Abstract
Microfluidic systems are under development to address a variety of medical problems. Key advantages of micrototal analysis systems based on microfluidic technology are the promise of small size and the integration of sample handling and measurement functions within a single, automated device having low mass-production costs. Here, we review the spectrum of methods currently used to detect malaria, consider their advantages and disadvantages, and discuss their adaptability towards integration into small, automated micro total analysis systems. Molecular amplification methods emerge as leading candidates for chip-based systems because they offer extremely high sensitivity, the ability to recognize malaria species and strain, and they will be adaptable to the detection of new genotypic signatures that will emerge from current genomic-based research of the disease. Current approaches to the development of chip-based molecular amplification are considered with special emphasis on flow-through PCR, and we present for the first time the method of malaria specimen preparation by dielectrophoretic field-flow-fractionation. Although many challenges must be addressed to realize a micrototal analysis system for malaria diagnosis, it is concluded that the potential benefits of the approach are well worth pursuing.
Keywords: Malaria diagnosis, Microfluidic systems, Dielectrophoresis, Polymerase chain reaction, Micrototal analysis systems
1. Introduction
Advances in microfluidics promise to revolutionize the detection of pathogens in vivo and in the environment through the development of lab-on-chip instruments (Huikko et al., 2003). In particular, instruments called micrototal analysis systems (μTAS) are under development that will be able to undertake all steps in analysis from sample collection and preparation to molecular detection and to integrate these into automated instruments that will be suitable for point-of-care applications even in environments that lack infrastructure. Once implemented, such instruments promise to provide rapid diagnosis based on molecular signatures. Furthermore, their potential for automation suggests that these devices will be suitable for use by individuals having only basic training. Malaria is one of many diseases that pose particular problems in regions lacking good infrastructure that could potentially benefit from the introduction of such automated diagnostic methods. In this article, we review current malaria detection methods, consider the most desirable features that might be made available through a μTAS, and discuss the methods we are currently pursuing to achieve microsystem integration.
2. Methods of diagnosis
The definitive diagnosis of malaria remains the detection of malarial parasites in the blood. Table 1 provides a “wish list” that defines what might be considered the ideal features of a generally-applicable diagnostic method. Ideally, a test for malaria can detect as few as 1 parasite/μl of blood, is applicable to all species of malaria and can identify which species is (are) present, provides no false negatives and very few false positives, yields definitive results in 20 min or less, provides a quantitative measure of the current level of infection, and is not influenced by previous malarial infections in the subject. Furthermore, because most malaria occurs in poorly developed regions, it should be portable, easy to perform in areas lacking infrastructure, and be extremely inexpensive. The “gold standard” test for malaria remains essentially the same microscopic method that was used over 100 years ago to describe parasite-infected erythrocytes in the blood. Both thin and thick blood smear microscopic methods are used with a variety of stains and these, administered by pathologists or technicians, remain the most commonly applied diagnostic approaches. A skilled technician can analyze specimens quickly, detect (with multiple smears) as few as four parasites/μl of blood, and identify the parasite species. The method is highly competitive with most other diagnostic techniques available today and, in underserved communities, remains cheaper than all of the others. Disadvantages of the approach are that microscopes are hard to transport to very remote areas, it takes considerable skill and time to achieve the limit of sensitivity, difficulty can exist in identifying malarial species at low infection levels, mixed infections can be hard to identify, and there is the need for an experienced microscopist.
Table 1.
Ideal properties for a general-purpose technology for the diagnosis of malaria
| Parameter | Ideal characteristic |
|---|---|
| Sensitivity | 1 Parasite/microliter blood |
| Specificity | Identifies the species and, ideally, the strain Detects all types of malaria: falciparum, vivax, etc. |
| Accuracy | No false negatives, few false positives |
| Speed of operation | Results in <30 min |
| Freedom from confounding factors | Accuracy is not compromised by prior infection or other patient characteristics |
| Portability | Operates from battery power, weighs <2 kg, uses minimal, durable supplies |
| Ease of Use | Requires minimal expertise to operate |
| Cost | Initial investment less than the cost of a microscope, running costs <30 cents per test |
| Adaptability | Able to take advantage of new genomic markers as they become available |
Several useful immunochromatographic “dipstick” methods have been developed for malaria detection and these rapid methods are extremely easy to use even in the most challenging of environments (Wongsrichanalai, 2001; Moody, 2002; Murray et al., 2003). Assays are commercially available based on the antigenic detection of malaria proteins, including malarial lactate dehydrogenase (pLDH) (Makler et al., 1993) and malarial histidine-rich protein (HRP II) (Birku et al., 1999; Wolday et al., 2001), and of sporozoites (Bangs et al., 2002). Proteins are released into the blood of the host by parasitized cells so that the methods detect evidence for the activity of malaria in the blood rather than definitively detecting the parasites themselves. Nevertheless, the tests are able to discriminate between the major malarial species through immunogenic differences in the proteins. At best, however, the sensitivity of the tests (most become reliable above 100 parasites/μl) is still below the threshold of the microscopic method and the proportionality between the strengths of the test responses and the degree of infection shows variation from patient to patient (John et al., 1998; Lema et al., 1999; Labbe et al., 2001; Cho et al., 2001; Iqbal et al., 2002, 2003; Richardson et al., 2002). The presence of the malarial proteins drops fairly quickly as the infection abates, so these methods are useful in detecting recurrent infections but not for tracking rapid responses to chemotherapy. Immunochromatographic and ELISA-dipstick tests for host antibodies to malaria proteins are also available (Schwick et al., 1998; Irion et al., 2002; Silvie et al., 2002). While this alternative dipstick approach can also differentiate malarial species based on antigenic differences, host antibodies remain after the infection has abated making the method unsuitable for regions where patients have a likelihood of having suffered prior infections (Irion et al., 2002; Iqbal et al., 2003). Despite their ability to discriminate between different species of malaria, the dipstick methods are poor at detecting mixed infections when one species is present at a significantly lower parasitemia than the other. Dipstick tests are generally easy to perform, requiring a single drop of blood and a development time up to half an hour. Nevertheless, they are relatively expensive, and offer no significant advantages over microscopic methods in areas having high infection rates. But obviously, they are the best recourse in regions lacking microscopic methods.
Flow cytometry is a popular approach to blood analysis in high throughput laboratories and can offer malaria diagnosis as well as blood differential analysis (Barkan et al., 2000; Saito-Ito et al., 2001). In principle, flow cytometers can offer extremely good cell analysis using multiple channels to analyze several cellular parameters simultaneously and this might, in principle, allow for speciation. Nevertheless, current cytometers and blood analyzers are expensive and delicate, demand skilled operators, and are not generally accessible. Until issues of speciation are solved and tiny cytometers based on chip-based fluidics are developed, it is unlikely that flow cytometry will provide a viable approach to widespread malaria diagnosis.
