Abstract
Dielectrophoretic field-flow fractionation (dFFF) was applied as a contact-free way to sense changes in the plasma membrane capacitances and conductivities of cultured human HL-60 cells in response to toxicant exposure. A micropatterned electrode imposed electric forces on cells in suspension in a parabolic flow profile as they moved through a thin chamber. Relative changes in the dFFF peak elution time, reflecting changes in cell membrane area and ion permeability, were measured as indices of response during the first 150 min of exposure to eight toxicants having different single or mixed modes of action (acrylonitrile, actinomycin D, carbon tetrachloride, endosulfan, N-nitroso-N-methylurea (NMU), paraquat dichloride, puromycin, and styrene oxide). The dFFF method was compared with the cell viability assay for all toxicants and with the mitochondrial potentiometric dye assay or DNA alkaline comet assay according to the mode of action of the specific agents. Except for low doses of nucleic acid-targeting agents (actinomycin D and NMU), the dFFF method detected all toxicants more sensitively than other assays, in some cases up to 105 times more sensitively than the viability approach. The results suggest the dFFF method merits additional study for possible applicability in toxicology.
All countries face increasing environmental pressures as the result of the release of agents having established or potential toxic effects in the aquatic milieu. Current methodologies for toxicant screening typically involve the collection, transportation, and subsequent chemical or biological investigation of field samples in a centralized laboratory. Although chemical analysis is fast, cost-effective, and quantitatively determines the composition of environmental samples, it does not reveal the health impact of the contaminants. Conversely, while laboratory animal tests may detect the health impact of toxicants, they are slow, expensive, suffer from large species variations that render them insufficiently sensitive for reliable analysis of human toxicity,1,2 and are subject to ethical concerns. In vitro viability testing using human-derived cell cultures is an alternative strategy for initial assessment of potential health effects without ethical concerns. However, to make testing with cultures effective, an array of rapid indicators of cell responses is desirable. Cell viability measurements usually reveal responses over hours or days, making them relatively slow. Flow cytometric tests can assess a number of bioindicators such as mitochondrial membrane potential and cell surface markers, but these are dependent on expensive infrastructure.
Here we explore the feasibility of using dielectrically detected changes in membrane capacitance and conductivity of mammalian cells in suspension as a rapid indicator of responses to toxicant exposures. It is well-known from painstaking electrophysiological measurements of single cells that changes in cell membrane capacitance and conductivity reveal not only gross damage to the membrane but also subtle variations reflecting alterations in cellular metabolic rate,3,4 transmembrane transport,5,6 and membrane signaling events.7–9 Such subtle changes occur long before (and usually in the absence of) disruption of the barrier function that is the hallmark of cell viability determinations, a cornerstone of many toxicity studies.
Dielectrophoresis (DEP)10 and the related effect of electrorotation (ROT)11 have been used for many years to manipulate cells according to their membrane capacitance and conductivity properties and to measure these without the need to make contact with the cells.12–15 Furthermore, DEP and ROT have been shown to respond sensitively to changes in these cell membrane dielectric characteristics.13,16–19 ROT revealed that, when cells are exposed to toxic metals, cell membrane capacitance falls and membrane conductivity increases.20 Using either DEP or ROT methods, similar dielectric changes have been observed in human mesothelial cells following crocidolite fiber exposure,16 within minutes of exposure to apoptosis-inducing compounds,18,21,22 and upon exposure to styrene oxide, paraquat dichloride, N-nitroso-N-methylurea (NMU), and puromycin.23
Although these DEP and ROT results, as well as more recent microchip-based cell dielectric measurements,24–26 show that dielectric approaches have the potential for enabling the detection of toxicants, those techniques remain impractical for routine screening. However, dielectrophoretic field-flow fractionation (dFFF) allows the rapid and convenient analysis of bulk cell suspensions while retaining single-cell discrimination of membrane dielectric properties.27,28 Over the past decade, we have developed this approach for the separation of different cell types from cell mixtures based on their membrane dissimilarities.19,29 Our focus was on the isolation of tumor cells.12,29 However, because dFFF also allows changes in the membrane capacitance and conductivity properties of 105 or more individual cells to be inferred from alterations in their elution characteristics, we undertook an initial study of the feasibility of using this technique for toxicity testing. We report here that the method was able to detect cell responses to toxic agents having different modes of action more quickly and sensitively than conventional cell viability assays and, in most cases, more sensitively than other assays designed to detect more directly the action of specific toxicants. We conclude that the dFFF technique merits further study as a potential in vitro toxicant screening tool.
MATERIALS AND METHODS
Chemicals
The selected chemicals were endosulfan (C9H6Cl6O3S, CAS No. 115-29-7) obtained from Supelco (Bellefonte, PA), paraquat dichloride (C12H14Cl2N2, CAS No. 1910-42-5), and acrylonitrile (C3H3N, CAS No. 107-13-1) purchased from Aldrich Chemical Co.; actinomycin D (C62H86N12O16, CAS No. 50-76-0) and carbon tetrachloride (CCl4, CAS No. 56-23-5) obtained from Merck (Darmstadt, Germany); N-nitroso-N-methylurea (C2H5N3O2, CAS No. 684-93-5) and puromycin (C22H29N7O5 2HCl, CAS No. 58-58-2) purchased from Sigma Chemical Co. (St. Louis, MO); and styrene oxide (SO, C8H8O, CAS No. 96-09-3) obtained from Fluka (Sigma-Aldrich Chemie GmbH). All chemicals were of analytical grade.
