to gather persuasive evidence to raise confidence and ultimately influence broader adoption.
References
- Anon (2002) Editorial. Nat Rev Drug Discov 1: 3–412119607 [Google Scholar]
- Anon (2004) The CMR International 2004 R&D Factbook. Epsom, UK: Centre for Medicines Research Ltd [Google Scholar]
- Anon (2005) Editorial. Models that take drugs. The Economist, 11 June [Google Scholar]
- Anon (2007) Editorial. Nat Rev Drug Discov 6: 317269156 [Google Scholar]
- Bangs (2005) Predictive biosimulation and virtual patients in pharmaceutical R&D. Stud Health Technol Inform 111: 37–42 [PubMed] [Google Scholar]
- Bangs A, Paterson T (2003) Finding value in in silico biology. BioSilico 1: 18–22 [Google Scholar]
- Chiswell D, Davies N, Ernest G, Felton T, Gennery BA, Hutton P, Murphy J, Roblin D (2007) Pharma 2020: The Vision. Which Path Will You Take? PricewaterhouseCoopers Report [Google Scholar]
- Dollery C, Kitney R (2007) Systems Biology: A Vision for Engineering and Medicine. Report of the Academy of Medical Sciences and Royal Academy of Engineering Working Group on Systems Biology. http://www.acmedsci.ac.uk/p99.html [Google Scholar]
- Frantz S (2007) Pharma faces major challenges after a year of failures and heated battles. Nat Rev Drug Discov 6: 5–7 [DOI] [PubMed] [Google Scholar]
- Garnier J-P (2008) Rebuilding the R&D engine in big pharma. Harv Bus Rev, 1 May [PubMed] [Google Scholar]
- Hendriks B et al. (2006a) Decreased internalisation of ErbB1 mutants in lung cancer is linked with a mechanism conferring sensitivity to gefitinib. Syst Biol 153: 457–466 [DOI] [PubMed] [Google Scholar]
- Hendriks BS, Cook J, Burke JM, Beusmans JM, Lauffenburger DA, de Graaf D (2006b) Computational modelling of ErbB family phosphorylation dynamics in response to transforming growth factor alpha and heregulin indicates spatial compartmentation of phosphatase activity. Syst Biol 153: 22–33 [DOI] [PubMed] [Google Scholar]
- Henney AM, Superti-Furga G (2008) A network solution. Nature 455: 730–731 [DOI] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF (1952) A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol 117: 500–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood L, Perlmutter RM (2004) The impact of systems approaches on biological problems in drug discovery. Nat Biotechnol 22: 1215–1217 [DOI] [PubMed] [Google Scholar]
- Jefferys D, Gennery BA, Manos S, Baxter G, Page C (2008) Pharma 2020: Virtual R&D. Which Path Will You Take? PricewaterhouseCoopers Report [Google Scholar]
- Kansal AR, Trimmer J (2005) Application of predictive biosimulation within pharmaceutical clinical development: examples of significance for translational medicine and clinical trial design. Syst Biol 152: 214–220 [DOI] [PubMed] [Google Scholar]
- Lynch TJ et al. (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139 [DOI] [PubMed] [Google Scholar]
- McGee P (2005) Modelling success with in silico tools. Drug Discov Devel 8: 24–28 [Google Scholar]
- Paez JG et al. (2004) EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304: 1497–1500 [DOI] [PubMed] [Google Scholar]
- Uehling M (2004) I, virtual patient. BioIT World, 18 August. http://www.bio-itworld.com/archive/081804/roche.html [Google Scholar]
- van der Greef J, McBurney RN (2005) Rescuing drug discovery: in vivo systems pathology and systems pharmacology. Nat Rev Drug Discov 4: 961–967 [DOI] [PubMed] [Google Scholar]