Abstract
Although brain tumors are classified as if their lineage were well understood, the relationship between the molecular events that specify neural cell lineage and brain tumors remains enigmatic. Traditionally, cell surface membrane antigens have served as biomarkers that distinguish brain tumor origin and malignancy. In this study, membrane proteins were identified from a terminally differentiated mouse astrocyte (AC) and CT-2A astrocytoma (CT-2A) cell line using liquid-chromatography coupled with tandem mass spectrometry (LC-MS/MS). A total of 321 and 297 protein groups with at least one unique peptide were identified in the AC and CT-2A cells. Using a label-free quantitative MS approach, 25 plasma membrane proteins in CT-2A were found significantly up- or down-regulated compared with those in AC. Three of the up-regulated proteins, chondroitin sulfate proteoglycan-4 (Cspg4), interferon induced transmembrane protein-2 (IFITM2) and -3 (IFITM3) were further validated by semi-quantitative RT-PCR analysis. In addition, a third member of the IFITM family, interferon induced transmembrane protein-1 (IFITM1) was also analyzed. Expression of Cspg4, IFITM1 and IFITM3 was significantly greater in the CT-2A cells than that in the AC cells. Interestingly, Cspg4, also known as neuronal/glial 2 (NG2) proteoglycan in human, is an oligodendrocyte progenitor marker. Therefore, our data suggests that the CT-2A tumor may be derived from NG2 glia rather than fully differentiated astrocytes. Moreover, the CT-2A cells also express a series of interferon-induced signature proteins that may be specific to this tumor. These data highlight the utility of LC-MS/MS for the identification of brain tumor membrane biomarkers.
Keywords: Angiogenesis, biomarker, invasion, inflammation, tumor
Introduction
Membrane proteins play crucial roles in cell-cell and cell-matrix interactions. For example, cell proliferation and differentiation are often mediated through membrane signaling events [1–4]. In the brain, these signaling pathways are crucial for normal neural development and for brain homeostasis. Therefore, it is not surprising that dysregulation of these pathways can contribute to the neoplastic transformation of mature glial, neural stem cells (NSCs), or their progeny [2,5]. Furthermore, over-expression of many cell surface membrane proteins contributes to aspects of glioma progression, such as angiogenesis and invasion [6].
Traditionally, neoplastic transformation in the brain was thought to arise from a progressive accumulation of genetic alterations in mature glia [2]. However, increasing evidence suggests that the cellular and molecular events that lead to glioma formation may originate from neural stem cells (NSCs) [2,5,7]. Although brain tumors are classified as if their lineage were well understood (i.e., astrocytoma derived from astrocytes), the relationship between the molecular events that specify neural cell lineage and brain tumors remain obscure [2,5,7]. Therefore, research tools that identify specific glial or NSC biomarkers are essential for glioma characterization. Traditionally, membrane antigens have served as excellent biomarkers that distinguish brain tumor origin and malignancy. For example, a population of cells expressing the cell surface marker CD133 (prominin-1) was recently identified and isolated from human brain tumors [8]. These cells exhibited NSC properties in vitro and initiated tumors in vivo. Termed as, cancer stem cells, they were found more resistant to radiation treatment compared with tumor cells lacking the CD133 [9]. These findings suggest that brain tumor initiation, progression, and treatment resistance may result in part from a subpopulation of cancerous neural progenitor cells.
Methods that can help identify biomarkers and receptors are important for the molecular classification of gliomas. One such method is mass spectrometry (MS), which has become an essential tool for the detection, identification, and characterization of proteins in cells and tissue [10]. Typically, liquid-chromatography coupled with tandem mass spectrometry (LC-MS/MS) is used to analyze complex protein mixtures for large-scale proteomic applications [10]. However, because of their hydrophobicity it is often difficult to analyze membrane proteins directly by LC-MS/MS without prior separation. Therefore, various separation strategies have been developed, such as two-dimensional (2D) gel electrophoresis and multi-dimensional protein identification technologies (MudPIT), to enhance protein and peptide separation, respectively, in order to improve identification coverage by MS analysis [11]. In addition, traditional protein separation methods, such as one-dimensional (1D) gel electrophoresis continue to provide excellent separation of membrane proteins prior to LC-MS/MS [11].
In this study we used a proteomics approach to assess differences in membrane protein expression between a terminally differentiated mouse astrocyte (AC) and a neoplastic astrocytoma (CT-2A) cell line. In particular, membrane proteins were enriched by sucrose density centrifugation, separated by 1D gel electrophoresis, and analyzed by LC-MS/MS. Using a label-free MS quantitation method we identified 25 plasma membrane proteins that showed significant changes in the CT-2A astrocytoma cells. Three of these proteins, chondroitin sulfate proteoglycan-4 (Cspg4), also known as neuronal/glial 2 (NG2) proteoglycan in human, interferon induced transmembrane protein-2 (IFITM2), and -3 (IFITM3) were found significantly up-regulated. The regulation of these proteins was further evaluated by semi-quantitative RT-PCR analysis. A third member of the IFITM protein family, interferon-induced transmembrane protein-1 (IFITM1) was also analyzed. Of these, Cspg4, IFITM1 and IFITM3 were found significantly up-regulated in CT-2A compared with AC cells. These data highlight the utility of LC-MS/MS for the identification of brain tumor specific membrane proteins that may lead to a better understanding of cell lineage and, ultimately, serve as potential therapeutic targets.