Phosphate buffer saline (PBS) solution was made by dissolving 0.8% (w/v) NaCl, 0.02% (w/v) KCl, 0.12% (w/v) Na2HPO4, and 0.02% (w/v) KH2PO4 (all from Sigma Chemical Co.) in ultrapure water (Millipore, Billerica, MA). The dFFF running buffer was an aqueous solution containing 8.5% (w/v) sucrose (MP Biomedicals, Aurora, OH) and 0.3% (w/v) dextrose (ICN Biomedicals, Costa Mesa, CA), adjusted to a conductivity of 30 mS·m−1 with the aid of a conductivity meter (Cole Parmer Instrument Co., Chicago, IL) using PBS. All chemicals were of analytical grade.
Chemicals for the alkaline comet assay were NuSieve GTG agarose LMP from FMC Bioproducts, agarose gel (NuSieve 3:1 agarose) from FMC Bioproducts, Sybr-Green from Molecular Probes Inc., and Vectashield mounting medium from Vector Laboratories Inc.
Cell Culture
Human leukemia HL-60 cells were used as an easy-to-culture, nonadherent human cell model for toxicity testing. Cells were grown in RPMI 1640 (Invitrogen Corp., Carlsbad, CA.) with 10% fetal bovine serum (Hyclone), 0.2 mol·L−1 l-glutamine (Sigma-Aldrich, St. Louis, MO), 0.02 mol·L−1 HEPES (Sigma-Aldrich), and 0.4% gentamicin sulfate (Cambrex Bio Science, Walkersville, MD) in 75-cm2 plastic flasks under a 5% CO2/95% air atmosphere at 37 °C in a humidified incubator. Cells were harvested at a density of 1.5 × 106 mL−1 in exponential growth phase (48 h). Counts and viability were measured after harvest and during toxicant exposure by vital staining with 0.4% trypan blue using a hemocytometer.30 Cell viability exceeded 98% at the beginning of every dFFF experiment and conventional assay.
Exposure to Toxicants
For testing of the dFFF methodology, eight agents were selected as being representative of toxicants having different modes of actions as shown in Table 1. Toxicants were applied by adding carrier vehicles containing appropriate concentrations of acrylonitrile, actinomycin D, carbon tetrachloride, endosulfan, NMU, paraquat dichloride, puromycin, or styrene oxide to cell culture to make a final volume of 1.5 mL (the maximum final concentration of the vehicle was 1%). The vehicles used were DMSO (for carbon tetrachloride and SO), ethanol (for endosulfan and actinomycin D), and ultrapure water (for acrylonitrile, paraquat dichloride, NMU, and puromycin). Control samples were treated with vehicle alone. Cell suspensions were incubated at 37 °C for exposure times ranging from 15 to 150 min.
Table 1.
Selected Chemicals Used in the Current Study and Their Modes of Action
| chemicals | sources of exposure | modes of action |
|---|---|---|
| acrylonitrile | emission from acrylonitrile, latex, resin production plants, packaging material for food | oxidative damage to cell including DNA, plasma membrane (IPCS, 1983; Whysner, at al., 1998; Zhang et al., 1998; Zhang et al., 2000; Zhang et al., 2002) |
| actinomycin D | anticancer drug | RNA synthesis inhibitor, DNA strand breaks, carcinogen (Fraschini, et al., 2005; Sobell 1985) |
| carbon tetrachloride | emission from carbon tetrachloride manufacturer, residual in foodstuffs and drinking-water, solvent in cleaning agent | lipid peroxidation resulting in necrosis or steatosis potentiated by cytochome P450 2E1 (IPCS, 1999; Plaa, 2000). |
| endosulfan | contaminated food, air and drinking water, emission from manufacturers | reduce cell viability and inhibit cell growth through cytochrome C release leading to apoptosis of cells (Kannan et al., 2000; IPCS, 1984) |
| N-nitroso-N-methylurea (NMU) | occupational exposure through inhalation or dermal contact where NMU is used in research | DNA damage via formation of alkylated bases(Huggins et al., 1981; IARC, 1998; Tominaga et al., 1997) |
| paraquat dichloride | contaminated soil or water, residual in foods, agricultural workers who spray paraquat | lipid peroxidation resulting in cellular membrane damage (Peter et al., 1992; WHO, 1984) |
| puromycin | workers in a production of puromycin, use in research | protein synthesis inhibition (Darken, 1964; Lehinger et al., 1993) |
| styrene oxide | migration from plastics and resins to food, occupational exposure in reinforced plastics, fabricated rubber products, paints, and allied products industry | DNA adducts, lipid peroxidation (Bastlová et al., 1995; Dypbukt et al., 1992; Vodicka et al., 1996) |
Conventional Assays of Toxicant Responses
Responses of cells to toxicants were assayed by the conventional cell viability approach. An aliquot of 40 µL was withdrawn from each sample and mixed with an equal volume of trypan blue dye solution. Live and dead cells were counted in a hemocytometer.
Responses to endosulfan, a mitochondrial poison,31 were evaluated using a mitochondrial activity assay, the MitoLight Apoptosis Detection Kit (Chemicon International, Temecula, CA). Control and endosulfan-treated HL-60 cells were incubated for different doses and times. Suspended cells were then washed with PBS and centrifuged at 1500 rpm for 10 min to form a pellet. Cells were resuspended in 1 mL of 0.1% MitoLight solution and then incubated at 37 °C in a 5% CO2 incubator for 20 min. They were then centrifuged at 1500 rpm for 10 min, and the supernatant was discarded. Prior to flow cytometric analysis, the pellet was resuspended in 1 mL of PBS. Stained cells were analyzed immediately by flow cytometry. MitoLight aggregates in healthy mitochondria cells were detectable in the PI channel (FL2) while MitoLight monomers in apoptotic cells were seen in the FITC channel (FL1). Fluorescence intensity histograms for treated samples were compared to those from control samples.