Another recent approach to malaria diagnosis has been the introduction of the polymerase chain reaction (PCR) and Taqman amplification methods for detecting the genetic signature of malarial parasites (Zhou et al., 1998; Zhong and Kain, 1999; Zhong et al., 2001; Blair et al., 2002; Lee et al., 2002). In principle, these amplification methods can provide quantitative signals that reflect the level of malaria infection and offer almost unlimited sensitivity. Furthermore, genetic signatures identify species uniquely. PCR was the first method to dramatically outperform microscopic analysis in terms of both sensitivity and speciation and the first to allow the limits of microscopic testing to be accurately quantified (Ohrt et al., 2002). For example, PCR has revealed that the microscope fails to detect mixed infections far more often than had been assumed. However, if they are to be quantitative, gene amplification methods require careful preparation of blood to remove inhibitors of amplification and PCR demands thermal cycling. Quantitative measurements of a molecular beacon (Blair et al., 2002; Lee et al., 2002) or some other detectable product are also required if real time measurements are to be made. Despite these challenges, gene amplification methods emerge as extremely attractive technologies because they offer superb sensitivity (Cox-Singh et al., 1997), greatly improved identification of species and strain, and, for the first time, the possibility of identifying additional disease characteristics. For example, the malarial genome has been sequenced recently and genotypic markers corresponding to drug resistance and other potentially important diagnostic and prognostic determinants are already identifiable by the methods (Labbe et al., 2001; Pillai et al., 2001; Durand et al., 2002; Aubouy et al., 2003; Pickard et al., 2003). Finally, and even better than gene amplification, methods are under development that offer extremely low detection limits for gene sequence detection without the need for amplification. At present, however, such amplification-free genetic detection methodologies are not sufficiently developed for serious consideration in malaria diagnostic applications.
Gene chips represent another technology for exploiting genotypic signatures as diagnostic and prognostic markers. In gene chips, multiple oligonucleotide probes that are complimentary to target gene sequences are spatially arrayed on a substrate (Ganesan et al., 2002; Rathod et al., 2002). These are exposed simultaneously to specimen nucleic acids, which hybridize with any complimentary probes that are present on the array to form short double stranded DNA segments that are stained and detected by fluorescence. Unfortunately, shotgun PCR is usually required to amplify the DNA from a small number of infected cells to provide the relatively large amount of target DNA that is needed. Sample preparation required for gene chip analysis is therefore very complex even compared to that needed for the PCR and Taqman gene amplification methods. The data reader and analyzer for gene chips also need to be sophisticated, especially if they are to provide interpretation of results, and gene chips cannot be reused, making it hard to imagine that they could become sufficiently easy to use or cost-effective for routine diagnosis in the foreseeable future. Similar arguments of complexity and cost apply to proteomic analysis (Florens et al., 2002) and mass spectrometry (Demirev et al., 2002). Thus while the level of discrimination provided by gene and protein chips are unparalleled, it seems likely that these will remain, for the foreseeable future, research laboratory-based technologies that will help provide much deeper understanding of malaria. Most likely, only a very limited subset of the information available from such refined analysis will become exploited for routine diagnostic purposes by less demanding technologies.
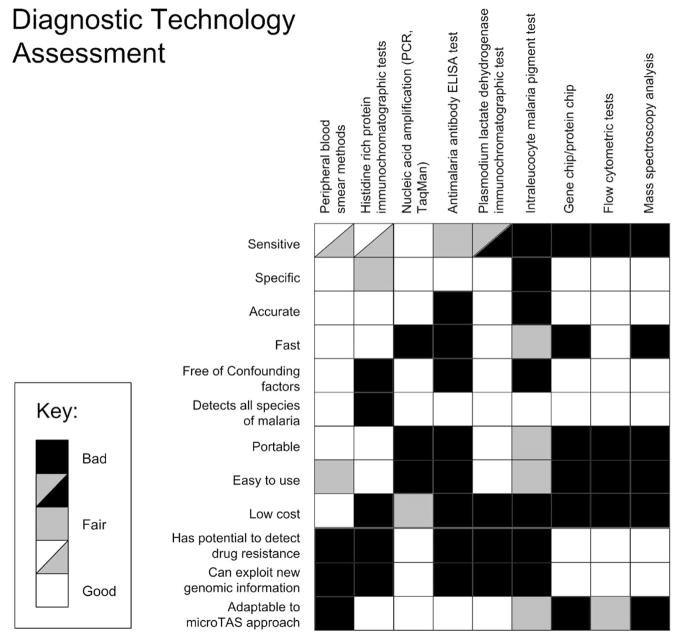
Fig. 1 summarizes the foregoing discussion of malaria diagnostic methods, highlighting in the top panels their strengths and weaknesses. In the bottom three panels of Fig. 1, we also address the question “what if each method could be automated and run inexpensively in the field?” Two characteristics that would have a major impact on malaria diagnosis, namely the ability to detect drug resistance and to utilize new genetic information as it becomes available as a diagnostic marker, are considered there.
Fig. 1.
Visual representation of some of the main methodologies used in the diagnosis of malaria. While microscopic analysis is among the most competitive methods in common use today, the diagram reveals that genetic methods offer significant advantages for the future if they can be realized in an inexpensive micro total analysis format.
Based on these criteria, the table shows that, of the current methods, microscopic analysis remains the most applicable to inexpensive field diagnosis of malaria. Of the methods that are adaptable to an automated microTAS environment, however, gene amplification offers the best sensitivity, species discrimination, ability to identify genetic variants, and adaptability to microTAS approaches. Each of the other methods falls short on one or more desirable criteria for diagnostic technology, including the microscopic techniques that are currently so dominant.
3. Micrototal analysis system approaches
Although gene amplification emerges from the foregoing analysis as a highly desirable approach for malaria diagnosis, implementing it in a fully automated system provides a significant technological challenge.
First, sample preparation needs to be carefully considered (Lantz et al., 2000; von Wintzingerode et al., 1997; Barker et al., 1992, 1994). Although, in principle, each cycle of gene amplification can double the number of target sequences so that the yield after N cycles in the absence of reagent exhaustion should be 2N, this efficiency is never achieved in practice. Rather, the actual yield approaches aN, where 1 ≤ a ≤ 2 and a depends critically on the reaction conditions. Variations in a are introduced by non-idealities of the reagent composition and physical environment and, especially, by any inhibitors that may be present in the mixture. Unfortunately, small changes in a produce very large changes in the yield aN because, typically, 20–30 amplification cycles are employed (N is between 20 and 30). For example, a 10% drop in reaction efficiency for N = 30 results in an 80% fall in yield. Put another way, for a given concentration of target nucleic acid, a five fold variation in detected signal will result from a change of only 0–10% in the efficiency of each amplification step. Inhibitors, such as trace metals are common in hematological specimens, and gene amplification of blood therefore demands sample preparation steps that will remove such confounding agents if the detected signal is to bear any reasonable quantitative relationship to the concentration of genetic target in the specimen. A successful micrototal analysis system for malaria detection by gene amplification therefore requires a sample preparation front end to remove reaction inhibitors.