Materials and Methods
Cell culture
The CT-2A astrocytoma cell line was established as previously described [12]. The astrocyte C8-D30 (Astrocyte type III clone) cell line was purchased from American Type Culture Collection (Manassas, VA) [13]. Both cell lines were derived from C57BL/6 mice and were expanded in DMEM (Dulbecco’s modified Eagle’s medium, Sigma Aldrich, St. Louis, MO) supplemented with high glucose (25 mM), 10% (v/v) fetal bovine serum (Sigma Aldrich), and 50 μg/ml penicillin-streptomycin (Sigma Aldrich). The cells were cultured in a CO2 incubator with a humidified atmosphere containing 95% air and 5% CO2 at 37 °C. After reaching confluence the cells were washed in phosphate buffered saline, pH 7.2 (PBS), scraped from a tissue culture dish (150 × 10 mm) and centrifuged to obtain a pellet. Three samples of either CT-2A or AC cells (~2.0 × 107 cells/sample), were pooled prior to membrane protein extraction.
Membrane protein enrichment
Crude membrane extracts were prepared essentially as described previously [14]. Briefly, cells were washed and centrifuged (6000 × g) three times in 2 mL ice cold PBS and resuspended in 3 mL lysis buffer (10 mM HEPES, 1 mM EDTA, pH 7.2) containing appropriate protease inhibitors according to the manufacturer’s instructions (P2714, Sigma Aldrich). Following incubation on ice for 10 min, cell membranes were disrupted with 10 strokes of a pre-cooled 7 mL Dounce tissue homogenizer (Bellco Biotechnology, Vineland, NJ). An equal volume (3 mL) of sucrose buffer (500 mM sucrose, 10 mM HEPES, 1 mM EDTA, pH 7.2) was added and the same number of strokes repeated. The samples were centrifuged (6000 × g) for 10 min at 4 °C to pellet cellular debris. The supernatant was collected (6 mL) and centrifuged at 100,000 × g (SW28 bucket rotor, Beckman Ultra Centrifuge, Fullerton, CA) for 1 h at 4 °C. The membrane pellet was then washed and centrifuged twice in 10 mL of 100 mM Na2CO3, pH 11.3 to remove residual peripheral membrane proteins [15]. The pellet was resuspended in PBS and protein concentration was determined using a bicinchoninic acid (BCA) protein assay (Pierce, Rockfort, IL). Membrane protein concentration was normalized between samples accordingly.
In-gel trypsin digestion
Lyophilized membrane extracts (15 μg) from both the AC and CT-2A astrocytoma cell lines were dissolved in protein sample buffer (125 mM Tris-HCl, 6% (v/v) SDS, 20% (v/v) glycerol, 10% (v/v) 2-mercaptoethanol, 0.02% (w/v) bromophenol blue, pH 6.8) boiled for 15 min and run on 10% acrylamide gels essentially as described [16]. Gels were either silver stained or cut into bands (nine in total) between pre-stained molecular weight standards (SeeBlue Plus2™; Invitrogen, La Jolla, CA). Gel bands were cut into cubes (1 × 1 mm), dehydrated with 50% (v/v) acetonitrile in 50 mM ammonium bicarbonate for 15 min and subsequently dried under vacuum. Slices were rehydrated and reduced with 10 mM dithiothreitol (DTT) in 100 mM ammonium bicarbonate and incubated at 56 °C for 45 min. The reducing solution was discarded and gel slices alkylated with 55 mM iodoacetamide in 100 mM ammonium bicarbonate for 30 min at room temperature. After the supernatant was discarded the gel slices were dehydrated with 50% acetonitrile (v/v) in 50 mM ammonium bicarbonate for 15 min and dried under vacuum as above. The slices were rehydrated with a solution of sequencing grade trypsin (12.5 ng/μl in 50 mM ammonium bicarbonate and 5 mM CaCl2; Promega, San Luis Obispo, CA) and incubated overnight (16 h) in 37 °C water bath. Peptides were extracted from the gel slices with consecutive washes of 2.5% (v/v) formic acid and 50% (v/v) acetonitrile and lyophilized to dryness.
Protein identification by LC-MS/MS
Prior to LC-MS/MS analysis, the trypsin digested samples were filtered through a 30 kDa molecular weight cut-off membrane (Millipore, Billerica, MA) at 6,000 × g for 5 min to remove any residual particulate and intact trypsin. The peptides were independently analyzed on an Agilent 1100 capillary LC (Palo Alto, CA) interfaced directly to a LTQ linear ion trap mass spectrometer (Thermo Electron, San Jose, CA). Mobile phase A and B were H2O/0.1% (v/v) formic acid and acetonitrile/0.1% (v/v) formic acid, respectively. Each fraction was loaded for 40 min onto a PicoFrit (8 cm × 50 μm) column (New Objective, Woburn, MA) packed with 5 μm diameter C18 beads using positive N2 pressure. The peptides were then desalted for 10 min with 0.1% (v/v) formic acid using positive N2 pressure, and were eluted from the column and electrosprayed into the mass spectrometer during a 70 min linear gradient from 5 to 45% (v/v) B at a flow rate of 200 nL/min. The instrument was set to acquire MS/MS spectra on the nine most abundant precursor ions from each MS scan.
Protein identification and validation
Raw tandem mass spectra were converted into mzXML format and then extracted into PKL files using ReAdW followed by mzXML2Other [17]. The PKL files were searched against a target and decoy National Center for Biotechnology Information (NCBI) house mouse (Mus musculus) database (www.ncbi.nih.gov) using Mascot v1.9 (Matrix Science, Boston, MA). The following search parameters were used: fully tryptic enzymatic cleavage, maximum two missed cleavages, peptide tolerance of 600 parts-per-million, a fragment ion tolerance of 0.6 Da and modification on cysteine residue due to carboxyamidomethylation (+57 Da). Peptide matches above discrete Mascot-ion scores were extracted from target and decoy database search results. Using ProValT, peptide redundancies were removed and the protein false-discovery rate (FDR) was calculated at each ion score threshold and peptide coverage level by comparing the distribution of protein identifications between the target and decoy database search results as previously described [18]. Proteins identified by single peptides were only considered significant at a 1% FDR if they exceeded a Mascot ion score of 45 and 52 for CT-2A and AC, respectively. The difference in threshold cutoffs between samples results from the insufficiency of the Mascot scoring system to estimate the FDR for peptide and protein identifications [18]. After analysis, peptides which corresponded to trypsin and keratin contaminants were removed from both AC and CT-2A protein lists. Gene identifications were acquired from the NCBI mouse database; cellular localization and functions were manually annotated from the gene ontology (www.geneontology.org) or the UniProtKB/Swiss-Prot (http://www.ebi.ac.uk/swissprot/) databases. Transmembrane spanning domains were predicted with the TMHMM 2.0 algorithm (www.cbs.dtu.dk/services/TMHMM/) [19].