The alkaline comet assay was used to evaluate responses to actinomycin D and acrylonitrile, agents known to damage cellular DNA. The assay was used to determine DNA strand breaks, as described by Tuntawiroon et al. 32 with a minor modification. Briefly, 50 µL of HL-60 cell culture was mixed with LMP agarose and embedded into an agarose-precoated slide. Slides were submerged in cold lysis solution for at least 1 h at 4 °C. Subsequently, slides were transferred to an electrophoresis chamber and immersed in alkaline solution (pH 13) for 20 min before electrophoresis at 300 mA, 24 V for 20 min. After electrophoresis, slides were neutralized with 1 mol·L−1 ammonium acetate and stained with 50 µL of Sybr-Green solution (1:5000) (Molecular Probes). A total of 50 cells from each of three duplicate slides were examined randomly under an epifluorescence microscope (Axioplan 2, Zeiss). The extent of DNA strand breaks was quantified using the CometScan image analysis software (MetaSystems) and expressed as olive tail moment (the product of the proportion of DNA in the tail by the distance between the centers of gravity of the head and tail).
dFFF Assay
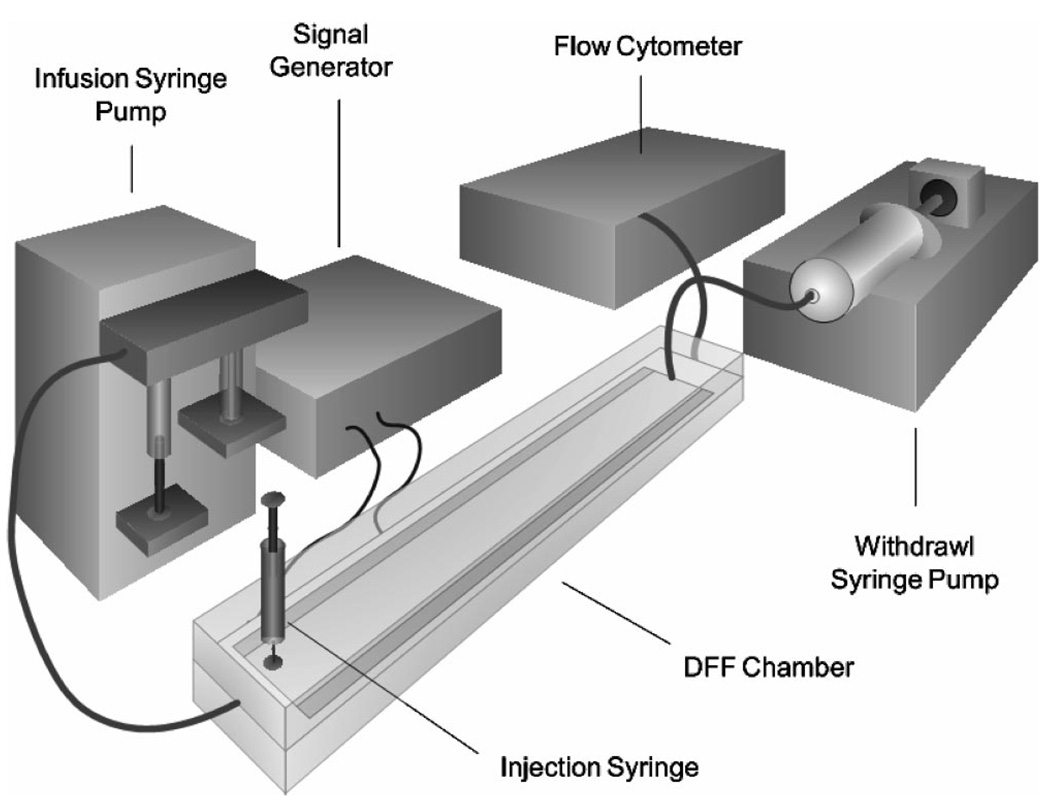
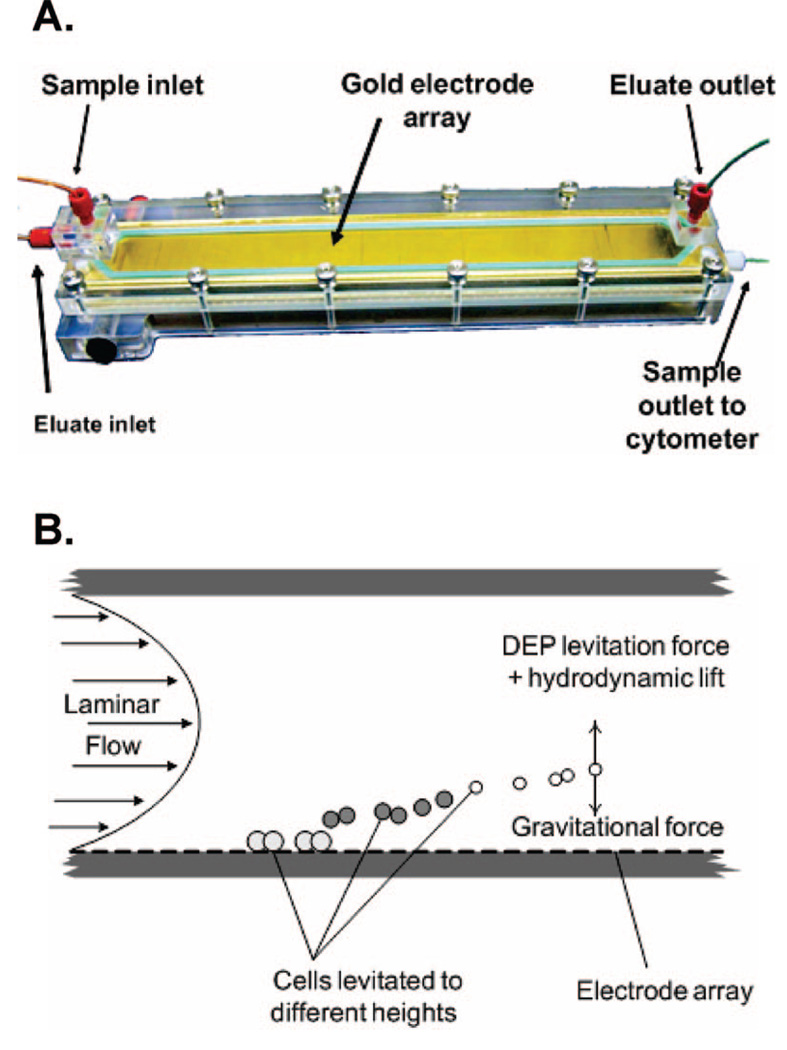
Figure 1 shows the dFFF configuration, which comprised a DFF-30 dielectrophoretic field-flow fractionation system (InGeneron, Inc., Houston, TX) with a chamber, computer-controlled signal generator, and digital syringe pumps (KD Scientific) for controlling eluate flow through the chamber. A flow cytometer (Bryte HS, Bio-Rad Laboratories, Hercules, CA) was used to observe cell elution from the dFFF chamber. The chamber (Figure 2A) contained an array of interdigitated, gold-plated microelectrodes (50 µm wide × 50 µm spacing) covering the floor of the flow channel (420 µm height × 25 mm width × 280 mm length) inside an acrylic polymer body. In dFFF, a balance of gravitational, hydrodynamic lift, and DEP forces positions cells in a laminar flow velocity profile and causes cells with different dielectric properties to elute at different rates. Agents that alter their dielectric properties will change their dFFF properties and modify their transit time in the chamber, as illustrated in Figure 2B. The principles and design of dFFF devices have been described in detail earlier.27
Figure 1.