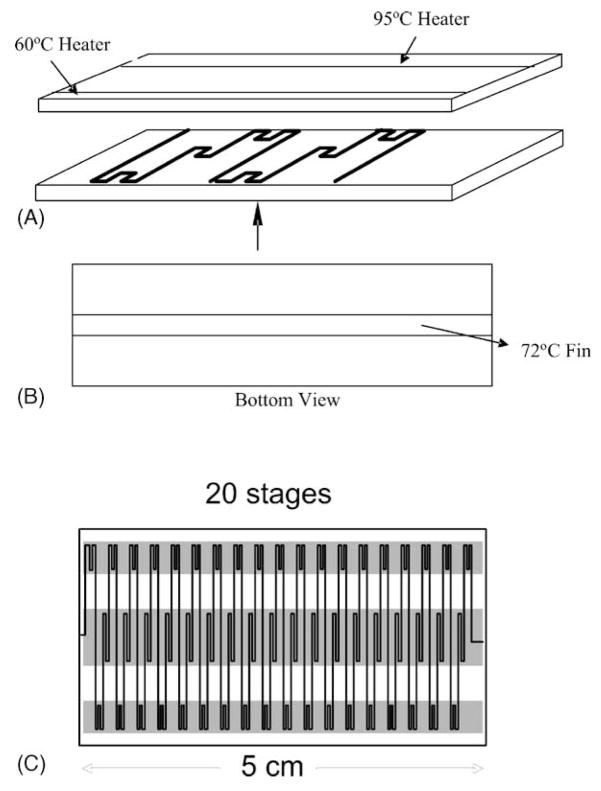
Once purified, real time PCR amplification in microfluidic systems is possible as has been published by Kopp et al. (1998) and, more recently, by several other groups (deMello, 2003). In collaboration with Shaochen Chen of the University of Texas in Austin, we have tested a similar flow-through PCR system adapted from these designs (see Fig. 2) in our own laboratory but have not yet applied it to malaria. The basic design for this flow-through system is shown in Fig. 2.
Fig. 2.
A. Exploded view of a flow-through PCR chip showing the configuration of flow channels in the first two temperature cycling stages. As the sample slowly flows through the channel is temperature cycled and nucleic acid amplification occurs. B. Bottom view showing the central temperature zone. C. Top view of a device to provide 20 thermal cycles. Such a device is capable of completing each temperature cycle in a matter of seconds, allowing rapid real-time PCR.
4. Microfluidic sample preparation by dielectrophoresis
To address the need for sample preparation, we have applied dielectrophoresis (DEP) to the problem of the isolation of malaria-parasitised cells. The DEP method is also applicable to other diseases characterised by the presence of abnormal cells including anomalous native cells, parasites and bacteria. Cells having dissimilar dielectric properties tend to move to a position of low energy if their suspending medium is subjected to an inhomogeneous electrical field because of the so-called dielectrophoretic force (Pohl, 1978, Wang et al., 1995). This is not related to the more familiar electrophoresis effect and is comprised of two distinct and independent components, F̄DEP = F̄in hom + F̄trav, that arise from interactions between field-induced charge polarization in the cells and inhomogeneneities in the electrical field and with movement of the electrical field pattern, respectively. F̄in hom is a force that attracts or repels cells to or from electrode edges, while F̄trav carries cells along parallel to electrode surfaces. The DEP force can be approximated in terms of dipolar effects as
where the first and second terms correspond to F̄in hom and F̄trav, respectively, v the particle volume, and εm the electrical permittivity of the suspending medium (Wang et al., 1995). The Claussius–Mossotti charge polarization factor, given by
describes the electrical polarization of the cell with respect to its suspending medium. and are the complex permittivities of the cell and its medium, respectively (Irimajiri et al., 1979). These take the form where εx is the real dielectric permittivity (dielectric constant), σx is the conductivity, f is the frequency of the applied electrical field, and . The real and imaginary components of fCM, Re[fCM] and Im[fCM], couple with the spatial inhomogeneity and travelling components of the applied electrical field to create the DEP force components. These forces depend not only on the geometrical configuration and excitation scheme of the electrode array but also on the dielectric properties of the cell and of its suspending medium (Chan et al., 1997; Yang et al., 1999; Ratanachoo et al., 2002).
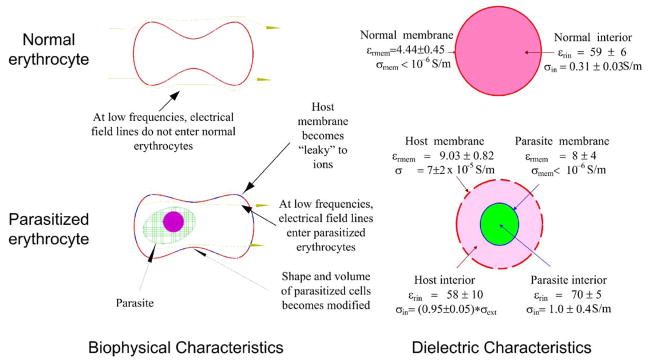
Although they may sound complicated, DEP forces are produced in cells by applying alternating (ac) electric fields of 10 kHz to 100 MHz in frequency from a simple signal generator to a small electrode patterned on the floor of a thin chamber. The magnitude, direction and frequency dependencies of cellular DEP responses depend on the composition, conformation and barrier function of the cell plasma membranes (Pethig and Kell, 1987; Wang et al., 1994). Because cells of different types and different physiological and pathologic states have unique morphological and structural properties (i.e. the features that are observed by a pathologist), it is possible to discriminate between them and differentially manipulate them by exploiting their DEP frequency dependencies (Gascoyne and Vykoukal, 2002; Fuhr et al., 1994). We and others have shown that the electrical conductivity of erythrocyte membranes increases sharply when they become hosts to malarial parasites (Aceti et al., 1990; Gascoyne et al., 1997, 2002), an effect that may be related to the introduction of membrane permeation pathways (Ginsburg, 1990; Kirk, 2000; Wagner et al., 2003) to membrane peroxidation damage (Huber et al., 2002; Mohan et al., 1992), and to changes in membrane fluidity (Sibmooh et al., 2000; Ginsburg, 1990) following infection. The dielectric differences between normal and parasitized cells determined by us are summarized in Fig. 3. Recently, we showed that these properties could be exploited in order to permit the discrimination and isolation of parasitized cells from uninfected cells on small electrode arrays (Gascoyne et al., 2002) to aid in the microscopic detection of malaria. While that isolation method provides spatial focusing of parasitized cells, it does not readily interface with follow-on stages, such as those needed to provide gene amplification and detection.