Quantitative proteomics by spectral counts
Many proteins identified in this study were found exclusively in CT-2A or AC. For those proteins not identified in either cell line, spectral counts of 0.5 were applied. For discrete frequency data, such as spectral counts, the likelihood ratio G-test was chosen to judge the significance of protein abundance as described previously [20]. Briefly, the G-value of each protein was calculated as shown in equation (1).
| (1) |
The p-value of each protein was subsequently calculated as the probability of observing a random variable larger than G from the Chi-square distribution (one degree of freedom). The frequency histogram of the p-values was afterward created and 0.01 was determined as the cutoff for p-values to detect significant changes (Supplemental Figure 1).
RT-PCR analysis
The AC and CT-2A astrocytoma cell lines were grown under identical culture conditions as described above. Total RNA was isolated from homogenized cell pellets of AC and CT-2A using the Trizol Reagent (Invitrogen, La Jolla, CA), according to the manufacturer’s protocol. Single strand cDNA was synthesized from total RNA and used for single-plex PCR amplification as previously described [21]. Primer sequences used for PCR were for β-actin, forward 5′-TGTGATGGTGGGAATGGGTCAG-3′ and reverse 5′-TTTGATGTCACGCACGATTTCC-3′; for Cspg4, forward 5′-AGCACGATGACTCTGAGACC-3′ and reverse 5′-TTGGCTACGTGAAGATAGGG-3′. For interferon-induced transmembrane proteins (IFITM) the primers used were reported previously [22]. For IFITM1 forward 5′-CCTTCCTTATTCTCACTCTG-3′ and reverse 5′-GTTGCAAGACATCTCACATC-3′; for IFITM2, forward 5′-GATCTTCAGCATCCTTATGGTC-3′ and reverse 5′ GAAGGTAACATTTGCATACGCG 3′ and for IFITM3 forward 5′-GTTATCACCATTGTT AGTGTCATC-3′ and reverse 5′-ATTGAGTGTTACACCTGCGTG-3′. Primers were optimized for annealing temperatures and cycle numbers as previously described [21]. PCR products were separated on 1 % agarose gels containing ethidium bromide, visualized by UV light, and analyzed using Kodak 1D software (Eastman Kodak Co., Rochester, NY). RT-PCR was performed on the total RNA of each sample in the absence of reverse transcriptase to control for possible DNA contamination.
Results and Discussion
The strategy used to identify proteins from purified mouse astrocyte (AC) and CT-2A astrocytoma (CT-2A) membrane extracts using LC-MS/MS is outlined in Figure 1. First, membrane extracts were purified using sucrose density centrifugation and separated by 1D SDS gel electrophoresis. To identify all protein, the gel lanes were divided into fractions, trypsin digested and the peptides were analyzed by LC-MS/MS (Fig. 1B). The bioinformatics software Mascot [23] was used to match the experimental MS/MS spectra with theoretical in silico generated MS/MS spectra of predicted peptides from the NCBI mouse protein database. The ProValT algorithm was used to filter peptide matches from Mascot results, eliminate peptide redundancy, cluster peptides into protein groups and calculate protein false discovery rates [18].
Figure 1.
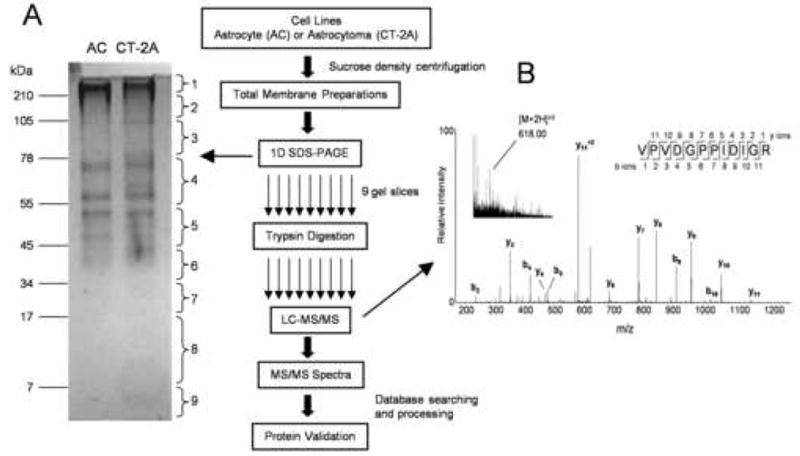
Membrane extracts for astrocytoma (CT-2A) and astrocyte (AC) cell lines were resolved on a 10% SDS polyacrylamide gel (A). The gel lanes were cut into nine gel slices followed by trypsin digestion. The peptides were analyzed by reverse-phase liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). A doubly charged [M+2H]2+ precursor ion (m/z = 618.0) from the AC membrane sample is shown in a MS survey scan (B, inset). This ion was selected for further sequence analysis by collision induced dissociation (CID) in the mass spectrometer. The MS/MS fragment ions (b- and y-ions) were later matched to the peptide sequence shown (VPVDGPPIDIGR) after searching against the mouse NCBI protein database using Mascot search algorithm. This sequence corresponded to amino acids 1707–1718 from the insulin-like growth factor-2 receptor (Supplemental Table 1).