Experimental arrangement for testing toxicant detection by the dFFF approach that combines DEP forces with field-flow fractionation.
Figure 2.
(A) dFFF chamber used for toxicological testing (B) In dFFF, the position of cells in a laminar flow stream is controlled by a balance of gravitational, DEP, and hydrodynamic lift forces causing cells to be transported at different speeds and to emerge at different times determined by their dielectric properties.
To provide fluid flow, a digital syringe pump infused the dFFF running buffer at a rate of 1.5 mL·min−1 through the inlet port of the chamber. A second pump was used to withdraw 1 mL·min−1 eluate from the topside of the outlet end of the channel. The remaining 0.5 mL·min−1 eluate flowed from a port at the bottom of the outlet end and directly through the flow cytometer for cell measurements (as shown in Figure 2A). Because cells traveled near the bottom of the dFFF chamber, they emerged from the bottom port, allowing them to be counted by the cytometer while its flow rate remained within specifications.
Cell samples were washed by centrifugation (1500 rpm for 10 min) and resuspended at ~4.5 × 106 cell·mL−1 in isotonic 8.5% (w/v) sucrose + 0.3% (w/v) dextrose that had been adjusted to a conductivity of 30 mS·m−1 with PBS verified with a conductivity meter (EC1481-61; Cole-Parmer Instruments). The chamber was preloaded with 30 mS·m−1 running buffer. Then, 300 µL of cell suspension containing 1.3 × 106 HL-60 cells was introduced into the inlet port using a 1-mL disposable syringe. An electrical signal of 80 kHz and 3.8 Vp-p was applied to the dFFF electrode array during sample injection—so cells were levitated by DEP forces and prevented from adhering to the chamber bottom.
After 3-min settling time, the majority of cells had sedimented to the chamber floor where their position could be controlled by a balance of DEP, sedimentation, and hydrodynamic lift forces, and eluate flow was initiated from the infusion pump at 1.5 mL·min−1. Simultaneously, the withdrawal pump was started at 1.0 mL·min−1. Elution time and forward and side light scatter were measured for each cell by the cytometer, and the cell elution concentration versus time was recorded. After 90% of the cells had eluted (~20 min), the DEP signal was switched to 15 kHz at 3.8 Vp-p to repel remaining cells far from the electrode and quickly elute them.28
The time of the dFFF elution peak for untreated cells, T0, was taken as the baseline measurement of their DEP characteristics. After treatment under each test condition, the modified time of the elution peak, Tt, was measured. The dFFF response, Di, was defined as the relative change in migration speed of the cells through the dFFF chamber, given by,
| (1) |
dFFF dose-time response characteristics were determined on the basis of this Di response index.
RESULTS
dFFF Responses
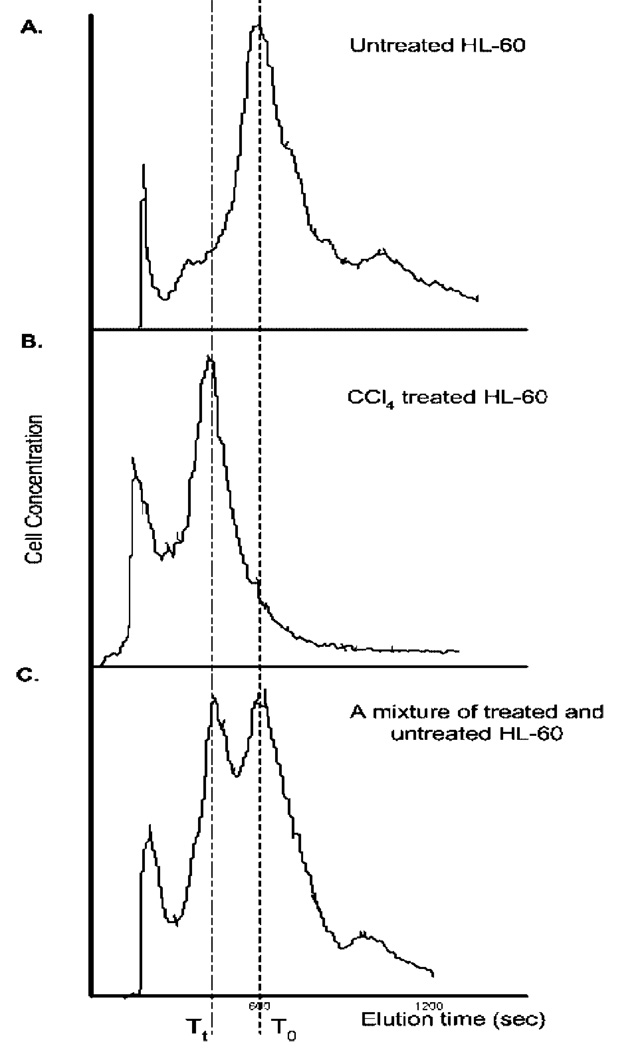
The parameter Di used to quantify dFFF responses of the cells to toxicant exposure is illustrated in Figure 3, which shows HL-60 elution profiles for untreated cells, for cells treated with 15 mmol·L−1 CCl4 for 30 min, and for a mixture of untreated and CCl4-treated cells. The values of Di for HL-60 cells exposed to endosulfan, acrylonitrile, and actinomycin D are shown plotted in Figure 4 in panel A as a function of doses and times. Results for viability responses are shown in panel B. Panel C shows responses to assays chosen to reflect the mode of action of these toxicants.