Fig. 3.
The biophysical and dielectric differences between normal and malarially-parasitized cells deduced from our previous studies (summarized from Gascoyne et al., 1997).
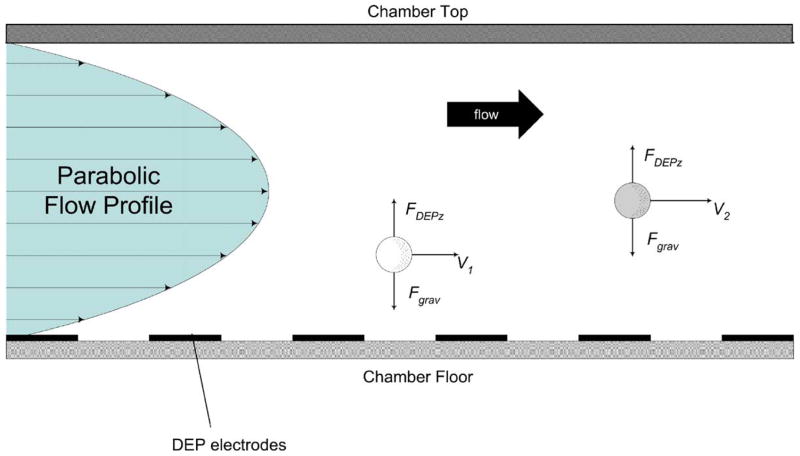
Here, we have modified our DEP approach to malaria isolation by applying another method called DEP-field-flow-fractionation in order to allow malarially-infected erythrocytes to be both isolated and passed on to a subsequent analysis stage. Field-flow fractionation (FFF) is a family of methods (Giddings, 1993) in which force fields are applied to particles to position them characteristically within the velocity profile of a fluid flow stream. Particles having different characteristics are then carried along at different velocities in accordance with their respective positions in the flow stream. A specimen containing a mixture of cell types can be introduced at one end of a microchannel FFF device and different cell types are fractionated as the specimen traverses the channel. Different cell types then emerge from the other end of the channel at different times. In DEP–FFF, a separation chamber is utilized that has electrodes patterned on its floor to which ac signals are applied (Gascoyne and Vykoukal, 2002; Wang et al., 2000). By choosing an applied frequency that provides DEP repulsion, cells are levitated to heights at which the levitation and sedimentation forces balance. This effect is illustrated in Fig. 4. Detailed analyses of the DEP levitation properties of particles having different dielectric properties, and the corresponding DEP–FFF separation equations, have been published by us elsewhere (Gascoyne and Vykoukal, 2002; Wang et al., 2000). Earlier dielectric studies of malaria showed that the dielectric properties of parasitized cells are different from those of uninfected cells (Aceti et al., 1990; Gascoyne et al., 1997, 2002). Consequently, with the application of an appropriate electrical signal, parasitized and normal cells may be levitated to different heights and these differential DEP levitation characteristics can be exploited to fractionate them by DEP–FFF. Because the different cell types emerge at different times, their isolation is facilitated.
Fig. 4.
Side view of a DEP–FFF particle fractionating chamber showing the parabolic hydrodynamic flow profile that forms spontaneously when fluid flow is initiated and the forces that are experienced by particles due to sedimentation and repulsive dielectrophoretic forces from electrodes on the chamber floor. These forces balance at different levitation heights for particles having different density and dielectric properties. Because they equilibriate at different heights, different particles are carried at different speeds by the hydrodynamic flow profile and emerge from the chamber at different times.
Cell samples from erythrocyte cultures infected with chloroquine-resistant P. falciparum strain T9/94 RC17 were derived by dilution, and subsequent micromanipulation-cloning of isolate T9 from a patient in Mae Sod, Tak Province, Thailand, by Dr. Sodsri Thaithong of the WHO Collaboratory Centre for Biological Characterization of Malarial Parasites, Chulalongkorn University, Bangkok, Thailand. Parasites were cultured in normal (type ‘O’) erythrocytes in human serum-supplemented-RPMI 1640 medium under a 95% air/5% CO2 atmosphere at 37 °C using the methods of Trager and Jensen (1976). Percent parasitemia was determined from Giemsa-stained thin smears and was maintained between 1 and 5% during culture. For DEP analysis, cell cultures were diluted with an isotonic buffer of 8.5% sucrose + 0.3% dextrose to provide suspensions having conductivities of 55 mS/m. The live cell DNA dye Sybro 14 (Molecular Probes, Eugene, OR, USA), was used to render viable parasites easily detectable within host cells by fluorescence microscopy and flow cytometry.
For these studies we used an existing DEP–FFF chamber that was 300 mm in length, 25 mm width, and 400 μm height and the electrode array consisted of 50 μm wide parallel electrodes spaced by 50 μm gaps. The electrode arrays were cut from approximately 300 m lengths that were mass produced by bulk micromachining 0.8 μm copper on a 35 μm thick by 50 mm wide Kaptan substrate. The resulting copper electrodes were then flash coated with 0.2 μm gold (Minnesota Mining and Manufacturing Company, Austin, TX). Sinusoidal signals up to 5 V p–p in the frequency range 40–250 kHz were used in different experiments and these were provided by a home-built function generator. Fluid flow of up to 1.5 ml/min was used to separate the cells in the DEP–FFF channel and fluid emerging from the channel was passed directly through a flow cytometer (Bryte cytometer, Bio Rad, USA), bypassing the cytometer sample pump, to detect the cells. The cytometer was operated in time-resolved mode, gated by forward light scatter, and set to measure Sybro 14 fluorescence. In this way, elution fractograms of both parasitized- and normal-erythrocytes could be resolved and parasitization could be inferred from the fluorescence signal of the Sybro 14. Relatively high parasitemias (range 1–10%) and small sample sizes (~105 cells) were used to facilitate flow cytometric evaluations of the separations because the cytometer was unable to count huge numbers of cells.