A total of 1,177 and 1,074 peptides, matching 599 and 587 proteins were identified from AC and CT-2A membrane extracts, respectively. Proteins from AC and CT-2A datasets were clustered into 321 and 297 groups in which the top match of each group was identified by at least one unique peptide (Supplemental Tables 1 and 2). The locations and functions of all proteins were manually annotated from either the gene ontology or Swiss/Prot databases. It should be noted that the categorization is not a strict assignment since many proteins can be found in multiple locations and can have various functions in the cell.
To assess membrane protein enrichment the number of transmembrane domains (TMDs) for each protein was predicted using the TMHMM 2.0 web server [19]. After analysis, 53% (169/321) and 45% (135/297) of proteins in AC and CT-2A datasets were predicted to have at least one TMD (Supplemental Tables 1 and 2). These proteins were further categorized according to their subcellular location (Fig. 2). Although many proteins were found localized to the plasma membrane, the majority were localized to intracellular organelles including the endoplasmic reticulum, mitochondria, nucleus, Golgi, endosomes and lysosomes (Fig. 2). This is not surprising since this preparation of crude membrane fractions will include a significant proportion of intracellular transmembrane proteins [14]. Those transmembrane proteins that had unknown or multiple locations were simply considered as membrane proteins (MP). In this study the function of plasma membrane (PM) proteins were of interest because receptors and antigens involved in cellular signaling and differentiation are often located on the cell surface (Fig. 2). Specifically, cell surface receptors represented 15% (9/59) and 27% (20/74) of the total number of PM proteins identified in AC cells and CT-2A cells (Supplemental Tables 1 and 2). In comparison, cell surface adhesion molecules represented 20% (12/59) and 14% (10/74) of proteins identified from these cell lines. However, the majority of transmembrane proteins, 44% (26/59) and 34% (25/74) in AC and CT-2A cells, were identified as solute ion-transporters, implicating the important role of these molecules in the cellular homeostasis.
Figure 2.
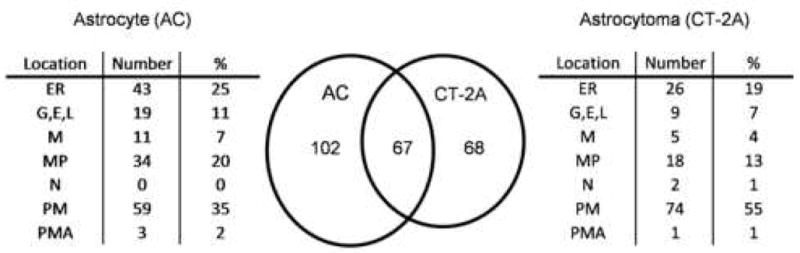
The locations of identified proteins with predicted transmembrane domains (TMDs) were analyzed in both AC (n=169) and CT-2A (n=135) cells, respectively. Of these 67 were shared between samples. Abbreviations are as follows: Endoplasmic reticulum (ER), Golgi, endosome, lysosome (G,E,L), mitochondria (M), membrane protein with unknown location (MP), nuclear (N), plasma membrane (PM), and plasma membrane associated (PMA).
The CT-2A cell line, derived from 20-methylcholanthrene implantation into the mouse brain, was initially classified as highly malignant, poorly differentiated anaplastic astrocytoma [12]. Currently the mechanisms underlying CT-2A differentiation and tumorigenesis remain unknown. Here a label-free quantitative proteomics approach was used to evaluate the differences in plasma membrane proteins from terminally differentiated astrocytes and CT-2A cells. In this study we employed G-test to assess significant differences in protein spectral counts between AC and CT-2A cell lines. After analysis, 13 and 12 plasma membrane proteins were found significantly up- or down-regulated in CT-2A cells (Table 1). Of these, chondroitin sulfate proteoglycan-4 (Cspg4) was further validated by semi-quantitative RT-PCR because it was among the most up-regulated receptors identified exclusively on CT-2A astrocytoma cells (Table 1). RT-PCR was used to validate MS quantitation results because of its high throughput and sensitivity over Western blotting. Consistent with LC-MS/MS results, the Cspg4 transcript was found up-regulated in CT-2A compared with AC cells (Fig. 3). The difference in Cspg4 expression between cell lines is interesting since Cspg4 is a purported tumor specific antigen on chemically induced rat chondrosarcoma cells [24]. Also referred to as neuronal/glial 2 (NG2) in human, this protein is a marker for oligodendrocyte progenitor cells [25], in which NG2 positive glia represents the fourth major population of glia in the brain, distinct from astrocytes, mature oligodendrocytes, microglia and neurons [26]. Therefore, from our data it would appear that CT-2A astrocytoma cells arise from NG2 glia rather than from a mature astrocyte or a generic neural progenitor cell. Moreover, the expression of NG2 has also been shown to be associated with glioma proliferation, metastasis and angiogenesis [27]. Although CT-2A is not metastatic, it is highly angiogenic [21]. It is thought that NG2 promotes blood vessel development via the engagement of galectin-3 and integrin α3β1 to form a complex on endothelial cell surface [28,29]. While we did not identify galectin-3 or investigate expression of the integrins α3 or β1 in CT-2A, we did exclusively identify galectin-1 in CT-2A cells (NP_032521.1; Supplemental Table 2). Galectin-1 is highly expressed in human glioma cells and is thought to increase their mobility [30,31]. However, whether galectin-1 can replace galectin-3 and mediate clustering of integrin α3β1 and NG2 on the CT-2A cell surface needs further investigation.
Table 1.