Figure 3.
(A) dFFF elution profile for untreated HL-60 cells. The time of emergence of the peak, T0, was taken to represent the baseline characteristics of the cells. (B). After treatment with toxicant, in this case CCl4 at 15 mmol·L−1 for 30 min, the cell elution peak emerged at a shorter time, Tt, because of the effect of the toxicant on the membrane conductivity and capacitance. (C). dFFF elution profile for a mixture of untreated and CCl4-treated HL-60 cell culture. This was to confirm a change in elution time of treated cells. The relative change in the speed with which cells transited the chamber was taken as the measure of the response to the toxicant.
Figure 4.
Summary of methods and their results. HL-60 cells were incubated with different doses of endosulfan (first row), acrylonitrile (second row), and actinomycin D (third row) for (◆) 15, (■) 30, (Δ) 45, (×) 60, (− −) 90, (●) 120, and (—) 150 min. HL-60 cells were then measured for Di, cell viability and DNA damage in which the results are shown on left, middle, and right columns. respectively.
For endosulfan, changes in Di were detectable at 10 µmol·L−1 after only 15-min exposure. The viability assay was less sensitive than the dFFF method over the 150-min exposure window used in this study, and the smallest concentration for which a detectable fall in cell viability occurred was 100 µmol·L−1. The MitoLight mitochondrial transmembrane potential assay revealed a detectable response to endosulfan at 50 µmol·L−1 after 15-min exposure, showing that the dFFF assay was slightly more sensitive.
For actinomycin D, an RNA synthesis inhibitor and DNA damaging agent, the dFFF and viability assays showed almost no response during the 150-min exposure window. However, the comet assay indicated DNA damage by this agent even after 15 min exposure at 5 µmol·L−1.
For acrylonitrile, an agent known to have mixed modes of action including reactive oxygen species generation and DNA damage, the dFFF assay (Figure 4, panel A) was more sensitive than the viability assay (panel B) for up to 90 min of exposure, but the cell viability assay increased in relative sensitivity at longer times and matched the dFFF assay after 150 min. The comet assay (panel C), revealing DNA strand breaks, was more sensitive than the dFFF and viability assays.
Relative Assay Sensitivities
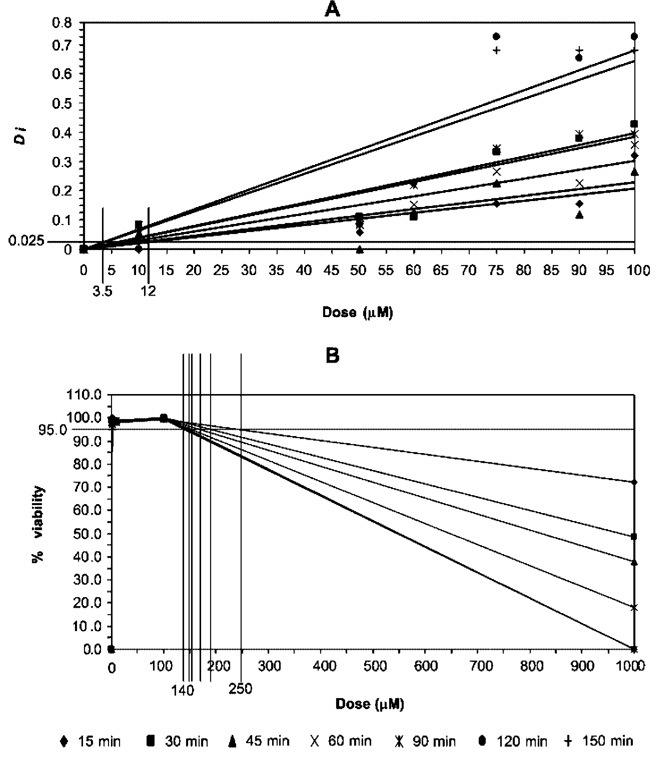
To determine the relative sensitivities of the dFFF and viability assays, the minimum dose thresholds at which the dose-response curves consistently exceeded the background noise was determined for each agent for each exposure time. For example, Figure 5 shows plots of Di and viability versus dose and time for endosulfan. The thresholds were found to be 0.025 for Di and 95% for viability, and the sensitivity of the assays was therefore taken to be the dose at which the dose-response curve crossed these values.
Figure 5.
Threshold determination for Di response index and percentage viability used in sensitivity comparison by isosensitivity locus chart.