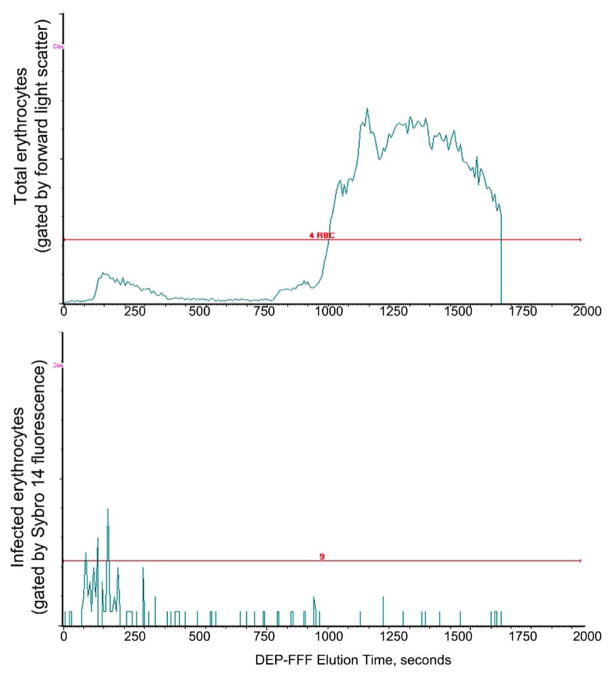
A typical elution profile for a parasitized cell sample is shown in Fig. 5. At the electric field frequency applied, parasitized cells were levitated more strongly than normal cells and they were therefore carried more swiftly by the eluate flow (since they further from the chamber floor) than the normal erythrocytes. Consequently, the parasitized cells emerged more rapidly from the DEP–FFF chamber than normal cells. During the separation the parasitized cells were washed by several hundred volumes of eluate so they emerged from the chamber free of serum proteins. Finally, under the conditions used, nucleated blood cells were retained in the DEP–FFF chamber and emerged after the normal erythrocyte fraction when the electric field was turned off. Consequently, no leukocytes contaminated the parasitized cell fraction. Therefore, the only DNA in the initial cell isolate was that in the parasites.
Fig. 5.
Elution profile for malarially-infected blood from a DEP–FFF chamber measured by time-resolved flow cytometry. The top panel shows all erythrocytes that emerge from the chamber. The bottom panel shows those erythrocytes that fluoresce because of staining of intracellular parasites with Sybro 14 live-cell DNA stain. Most of the parasitized cells emerge quickly from the DEP–FFF chamber, while normal cells emerge later. Leucocytes emerge after the normal erythrocytes (not shown).
5. Discussion
These results show that the dielectric differences between parasitized and normal erythrocytes that were reported previously (Aceti et al., 1990; Gascoyne et al., 1997) and were used by us for microscopic analysis of both infected cultures and clinical specimens (Gascoyne et al., 2002) can also be exploited by the DEP–FFF method to provide the means for both thoroughly washing parasitized cells and for separating them from normal cells and nucleocytes. For a micro-TAS system, the size of the DEP–FFF chamber could be significantly decreased without loss of performance and the chamber could be integrated within the same chip as a subsequent flow-through PCR stage. Despite the apparent sophistication of a microTAS containing integrated DEP–FFF and gene amplification components, such a device can be constructed from a bottom substrate having electrodes patterned on one side that is bonded to a top substrate having a single, serpentine channel. The electronics for this approach are simple and driven by a single controller. Release of nucleic acid from parasitized cells may be accomplished by electrobursting (Pawlowski et al., 1993, 1996), sonication (Fykse et al., 2003) or lysis, all readily adaptable to an integrated microsystem. Optical detection can be achieved by simple LED-based optics. Perhaps the greatest degree of complexity lies in the means to control molecular amplification reagents. In terms of expense, if it were possible to produce a durable, multi-use microTAS, the device should cost less than a microscope and the cost per test would be driven by the molecular amplification reagents.
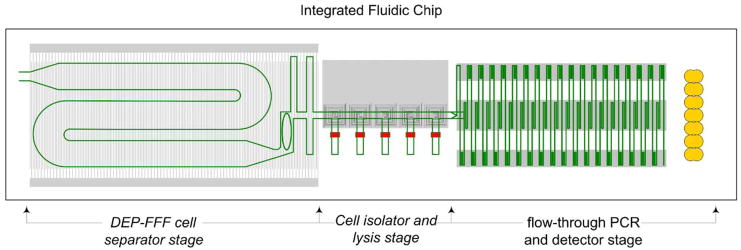
Based on our findings, it is now our aim to attempt to realize a small, self-contained system to demonstrate integrated separation and PCR analysis of malarially-parasitized cultures. Clearly, if this is successful, positive results would then need to be demonstrated not only for clinical samples but also for other plasmodium species and strains before the approach could be considered generally applicable for clinical applications. A design for the microfluidics for such a system is shown in Fig. 6; such a design could be cheaply mass-produced.
Fig. 6.
Design for an integrated DEP–FFF front end, cell lysis stage, and flow-through, real-time PCR system. Such a system could be inexpensively mass produced and could, in principle, be made highly portable for diagnosis of malaria and other diseases in laboratories, clinics, and in the field.
6. Conclusions
Of the currently available approaches to malaria detection, gene amplification appears to offer the most significant potential for improved diagnosis but will become generally applicable only if it can be embodied in a portable, fully automated format that can be operated at a low cost per specimen. Gene amplification methods, such as the PCR approach described here, and sample preparation, as demonstrated above for the DEP–FFF method for washing and isolating malarially-infected cells, are adaptable to integration within micro total analysis systems. Because such automated systems could potentially deliver high sensitivity diagnostic capabilities, accurately diagnose mixed infections, detect drug-resistant strains, and render other genetic markers available as diagnostic tools for laboratory and field environments alike, we believe that further efforts to explore microTAS approaches to malaria detection are an important priority.
Micro total analysis system sample preparation and gene amplification methodologies are potentially applicable not only to malaria but also to other pathogens as well. For example, the combined DEP–FFF-gene amplification approach described here is highly adaptable because DEP–FFF conditions can be electronically programmed to identify and isolate a wide variety of different cell types (Gascoyne and Vykoukal, 2002) and PCR can be attuned to any target molecular signature by the use of appropriate probes. Therefore, it is conceivable that a well-designed microTAS instrument could address not only malaria but also a variety of other tropical diseases in developed and underserved regions alike.
Acknowledgments
We thank the staff of the Chulabhorn Research Institute for the culture, preparation and experiments with malarial cultures and clinical samples, Shaochen Chen and his students for the construction of the microfluidic PCR devices and Tom Anderson for building the DEP–FFF chambers. This work was supported by the Chulabhorn Research Institute and the National Institute for Diabetes and Digestive and Kidney Diseases (NIDDK).