The list of plasma membrane proteins displaying significant differences between AC and CT-2A cells
| Protein name | GenBank™ accession number | TMDa | Spectral counts CT-2A | Spectral counts AC | log2(CT-2A/AC)b | pc |
|---|---|---|---|---|---|---|
|
Adhesion | ||||||
| stomatin | NP_038543.1 | 1 | 0 | 13 | −4.67 | 1.49E-04 |
| integrin alpha 7 | NP_032424.1 | 2 | 1 | 10 | −3.29 | 3.59E-03 |
|
| ||||||
|
Antigen Processing | ||||||
| histocompatibility 2, K1, K region | XP_619453.1 | 1 | 14 | 0 | 4.83 | 5.99E-05 |
| similar to MHC class I antigen precursor | XP_622849.1 | 1 | 7 | 0 | 3.83 | 8.66E-03 |
|
| ||||||
|
Metabolism | ||||||
| leucyl/cystinyl aminopeptidase | NP_766415.1 | 1 | 1 | 13 | −3.67 | 4.96E-04 |
| acyl-CoA synthetase long-chain family member 3 | NP_083093.1 | 1 | 0 | 12 | −4.56 | 2.98E-04 |
|
| ||||||
|
Poliferation & Signaling | ||||||
| odd Oz/ten-m homolog 3 | NP_035987.2 | 1 | 0 | 11 | −4.43 | 5.98E-04 |
| ectonucleoside triphosphate diphosphohydrolase 2 | NP_033979.1 | 2 | 0 | 10 | −4.29 | 1.20E-03 |
|
| ||||||
|
Receptors | ||||||
| DAMP-1 protein | NP_932763.1 | 1 | 23 | 0 | 5.55 | 1.02E-07 |
| CD44 | NP_033981.1 | 1 | 38 | 1 | 5.28 | 1.35E-11 |
| transferrin receptor | NP_035768.1 | 1 | 48 | 12 | 2.03 | 9.64E-07 |
| chondroitin sulfate proteoglycan 4 (NG2) | NP_620570.1 | 1 | 19 | 0 | 5.28 | 1.73E-06 |
| scavenger receptor class B, member 2 | NP_031670.1 | 2 | 1 | 21 | −4.36 | 2.37E-06 |
| interferon induced transmembrane protein 2 | NP_109619.1 | 2 | 9 | 0 | 4.20 | 2.08E-03 |
| interferon induced transmembrane protein 3 | NP_079654.1 | 2 | 14 | 0 | 4.83 | 5.99E-05 |
| Eph receptor A2 | NP_034269.2 | 1 | 7 | 0 | 3.83 | 8.66E-03 |
| interleukin 1 receptor accessory protein isoform a | NP_032390.1 | 1 | 0 | 7 | −3.78 | 9.72E-03 |
| nerve growth factor receptor | NP_150086.1 | 1 | 0 | 7 | −3.78 | 9.72E-03 |
|
| ||||||
|
Transporters | ||||||
| Na+/K+-ATPase alpha 3 subunit | NP_659170.1 | 8 | 0 | 22 | −5.43 | 2.87E-07 |
| Na+/K+-ATPase alpha 1 subunit | NP_659149.1 | 10 | 33 | 82 | − 1.29 | 4.93E-06 |
| ATPase, Na+/K+ transporting, alpha 2 polypeptide | NP_848492.1 | 8 | 12 | 0 | 4.61 | 2.48E-04 |
| solute carrier family 1, member 5 | NP_033227.1 | 9 | 12 | 0 | 4.61 | 2.48E-04 |
| solute carrier family 12, member 7 | NP_035520.1 | 11 | 10 | 0 | 4.35 | 1.02E-03 |
|
| ||||||
|
Others | ||||||
| membrane bound C2 domain containing protein | NP_035973.1 | 1 | 16 | 2 | 3.03 | 3.56E-04 |
| glycoprotein, synaptic 2 | NP_598879.1 | 4 | 5 | 21 | −2.04 | 1.26E-03 |
Number of predicted transmembrane domains.
The values were derived from normalilzed spectral counts of identified proteins in CT-2A and AC samples.
Figure 3.
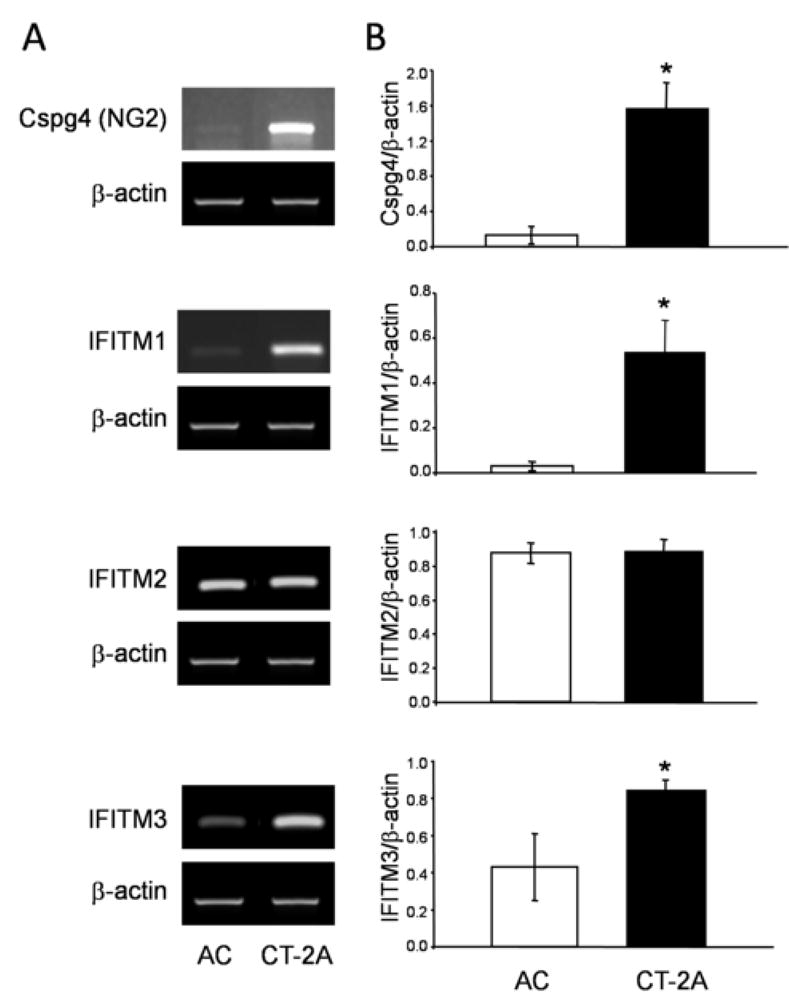
Detection and quantification of gene expression by RT-PCR analysis of chondroitin sulfate proteoglycan-4 (Cspg4), also known as NG2 in human, and interferon-induced transmembrane protein-1, -2, and -3 (IFITM1, IFITM2, and IFITM3) (A). The gene products were detected for both AC and CT-2A cells. PCR was performed using β-actin primers as internal control and conditions as described in experimental methods. (B) The ratio of RT-PCR gene expression for each protein was compared to the β-actin internal control. The ratios of gene/β-actin are expressed as means (± 95% confidence interval) and three independent samples were analyzed for each cell line. The transcription level of each gene was compared between AC and CT-2A and statistical analysis was performed using two-tailed Student’s t-test (*p<0.01).