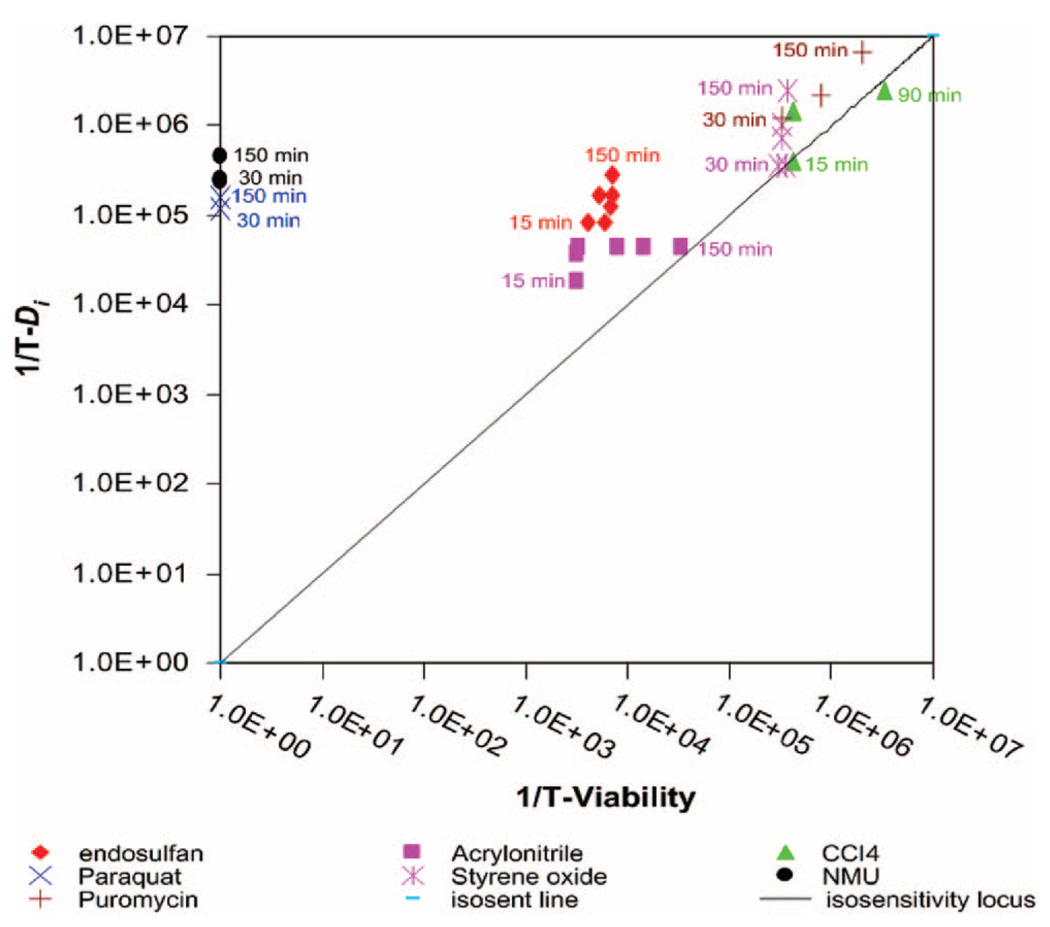
To compare the effectiveness of the assays for detecting the various agents, Figure 6 shows a plot of the reciprocal threshold sensitivities as a function of time. In this plot, points fall on a line with slope = 1 if the dFFF and viability assays have equal sensitivity. Otherwise, the points fall closer to the axis of the more effective assay. Figure 6 shows that the dFFF test was more responsive than the viability test for all agents in the 150-min exposure window except carbon tetrachloride, for which the assay sensitivities were essentially similar. Possible reasons for this will be discussed later. Results for actinomycin D, for which responses were not seen for either test during the 150-min exposure window, cannot be shown in this plot. Figure 6 reveals that the dFFF assay was more than 5 orders of magnitude more sensitive than the viability assay for NMU and paraquat, 30 times more effective for endosulfan, and up to 10 times more responsive for acrylonitrile, styrene oxide, and puromycin.
Figure 6.
Threshold sensitivity plots comparing the effectiveness of the dFFF method and viability assays. Assays lying on the isosensitivity locus have an identical sensitivity.
DISCUSSION
The aim of this study was to establish whether dFFF, a dielectrophoresis-based chromatrographic technique exploiting a metal nanofilm microelectrode array and microfluidic principles, could be adapted to detect toxicity of chemicals in an aqueous environment using a cultured mammalian cell model. HL-60 was selected because it is a human-derived cell line that is easy to culture reproducibly and because it provides a continuous source of cells. Furthermore, it grows in suspension culture allowing aliquots to be harvested without trypsinization whenever they are needed for experimentation.