References
- Aceti A, Bonincontro A, Cametti C, Celestino D, Leri O. Electrical conductivity of human erythrocytes infected with Plasmodium falciparum and its modification following quinine therapy. Trans R Soc Trop Med Hyg. 1990;84 (5):671–672. doi: 10.1016/0035-9203(90)90140-a. [DOI] [PubMed] [Google Scholar]
- Aubouy A, Jafari S, Huart V, Migot-Nabias F, Mayombo J, Durand R, Bakary M, Le Bras J, Deloron P. DHFR and DHPS genotypes of Plasmodium falciparum isolates from Gabon correlate with in vitro activity of pyrimethamine and cycloguanil, but not with sulfadoxine–pyrimethamine treatment efficacy. J Antimicrob Chemother. 2003;52 (1):43–49. doi: 10.1093/jac/dkg294. [DOI] [PubMed] [Google Scholar]
- Bangs MJ, Rusmiarto S, Gionar YR, Chan AS, Dave K, Ryan JR. Evaluation of a dipstick malaria sporozoite panel assay for detection of naturally infected mosquitoes. J Med Entomol. 2002;39 (2):324–330. doi: 10.1603/0022-2585-39.2.324. [DOI] [PubMed] [Google Scholar]
- Barkan D, Ginsburg H, Golenser J. Optimisation of flow cytometric measurement of parasitaemia in plasmodium-infected mice. Int J Parasitol. 2000;30 (5):649–653. doi: 10.1016/s0020-7519(00)00035-7. [DOI] [PubMed] [Google Scholar]
- Barker RH, Jr, Banchongaksorn T, Courval JM, Suwonkerd W, Rimwungtragoon K, Wirth DF. Plasmodium falciparum and P. vivax: factors affecting sensitivity and specificity of PCR-based diagnosis of malaria. Exp Parasitol. 1994;79 (1):41–49. doi: 10.1006/expr.1994.1057. [DOI] [PubMed] [Google Scholar]
- Barker RH, Jr, Banchongaksorn T, Courval JM, Suwonkerd W, Rimwungtragoon K, Wirth DF. A simple method to detect Plasmodium falciparum directly from blood samples using the polymerase chain reaction. Am J Trop Med Hyg. 1992;46 (4):416–426. doi: 10.4269/ajtmh.1992.46.416. [DOI] [PubMed] [Google Scholar]
- Birku Y, Welday D, Ayele D, Shepherd A. Rapid diagnosis of severe malaria based on the detection of Pf-Hrp-2 antigen. Ethiop Med J. 1999;37 (3):173–179. [PubMed] [Google Scholar]
- Blair PL, Witney A, Haynes JD, Moch JK, Carucci DJ, Adams JH. Transcripts of developmentally regulated Plasmodium falciparum genes quantified by real-time RT-PCR. Nucleic Acids Res. 2002;30 (10):2224–2231. doi: 10.1093/nar/30.10.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox-Singh J, Mahayet S, Abdullah MS, Singh B. Increased sensitivity of malaria detection by nested polymerase chain reaction using simple sampling and DNA extraction. Int J Parasitol. 1997;27 (12):1575–1577. doi: 10.1016/s0020-7519(97)00147-1. [DOI] [PubMed] [Google Scholar]
- Chan KL, Gascoyne PR, Becker FF, Pethig R. Electrorotation of liposomes: verification of dielectric multi-shell model for cells. Biochim Biophys Acta. 1997;1349 (2):182–196. doi: 10.1016/s0005-2760(97)00092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho D, Kim KH, Park SC, Kim YK, Lee KN, Lim CS. Evaluation of rapid immunocapture assays for diagnosis of Plasmodium vivax in Korea. Parasitol Res. 2001;87 (6):445–448. doi: 10.1007/s004360000360. [DOI] [PubMed] [Google Scholar]
- Demirev PA, Feldman AB, Kongkasuriyachai D, Scholl P, Sullivan D, Jr, Kumar N. Detection of malaria parasites in blood by laser desorption mass spectrometry. Anal Chem. 2002;74 (14):3262–3266. doi: 10.1021/ac025621k. [DOI] [PubMed] [Google Scholar]
- Durand R, Huart V, Jafari S, Le Bras J. Rapid detection of a molecular marker for chloroquine-resistant falciparum malaria. Antimicrob Agents Chemother. 2002;46 (8):2684–2686. doi: 10.1128/AAC.46.8.2684-2686.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, Witney AA, Wolters D, Wu Y, Gardner MJ, Holder AA, Sinden RE, Yates JR, Carucci DJ. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419 (6906):520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- Fuhr G, Muller T, Schnelle T, Hagedorn R, Voigt A, Fiedler S, Arnold WM, Zimmermann U, Wagner B, Heuberger A. Radio-frequency microtools for particle and liver cell manipulation. Naturwissenschaften. 1994;81 (12):528–535. doi: 10.1007/BF01139998. [DOI] [PubMed] [Google Scholar]
- Fykse EM, Olsen JS, Skogan G. Application of sonication to release DNA from Bacillus cereus for quantitative detection by real-time PCR. J Microbiol Methods. 2003;55 (1):1–10. doi: 10.1016/s0167-7012(03)00091-5. [DOI] [PubMed] [Google Scholar]
- Ganesan K, Jiang L, Rathod PK. Stochastic versus stable transcriptional differences on Plasmodium falciparum DNA microarrays. Int J Parasitol. 2002;32 (13):1543–1550. doi: 10.1016/s0020-7519(02)00182-0. [DOI] [PubMed] [Google Scholar]
- Gascoyne P, Pethig R, Satayavivad J, Becker FF, Ruchirawat M. Dielectrophoretic detection of changes in erythrocyte membranes following malarial infection. Biochim Biophys Acta. 1997;1323 (2):240–252. doi: 10.1016/s0005-2736(96)00191-5. [DOI] [PubMed] [Google Scholar]
- Gascoyne P, Mahidol C, Ruchirawat M, Satayavivad J, Watcharasit P, Becker F. Microsample preparation by dielectrophoresis: isolation of malaria. Lab Chip. 2002;2 (2):70–75. doi: 10.1039/b110990c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne PR, Vykoukal J. Particle separation by dielectrophoresis. Electrophoresis. 2002;23 (13):1973–1983. doi: 10.1002/1522-2683(200207)23:13<1973::AID-ELPS1973>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings JC. Field-flow fractionation: analysis of macromolecular, colloidal, and particulate materials. Science. 1993;260 (5113):1456–1465. doi: 10.1126/science.8502990. [DOI] [PubMed] [Google Scholar]
- Ginsburg H. Alterations caused by the intraerythrocytic malaria parasite in the permeability of its host cell membrane. Comp Biochem Physiol A. 1990;95 (1):31–39. doi: 10.1016/0300-9629(90)90006-e. [DOI] [PubMed] [Google Scholar]