Two additional up-regulated CT-2A plasma membrane proteins identified by LC-MS/MS analysis, IFITM2 and IFITM3, were also evaluated by RT-PCR analysis (Table 1). These proteins are members of the interferon-induced transmembrane family that are involved in interferon-mediated differentiation and adhesion events [32,33]. Although IFITM3 transcript expression was significantly up-regulated in the CT-2A cells, which is consistent with MS results, no difference in IFITM2 transcript expression was observed between CT-2A and AC (Fig. 3 and Table 1). This inconsistency might represent differences between transcriptional and translational level of regulation [34]. Alternatively, the lack of IFITM2 in the membrane fraction could be due to its translocation [35]. In this regard, it is possible that IFITM2 was preferentially localized to a different protein fraction after purification of AC membrane extracts and subsequently missed during MS/MS analysis. Since both IFITM2 and IFITM3 were found specifically on the CT-2A astrocytoma plasma membrane a third member of the IFITM protein family, interferon-induced transmembrane protein-1 (IFITM1), was also analyzed by RT-PCR. Consistent with IFITM3 results, IFITM1 was found up-regulated in CT-2A astrocytoma cells compared with AC cells (Fig. 3). Recently, it was suggested that all three IFITM genes, which neighbor each other on mouse chromosome 7, share regulatory elements leading to a common mechanism of transcriptional regulation [22]. However, at least with AC cells, this does not appear to be the case. Therefore, the expression of IFITM1, 2 and 3 in CT-2A astrocytoma cells, compared with only IFITM2 in AC cells, may reflect a difference in cellular differentiation. The IFITM gene family is also up-regulated specifically in human colorectal tumors, suggesting that IFITM proteins may be specific biomarkers for these tumors [22]. Interestingly, another interferon-induced membrane protein, DAMP-1 (known as bone marrow stromal cell antigen 2 in human), was observed up-regulated in CT-2A (Table 1). This protein is a specific marker of Type 1 interferon-producing cells in mouse [36] and is also over-expressed on multiple myeloma cells [37]. Overall, our data indicate that CT-2A express a series of interferon-induced signature proteins that may be either specific to this tumor or similar to those observed in certain breast and ovarian carcinomas [38]. However, whether interferon-induced differentiation contributes to neoplastic formation in mouse astrocytes or their progenitors remains to be determined.
Many of the other up-regulated proteins identified here have also been previously reported to have important roles in tumor migration, angiogenesis and proliferation (Table 1). These include the transferrin receptor [39,40], the epherin (Eph) receptor A2 [41], the receptor CD44 [42] and the antigen presentation MHC class proteins [43]. For example, the transferrin receptor has been shown to be significantly up-regulated in human glioma cells and may play a role in tumor malignancy [44]. In addition, the hyaluronan receptor CD44 together with NG2 are biomarkers for invasive behavior in human glioma cell lines [42,45]. Finally, there is evidence that indicates EphA2 receptor activation might increase glioma proliferation via a mitogen-activated protein kinase-dependent pathway [41]. Overall, CT-2A shares several common cell surface receptors with human glioma that may exacerbate tumor proliferation and invasion.
Conclusion
In sum, a tandem mass spectrometry proteomic approach was used to profile membrane proteins from terminally differentiated mouse astrocyte and CT-2A astrocytoma cell lines. The up-regulation of mouse IFITM1, IFITM3 and Cspg4, the homolog of human proteoglycan NG2, in CT-2A astrocytoma cells is also reported. Our data suggests that CT-2A astrocytoma cells might arise from NG2 glia. In addition, the presence of interferon-induced signature proteins in CT-2A may expand our knowledge of brain tumor etiology, progression and, potentially, serve as therapeutic targets
Supplementary Material
Acknowledgments
This work was funded by the National Institutes of Health/NCRR-funded Integrated Technology Resource for Biomedical Glycomics (P41 RR018502) and the National Institutes of Health/NCRR-funded Research Resource for Integrated Glycotechnology (P41 RR005351).