Untreated HL-60 cells have a specific membrane capacitance Cmem of 16.5 mF·m−2 as determined by DEP crossover frequency measurements. 23 It is accepted that Cmem for a smooth biological membrane is ~9 mF·m−2 13,18 showing that surface features like microvilli and ruffles increase the membrane surface area of HL-60 cells by a factor of 16.5/9 = 1.83. Therefore, the HL-60 membrane may be considered to have an “excess” of 83% membrane area compared with a perfectly smooth cell of the same diameter. Processes that result in alterations in the membrane area will change Cmem. For example, endo- and exocytosis, respectively, deplete and add net membrane area as a function of their rates.6,9,33 Also, the cell surface may shed membrane through budding and blebbing in response to free-radical damage34 and to apoptosis-inducing agents.35 In the case of HL-60, membrane loss could in principle lead to a maximum reduction in Cmem of 83% without requiring a change in cell diameter. The HL-60 cell membrane normally presents a good barrier to ion flow so that the membrane conductivity Gmem is low for healthy, intact cells. Stress can increase the ion permeability, thereby increasing Gmem. In the extreme case of necrosis, membrane barrier function collapses altogether and Gmem rises by many orders of magnitude.36
We and others have shown that values for the dielectric parameters Cmem and Gmem may be derived from measurements of DEP-induced motion of cells.13,15,17,18,23,36–39 For toxicological screening, it is less important to deduce explicit values for these membrane dielectric parameters than to quickly and sensitively detect changes in them. HL-60 cells (diameter ~12 µm, density 1065 kg·m−3) were allowed to sediment to the chamber floor in the running buffer (density 1032 kg·m−3) before flow was initiated. Once flow was started with the DEP field on, the height of cells was controlled by the balance of hydrodynamic lift, DEP, and sedimentation forces. The equations relating these parameters have been described in our earlier work.19,27,28 Depending on the applied electric field frequency and cell dielectric properties, the DEP force, which resulted from interaction of the induced dielectric polarization of the cells with the applied electric field, could be made repulsive (augmenting hydrodynamic lift), attractive (augmenting sedimentation), or zero (at the crossover frequency at which the DEP force changed direction). In combination with our eluate conductivity of 30 mS·m−1, we chose to use a field frequency of 80 kHz at which the DEP force was zero for HL-60 cells unperturbed by toxicants. Under these conditions, the hydrodynamic lift and sedimentation forces balanced when cells were ~10 µm above the chamber floor. Reductions in Cmem and increases in Gmem induced by toxicant exposure resulted in repulsive DEP forces that increased the cell equilibrium height to between 10 and 50 µm above the chamber floor, resulting in a reduced elution time (cells moved up to 4 times faster). In this way, toxicant-induced changes in cell elution times could be used to infer responses of membrane dielectric properties and, to a lesser extent, changes in cell density. Because the dFFF approach allowed 105 or more individual cells to be profiled in minutes within a bulk suspension, it permitted good statistical information about toxicant-induced changes in the distribution of cell dielectric properties to be determined quickly.
In eq 1, we have defined a convenient response index Di in terms of alterations in the cell peak elution time. Other response indices could be defined, for example, in terms of changes in the width of the elution peak profile. The DEP results for single cells in our previous study23 revealed decreasing Cmem and increasing Gmem values for both higher toxicant doses and longer exposure times. Because both types of changes result in shorter elution times in dFFF, we would expect cell responses in these parameters to be indicated by increasing values of Di, as we observed for all the agents we tested here.
Of importance for screening applications are the toxicant modes of action to which an assay responds and its threshold sensitivity. We tested the dFFF method with agents having different specific and mixed modes of action as summarized in Table 1. Agents that cause damage to the cell membrane are likely to bring about changes in Di that reflect their direct impact on the membrane dielectric properties. Paraquat, which causes damage to the cell membrane through lipid peroxidation, is in this class of toxicants, and it is not surprising that the dFFF assay for this agent was 5 orders of magnitude more sensitive than the viability assay.
Endosulfan, actinomycin D, NMU, and puromycin interfere with intracellular processes and are thought not to affect the plasma membrane directly. Changes in Di for these compounds reflect cell plasma membrane responses that are secondary to the initial toxicant modes of action and that must depend on how closely the relevant intracellular and membrane effects are coupled. Di showed slightly higher dose and time sensitivities to endosulfan than the flow cytometric mitochondrial potential assay, indicating that there must be an extremely close coupling between the HL-60 cell plasma membrane properties and the mitochondrial dysfunction that endosulfan elicits. Earlier studies also showed that agents that induce apoptosis in HL-60 cells can be detected by dielectric membrane responses even more quickly and sensitively than by the annexin V flow cytometric assay18 and within minutes of caspase expression, suggesting that there is a very close coupling between intracellular apoptotic and membrane processes as well. On the other hand, Di showed relatively low sensitivity for detecting actinomycin D and NMU compared with the alkaline comet assay, which directly reflects DNA strand breaks. This may suggest that the coupling between nucleic acid-based damage mechanisms and the HL-60 cell membrane properties is weaker than for cellular metabolic and apoptotic processes. Alternatively, it may reveal a limitation of using Di as the response index. DNA damaging agents affect genes randomly, and it is possible that only a minority of the HL-60 cells suffered DNA lesions that resulted in membrane changes within 150-min exposure. In that case, the position of the dFFF peak would be expected to remain unchanged after carcinogen treatment, while the shape of the elution peak would be expected to alter to reflect the presence of new minority cell subpopulations in which DNA damage had caused cell membrane or metabolic responses. On reviewing our data, we found a broadening of the dFFF elution peak that increased with dose and time of exposure to actinomycin D (data not shown), and this merits further investigation as a possible means for detecting cell subpopulations that are indicative of DNA damage. Nevertheless, even using our simple response index, dFFF was found to be 5 orders of magnitude more sensitive for detecting NMU toxicity than the viability assay (Figure 6) over our 150-min exposure window.
Carbon tetrachloride showed relatively low toxicity in both the dFFF and viability assays in accordance with the fact it owes its toxicity to activation by cytochrome P450 2E1 (CYP2E1),40 an enzyme that HL-60 cells lack.41 The dFFF method using HL-60 cells shares this deficiency of activation capabilities with other toxicological assays that depend on cultured cells. This shortcoming could be addressed by coculturing HL-60 cells and hepatocytes, which would activate toxicants through their cytochrome P450 pathways.42,43 Hepatocytes would be left behind as an adherent monolayer when the HL-60 cells (which would lie on top during culture) were withdrawn for dFFF analysis.
The dFFF assay was twice as sensitive, but followed the same time course, as the viability assay in detecting puromycin, an agent that specifically inhibits protein synthesis. dFFF assays for acrylonitrile and styrene oxide were up to 1 order of magnitude more sensitive than the viability assay and followed different time courses. Overall, with the exception of carbon tetrachloride, the dFFF assay was more sensitive than the cell viability assay for all agents listed in Table 1 over the 150-min time course of our experiments (see Figure 6). Our results are consistent with earlier findings for the agents we tested by the single-cell-based DEP method,23 yet the dFFF method used here profiled thousands of times more cells 20 times faster and far less laboriously than was possible with the DEP technique.