- Huber SM, Uhlemann AC, Gamper NL, Duranton C, Kremsner PG, Lang F. Plasmodium falciparum activates endogenous Cl(−) channels of human erythrocytes by membrane oxidation. EMBO J. 2002;21 (1–2):22–30. doi: 10.1093/emboj/21.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huikko K, Kostiainen R, Kotiaho T. Introduction to micro-analytical systems: bioanalytical and pharmaceutical applications. Eur J Pharm Sci. 2003;20 (2):149–171. doi: 10.1016/s0928-0987(03)00147-7. [DOI] [PubMed] [Google Scholar]
- Iqbal J, Khalid N, Hira PR. Comparison of two commercial assays with expert microscopy for confirmation of symptomatically diagnosed malaria. J Clin Microbiol. 2002;40 (12):4675–4678. doi: 10.1128/JCM.40.12.4675-4678.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J, Khalid N, Hira PR. Performance of rapid malaria Pf antigen test for the diagnosis of malaria and false-reactivity with autoantibodies. Adv Exp Med Biol. 2003;531:135–148. doi: 10.1007/978-1-4615-0059-9_10. [DOI] [PubMed] [Google Scholar]
- Irimajiri A, Hanai T, Inouye A. A dielectric theory of “multi-stratified shell” model with its application to a lymphoma cell. J Theor Biol. 1979;78 (2):251–269. doi: 10.1016/0022-5193(79)90268-6. [DOI] [PubMed] [Google Scholar]
- Irion A, Beck HP, Smith T. Assessment of positivity in immunoassays with variability in background measurements: a new approach applied to the antibody response to Plasmodium falciparum MSP2. J Immunol Methods. 2002;259 (1–2):111–118. doi: 10.1016/s0022-1759(01)00500-2. [DOI] [PubMed] [Google Scholar]
- John SM, Sudarsanam A, Sitaram U, Moody AH. Evaluation of OptiMAL, a dipstick test for the diagnosis of malaria. Ann Trop Med Parasitol. 1998;92 (5):621–622. doi: 10.1080/00034989859320. [DOI] [PubMed] [Google Scholar]
- Kirk K. Malaria channelling nutrients. Nature. 2000;406 (6799):949–951. doi: 10.1038/35023209. [DOI] [PubMed] [Google Scholar]
- Kopp MU, Mello AJ, Manz A. Chemical amplification: continuous-flow PCR on a chip. Science. 1998;280 (5366):1046–1048. doi: 10.1126/science.280.5366.1046. [DOI] [PubMed] [Google Scholar]
- Labbe AC, Bualombai P, Pillai DR, Zhong KJ, Vanisaveth V, Hongvanthong B, Looareesuwan S, Kain KC. Molecular markers for chloroquine-resistant Plasmodium falciparum malaria in Thailand and Laos. Ann Trop Med Parasitol. 2001;95 (8):781–788. doi: 10.1080/00034980120103414. [DOI] [PubMed] [Google Scholar]
- Labbe AC, Pillai DR, Hongvangthong B, Vanisaveth V, Pomphida S, Inkathone S, Hay Burgess DC, Kain KC. The performance and utility of rapid diagnostic assays for Plasmodium falciparum malaria in a field setting in the Lao People’s Democratic Republic. Ann Trop Med Parasitol. 2001;95 (7):671–677. doi: 10.1080/00034980120103243. [DOI] [PubMed] [Google Scholar]
- Lantz PG, Abu al-Soud W, Knutsson R, Hahn-Hagerdal B, Radstrom P. Biotechnical use of polymerase chain reaction for microbiological analysis of biological samples. Biotechnol Annu Rev. 2000;5:87–130. doi: 10.1016/s1387-2656(00)05033-x. [DOI] [PubMed] [Google Scholar]
- Lee MA, Tan CH, Aw LT, Tang CS, Singh M, Lee SH, Chia HP, Yap EP. Real-time fluorescence-based PCR for detection of malaria parasites. J Clin Microbiol. 2002;40 (11):4343–4345. doi: 10.1128/JCM.40.11.4343-4345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lema OE, Carter JY, Nagelkerke N, Wangai MW, Kitenge P, Gikunda SM, Arube PA, Munafu CG, Materu SF, Adhiambo CA, Mukunza HK. Comparison of five methods of malaria detection in the outpatient setting. Am J Trop Med Hyg. 1999;60 (2):177–182. doi: 10.4269/ajtmh.1999.60.177. [DOI] [PubMed] [Google Scholar]
- Makler MT, Ries JM, Williams JA, Bancroft JE, Piper RC, Gibbins BL, Hinrichs DJ. Parasite lactate dehydrogenase as an assay for Plasmodium falciparum drug sensitivity. Am J Trop Med Hyg. 1993;48 (6):739–741. doi: 10.4269/ajtmh.1993.48.739. [DOI] [PubMed] [Google Scholar]
- deMello AJ. Microfluidics: DNA amplification moves on. Nature. 2003;422 (6927):28–29. doi: 10.1038/422028a. [DOI] [PubMed] [Google Scholar]
- Mohan K, Ganguly NK, Dubey ML, Mahajan RC. Oxidative damage of erythrocytes infected with Plasmodium falciparum: an in vitro study. Ann Hematol. 1992;65 (3):131–134. doi: 10.1007/BF01695812. [DOI] [PubMed] [Google Scholar]
- Moody A. Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev. 2002;15 (1):66–78. doi: 10.1128/CMR.15.1.66-78.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CK, Bell D, Gasser RA, Wongsrichanalai C. Rapid diagnostic testing for malaria. Trop Med Int Health. 2003;8 (10):876–883. doi: 10.1046/j.1365-3156.2003.01115.x. [DOI] [PubMed] [Google Scholar]
- Ohrt C, Sutamihardja MA, Tang D, Kain KC. Purnomo, impact of microscopy error on estimates of protective efficacy in malaria-prevention trials. J Infect Dis. 2002;186 (4):540–546. doi: 10.1086/341938. [DOI] [PubMed] [Google Scholar]
- Pawlowski P, Szutowicz I, Marszalek P, Fikus M. Bioelectrorheological model of the cell. Part 5 Electrodestruction of cellular membrane in alternating electric field. Biophys J. 1993;65 (1):541–549. doi: 10.1016/S0006-3495(93)81056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski P, Szutowicz I, Rozycki S, Zielinski J, Fikus M. Bioelectrorheological model of the cell. Part VI Experimental verification of the rheological model of cytoplasmic membrane. Biophys J. 1996;70 (2):1024–1026. doi: 10.1016/S0006-3495(96)79647-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pethig R, Kell DB. The passive electrical properties of biological systems: their significance in physiology, biophysics and biotechnology. Phys Med Biol. 1987;32 (8):933–970. doi: 10.1088/0031-9155/32/8/001. [DOI] [PubMed] [Google Scholar]