Abbreviations
- AC
Astrocyte
- CD
cluster of differentiation
- Cspg4
chondroitin sulfate proteoglycan-4
- FDR
false discovery rate
- IFITM
interferon induced transmembrane protein
- LC-MS/MS
liquid chromatography coupled with tandem mass spectrometry
- NG2
neuronal/glial 2
- TMD
transmembrane domain
- NSC
neural stem cell
- RT-PCR
reverse transcriptase polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhang J, Li L. BMP signaling and stem cell regulation. Dev Biol. 2005;284:1–11. doi: 10.1016/j.ydbio.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353:811–22. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 3.McKay R. Stem cell biology and neurodegenerative disease. Philosophical Transactions of the Royal Society B: Biological Sciences. 2004;359:851–856. doi: 10.1098/rstb.2004.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emsley JG, Mitchell BD, Kempermann G, Macklis JD. Adult neurogenesis and repair of the adult CNS with neural progenitors, precursors, and stem cells. Prog Neurobiol. 2005;75:321–41. doi: 10.1016/j.pneurobio.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Bjerkvig R, Tysnes BB, Aboody KS, Najbauer J, Terzis AJ. Opinion: the origin of the cancer stem cell: current controversies and new insights. Nat Rev Cancer. 2005;5:899–904. doi: 10.1038/nrc1740. [DOI] [PubMed] [Google Scholar]
- 6.Bello L, Giussani C, Carrabba G, Pluderi M, Costa F, Bikfalvi A. Angiogenesis and invasion in gliomas. Cancer Treat Res. 2004;117:263–84. doi: 10.1007/978-1-4419-8871-3_16. [DOI] [PubMed] [Google Scholar]
- 7.Seyfried TN. Perspectives on brain tumor formation involving macrophages, glia, and neural stem cells. Perspect Biol Med. 2001;44:263–82. doi: 10.1353/pbm.2001.0035. [DOI] [PubMed] [Google Scholar]
- 8.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 9.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 10.Aebersold R, Goodlett DR. Mass spectrometry in proteomics. Chem Rev. 2001;101:269–95. doi: 10.1021/cr990076h. [DOI] [PubMed] [Google Scholar]
- 11.Wu CC, Yates JR., 3rd The application of mass spectrometry to membrane proteomics. Nat Biotechnol. 2003;21:262–7. doi: 10.1038/nbt0303-262. [DOI] [PubMed] [Google Scholar]
- 12.Seyfried TN, el-Abbadi M, Roy ML. Ganglioside distribution in murine neural tumors. Mol Chem Neuropathol. 1992;17:147–67. doi: 10.1007/BF03159989. [DOI] [PubMed] [Google Scholar]
- 13.Alliot F, Pessac B. Astrocytic cell clones derived from established cultures of 8-day postnatal mouse cerebella. Brain Research. 1984;306:283–291. doi: 10.1016/0006-8993(84)90377-9. [DOI] [PubMed] [Google Scholar]
- 14.Miguet L, Pacaud K, Felden C, Hugel B, Martinez MC, Freyssinet JM, Herbrecht R, Potier N, van Dorsselaer A, Mauvieux L. Proteomic analysis of malignant lymphocyte membrane microparticles using double ionization coverage optimization. Proteomics. 2006;6:153–71. doi: 10.1002/pmic.200500133. [DOI] [PubMed] [Google Scholar]
- 15.Wu CC, MacCoss MJ, Howell KE, Yates JR., 3rd A method for the comprehensive proteomic analysis of membrane proteins. Nat Biotechnol. 2003;21:532–8. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 16.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–79. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 17.Pedrioli PG, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, Pratt B, Nilsson E, Angeletti RH, Apweiler R, Cheung K, Costello CE, Hermjakob H, Huang S, Julian RK, Kapp E, McComb ME, Oliver SG, Omenn G, Paton NW, Simpson R, Smith R, Taylor CF, Zhu W, Aebersold R. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 2004;22:1459–66. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 18.Weatherly DB, Astwood JA, 3rd, Minning TA, Cavola C, Tarleton RL, Orlando RA. Heuristic method for assigning a false-discovery rate for protein identifications from Mascot database search results. Mol Cell Proteomics. 2005;4:762–72. doi: 10.1074/mcp.M400215-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. Journal of Molecular Biology. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 20.Xia Q, Wang T, Park Y, Lamont RJ, Hackett M. Differential quantitative proteomics of Porphyromonas gingivalis by linear ion trap mass spectrometry: Non-label methods comparison, q-values and LOWESS curve fitting. International Journal of Mass Spectrometry. 2007;259:105–116. doi: 10.1016/j.ijms.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abate LE, Mukherjee P, Seyfried TN. Gene-linked shift in ganglioside distribution influences growth and vascularity in a mouse astrocytoma. J Neurochem. 2006;98:1973–84. doi: 10.1111/j.1471-4159.2006.04097.x. [DOI] [PubMed] [Google Scholar]
- 22.Andreu P, Colnot S, Godard C, Laurent-Puig P, Lamarque D, Kahn A, Perret C, Romagnolo B. Identification of the IFITM family as a new molecular marker in human colorectal tumors. Cancer Res. 2006;66:1949–55. doi: 10.1158/0008-5472.CAN-05-2731. [DOI] [PubMed] [Google Scholar]
- 23.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Leger O, Johnson-Leger C, Jackson E, Coles B, Dean C. The chondroitin sulfate proteoglycan NG2 is a tumour-specific antigen on the chemically induced rat chondrosarcoma HSN. Int J Cancer. 1994;58:700–5. doi: 10.1002/ijc.2910580514. [DOI] [PubMed] [Google Scholar]
- 25.Stallcup WB. The NG2 proteoglycan: past insights and future prospects. J Neurocytol. 2002;31:423–35. doi: 10.1023/a:1025731428581. [DOI] [PubMed] [Google Scholar]
- 26.Nishiyama A, Watanabe M, Yang Z, Bu J. Identity, distribution, and development of polydendrocytes: NG2-expressing glial cells. J Neurocytol. 2002;31:437–55. doi: 10.1023/a:1025783412651. [DOI] [PubMed] [Google Scholar]
- 27.Chekenya M, Pilkington GJ. NG2 precursor cells in neoplasia: functional, histogenesis and therapeutic implications for malignant brain tumours. J Neurocytol. 2002;31:507–21. doi: 10.1023/a:1025795715377. [DOI] [PubMed] [Google Scholar]
- 28.Fukushi J, Makagiansar IT, Stallcup WB. NG2 proteoglycan promotes endothelial cell motility and angiogenesis via engagement of galectin-3 and alpha3beta1 integrin. Mol Biol Cell. 2004;15:3580–90. doi: 10.1091/mbc.E04-03-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen Y, Makagiansar IT, Fukushi J, Liu FT, Fukuda MN, Stallcup WB. Molecular basis of interaction between NG2 proteoglycan and galectin-3. J Cell Biochem. 2006;98:115–27. doi: 10.1002/jcb.20768. [DOI] [PubMed] [Google Scholar]
- 30.Camby I, Decaestecker C, Lefranc F, Kaltner H, Gabius HJ, Kiss R. Galectin-1 knocking down in human U87 glioblastoma cells alters their gene expression pattern. Biochem Biophys Res Commun. 2005;335:27–35. doi: 10.1016/j.bbrc.2005.07.037. [DOI] [PubMed] [Google Scholar]
- 31.Camby I, Le Mercier M, Lefranc F, Kiss R. Galectin-1: a small protein with major functions. Glycobiology. 2006;16:137R–157R. doi: 10.1093/glycob/cwl025. [DOI] [PubMed] [Google Scholar]
- 32.Lewin AR, Reid LE, McMahon M, Stark GR, Kerr IM. Molecular analysis of a human interferon-inducible gene family. Eur J Biochem. 1991;199:417–23. doi: 10.1111/j.1432-1033.1991.tb16139.x. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka SS, Yamaguchi YL, Tsoi B, Lickert H, Tam PP. IFITM/Mil/fragilis family proteins IFITM1 and IFITM3 play distinct roles in mouse primordial germ cell homing and repulsion. Dev Cell. 2005;9:745–56. doi: 10.1016/j.devcel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 34.Cox B, Kislinger T, Wigle DA, Kannan A, Brown K, Okubo T, Hogan B, Jurisica I, Frey B, Rossant J, Emili A. Integrated proteomic and transcriptomic profiling of mouse lung development and Nmyc target genes. Mol Syst Biol. 2007;3:109. doi: 10.1038/msb4100151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gurkan C, Koulov AV, Balch WE. An evolutionary perspective on eukaryotic membrane trafficking. Adv Exp Med Biol. 2007;607:73–83. doi: 10.1007/978-0-387-74021-8_6. [DOI] [PubMed] [Google Scholar]
- 36.Blasius AL, Giurisato E, Cella M, Schreiber RD, Shaw AS, Colonna M. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J Immunol. 2006;177:3260–5. doi: 10.4049/jimmunol.177.5.3260. [DOI] [PubMed] [Google Scholar]
- 37.Ohtomo T, Sugamata Y, Ozaki Y, Ono K, Yoshimura Y, Kawai S, Koishihara Y, Ozaki S, Kosaka M, Hirano T, Tsuchiya M. Molecular cloning and characterization of a surface antigen preferentially overexpressed on multiple myeloma cells. Biochem Biophys Res Commun. 1999;258:583–91. doi: 10.1006/bbrc.1999.0683. [DOI] [PubMed] [Google Scholar]
- 38.Einav U, Tabach Y, Getz G, Yitzhaky A, Ozbek U, Amariglio N, Izraeli S, Rechavi G, Domany E. Gene expression analysis reveals a strong signature of an interferon-induced pathway in childhood lymphoblastic leukemia as well as in breast and ovarian cancer. Oncogene. 2005;24:6367–75. doi: 10.1038/sj.onc.1208797. [DOI] [PubMed] [Google Scholar]
- 39.Daniels TR, Delgado T, Helguera G, Penichet ML. The transferrin receptor part II: Targeted delivery of therapeutic agents into cancer cells. Clinical Immunology. 2006;121:159–176. doi: 10.1016/j.clim.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 40.Daniels TR, Delgado T, Rodriguez JA, Helguera G, Penichet ML. The transferrin receptor part I: Biology and targeting with cytotoxic antibodies for the treatment of cancer. Clinical Immunology. 2006;121:144–158. doi: 10.1016/j.clim.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 41.Liu F, Park PJ, Lai W, Maher E, Chakravarti A, Durso L, Jiang X, Yu Y, Brosius A, Thomas M, Chin L, Brennan C, DePinho RA, Kohane I, Carroll RS, Black PM, Johnson MD. A Genome-Wide Screen Reveals Functional Gene Clusters in the Cancer Genome and Identifies EphA2 as a Mitogen in Glioblastoma. Cancer Res. 2006;66:10815–10823. doi: 10.1158/0008-5472.CAN-06-1408. [DOI] [PubMed] [Google Scholar]
- 42.Pilkington GJ. Cancer stem cells in the mammalian central nervous system. Cell Prolif. 2005;38:423–33. doi: 10.1111/j.1365-2184.2005.00358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engelhard VH, Altrich-Vanlith M, Ostankovitch M, Zarling AL. Post-translational modifications of naturally processed MHC-binding epitopes. Current Opinion in Immunology. 2006;18:92–97. doi: 10.1016/j.coi.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 44.Recht L, Torres CO, Smith TW, Raso V, Griffin TW. Transferrin receptor in normal and neoplastic brain tissue: implications for brain-tumor immunotherapy. J Neurosurg. 1990;72:941–5. doi: 10.3171/jns.1990.72.6.0941. [DOI] [PubMed] [Google Scholar]
- 45.Wiranowska M, Ladd S, Smith SR, Gottschall PE. CD44 adhesion molecule and neuro-glial proteoglycan NG2 as invasive markers of glioma. Brain Cell Biol. 2006;35:159–72. doi: 10.1007/s11068-007-9009-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