- Pickard AL, Wongsrichanalai C, Purfield A, Kamwendo D, Emery K, Zalewski C, Kawamoto F, Miller RS, Meshnick SR. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob Agents Chemother. 2003;47 (8):2418–2423. doi: 10.1128/AAC.47.8.2418-2423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai DR, Labbe AC, Vanisaveth V, Hongvangthong B, Pomphida S, Inkathone S, Zhong K, Kain KC. Plasmodium falciparum malaria in Laos: chloroquine treatment outcome and predictive value of molecular markers. J Infect Dis. 2001;183 (5):789–795. doi: 10.1086/318836. [DOI] [PubMed] [Google Scholar]
- Pohl HA. Dielectrophoresis: The behavior of neutral matter in nonuniform electric fields. Cambridge University Press; New York: 1978. [Google Scholar]
- Ratanachoo K, Gascoyne PR, Ruchirawat M. Detection of cellular responses to toxicants by dielectrophoresis. Biochim Biophys Acta. 2002;1564 (2):449–458. doi: 10.1016/s0005-2736(02)00494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathod PK, Ganesan K, Hayward RE, Bozdech Z, DeRisi JL. DNA microarrays for malaria. Trends Parasitol. 2002;18 (1):39–45. doi: 10.1016/s1471-4922(01)02153-5. [DOI] [PubMed] [Google Scholar]
- Richardson DC, Ciach M, Zhong KJ, Crandall I, Kain KC. Evaluation of the Makromed dipstick assay versus PCR for diagnosis of Plasmodium falciparum malaria in returned travelers. J Clin Microbiol. 2002;40 (12):4528–4530. doi: 10.1128/JCM.40.12.4528-4530.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito-Ito A, Akai Y, He S, Kimura M, Kawabata M. A rapid, simple and sensitive flow cytometric system for detection of Plasmodium falciparum. Parasitol Int. 2001;50 (4):249–257. doi: 10.1016/s1383-5769(01)00091-5. [DOI] [PubMed] [Google Scholar]
- Schwick P, Eggelte TA, Hess F, Tueumuna TT, Payne D, Nothdurft HD, von Sonnenburg F, Loscher T. Sensitive ELISA dipstick test for the detection of chloroquine in urine under field conditions. Trop Med Int Health. 1998;3 (10):828–832. doi: 10.1046/j.1365-3156.1998.00307.x. [DOI] [PubMed] [Google Scholar]
- Sibmooh N, Pipitaporn B, Wilairatana P, Dangdoungjai J, Udomsangpetch R, Looareesuiwan S, Chantharaksri U. Effect of artemisinin on lipid peroxidation and fluidity of the erythrocyte membrane in malaria. Biol Pharm Bull. 2000;23 (11):1275–1280. doi: 10.1248/bpb.23.1275. [DOI] [PubMed] [Google Scholar]
- Silvie O, Thellier M, Rosenheim M, Datry A, Lavigne P, Danis M, Mazier D. Potential value of Plasmodium falciparum-associated antigen and antibody detection for screening of blood donors to prevent transfusion-transmitted malaria. Transfusion. 2002;42 (3):357–362. doi: 10.1046/j.1537-2995.2002.00050.x. [DOI] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193 (4254):673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Wagner MA, Andemariam B, Desai SA. A two-compartment model of osmotic lysis in Plasmodium falciparum-infected erythrocytes. Biophys J. 2003;84 (1):116–123. doi: 10.1016/S0006-3495(03)74836-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Huang Y, Gascoyne PR, Becker FF, Holzel R, Pethig R. Changes in Friend murine erythroleukaemia cell membranes during induced differentiation determined by electrorotation. Biochim Biophys Acta. 1994;1193 (2):330–344. doi: 10.1016/0005-2736(94)90170-8. [DOI] [PubMed] [Google Scholar]
- Wang XB, Hughes MP, Huang Y, Becker FF, Gascoyne PR. Non-uniform spatial distributions of both the magnitude and phase of ac electric fields determine dielectrophoretic forces. Biochim Biophys Acta. 1995;1243 (2):185–194. doi: 10.1016/0304-4165(94)00146-o. [DOI] [PubMed] [Google Scholar]
- Wang XB, Yang J, Huang Y, Vykoukal J, Becker FF, Gascoyne PR. Cell separation by dielectrophoretic field-flow-fractionation. Anal Chem. 2000;72 (4):832–839. doi: 10.1021/ac990922o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wintzingerode F, Gobel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21 (3):213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- Wolday D, Balcha F, Fessehaye G, Birku Y, Shepherd A. Field trial of the RTM dipstick method for the rapid diagnosis of malaria based on the detection of Plasmodium falciparum HRP-2 antigen in whole blood. Trop Doct. 2001;31 (1):19–21. [PubMed] [Google Scholar]
- Wongsrichanalai C. Rapid diagnostic techniques for malaria control. Trends Parasitol. 2001;17 (7):307–309. doi: 10.1016/s1471-4922(01)01925-0. [DOI] [PubMed] [Google Scholar]
- Yang J, Huang Y, Wang X, Wang XB, Becker FF, Gascoyne PR. Dielectric properties of human leukocyte subpopulations determined by electrorotation as a cell separation criterion. Biophys J. 1999;76 (6):3307–3314. doi: 10.1016/S0006-3495(99)77483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong KJ, Kain KC. Evaluation of a colorimetric PCR-based assay to diagnose Plasmodium falciparum malaria in travelers. J Clin Microbiol. 1999;37 (2):339–341. doi: 10.1128/jcm.37.2.339-341.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong KJ, Salas CJ, Shafer R, Gubanov A, Gasser RA, Jr, Magill AJ, Forney JR, Kain KC. Comparison of IsoCode STIX and FTA Gene Guard collection matrices as whole-blood storage and processing devices for diagnosis of malaria by PCR. J Clin Microbiol. 2001;39 (3):1195–1196. doi: 10.1128/JCM.39.3.1195-1196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Liu Q, Wongsrichanalai C, Suwonkerd W, Panart K, Prajakwong S, Pensiri A, Kimura M, Matsuoka H, Ferreira MU, Isomura S, Kawamoto F. High prevalence of Plasmodium malariae and Plasmodium ovale in malaria patients along the Thai-Myanmar border. Trop Med Int Health. 1998;3 (4):304–312. doi: 10.1046/j.1365-3156.1998.00223.x. [DOI] [PubMed] [Google Scholar]