By spiking field-sampled canal water (running alongside a highway in Bangkok) with endosulfan, sterile-filtering it into concentrated media, and then incubating HL-60 cells in this mixture, we were able to detect toxicity with the same sensitivity as reported above. This approach would be feasible for testing environmental samples by the dFFF method. Other improvements were also identified in the course of this study to make the system more convenient. For example, we found that a laser light scatter instrument (LaserTrac PC2400D, Chemtrac Systems) provided a small, far less expensive detector than the cytometer that not only permitted higher flow rates for faster dFFF runs but that also allowed all the pumps to be replaced by a single gravity feed from an eluant reservoir. Finally, because we discovered that small variations in sample injection conditions affected the background noise in Di, an automated injector should make possible a dFFF system capable of higher threshold detection sensitivities. By arranging for this system to inject cell samples close to the chamber floor using frit-inlet methodology developed for FFF,64 the delay of waiting for cells to settle could also be eliminated completely.
Overall, our results show that the dFFF method with HL-60 as the mammalian cell response model was able to sensitively and conveniently detect, via cell membrane capacitance and conductivity, toxicant activities directed toward the cell membrane, cell metabolism, and that trigger apoptosis. Furthermore, the method detected activities of all the toxicant classes we studied within tens of minutes of exposure at concentrations significantly lower than cell viability and potentiometric flow cytometric assays though it proved to be less sensitive than the comet assay for the rapid detection of direct DNA-damaging agents. Because thousands of cells were examined simultaneously in each dFFF run, cell elution profiles provided good statistical information over the whole population of cells. We believe the dFFF technique demonstrates potential as a rapid in vitro toxicity-screening tool that may have wide applicability for detecting agents that perturb cell metabolic or membrane activities though perhaps less relevance for detecting DNA damaging agents that do not cause cell physiological responses.
ACKNOWLEDGMENT
This project was supported by research grants from Chulabhorn Research Institute and The Center on Enviromental Health, Toxicology and Management of Toxic Chemicals under Science and Technology Postgraduate Education and Research Development Office (PERDO) of the Ministry of Education, Thailand. Development of the dFFF technology was supported by the Defense Advance Research Projects Agency (Office of Naval Research Contract N6601-97-C-8608) and by the National Cancer Institute (grant CA88364).
References
- 1.Ferńandez A, Tejedor C, Cabrera F, Chordi A. Water Res. 1995;29(5):1281–1286. [Google Scholar]
- 2.Tahedl H, Hader DP. Water Res. 1999;33(2):426–432. [Google Scholar]
- 3.Chowdhury HH, Grilc S, Zorec R. Ann. N.Y. Acad. Sci. 2005;1048:281–286. doi: 10.1196/annals.1342.025. [DOI] [PubMed] [Google Scholar]
- 4.Rottensteiner H, Theodoulou FL. Biochim. Biophys. Acta. 2006;1763:1527–1540. doi: 10.1016/j.bbamcr.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Danahay H, Atherton HC, Jackson AD, Kreindler JL, Poll CT, Bridges RJ. Am. J. Physiol.: Lung Cell. Mol. Physiol. 2006;290:L558–L569. doi: 10.1152/ajplung.00351.2005. [DOI] [PubMed] [Google Scholar]
- 6.Gillis KD, Pun RY, Misler S. Pflugers Arch. Eur. J. Physiol. 1991;418(6):611–613. doi: 10.1007/BF00370579. [DOI] [PubMed] [Google Scholar]
- 7.Bertrand CA, Laboisse CL, Hopfer U. Am. J. Physiol.: Cell Physiol. 1999;276:907–914. doi: 10.1152/ajpcell.1999.276.4.C907. [DOI] [PubMed] [Google Scholar]
- 8.Engisch KL, Nowycky MC. J. Physiol. 1998;506(3):591–608. doi: 10.1111/j.1469-7793.1998.591bv.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallermann S, Pawlu C, Jonas P, Heckmann M. Proc. Natl. Acad. Sci. U. S. A. 2003;100(15):8975–8980. doi: 10.1073/pnas.1432836100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pohl HA. Dielectrophoresis. The behavior of neutral matter in nonunifrom electric fields. Cambridge: Cambridge Univ. Press; 1978. p. 7. [Google Scholar]
- 11.Fuhr G, Gimsa J, Glaser R. Stud. Biophys. 1985;108:149–164. [Google Scholar]
- 12.Becker FF, Wang XB, Huang Y, Pethig R, Vykoukal J, Gascoyne PRC. J. Phys. D: Appl. Phys. 1994;27:2659–2662. [Google Scholar]
- 13.Gascoyne PRC, Pethig R, Satayavivad J, Becker FF, Ruchirawat M. Biochim. Biophys. Acta. 1997;1323:240–252. doi: 10.1016/s0005-2736(96)00191-5. [DOI] [PubMed] [Google Scholar]
- 14.Markx GH, Pethig R. Biotechnol. Bioeng. 1995;45:337–343. doi: 10.1002/bit.260450408. [DOI] [PubMed] [Google Scholar]
- 15.Wang XB, Huang Y, Gascoyne PRC, Becker FF. IEEE Trans. Ind. Appl. 1995;33(3):1358–1365. doi: 10.1109/28.585856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dopp E, Jonas L, Nebe B, Budde A, Knippel E. Environ. Health Perspect. 2000;108(2):153–158. doi: 10.1289/ehp.00108153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang XB, Huang Y, Gascoyne PRC, Becker FF, Hölzel R, Pethig R. Biochim. Biophys. Acta. 1994;1193:330–344. doi: 10.1016/0005-2736(94)90170-8. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Becker FF, Gascoyne PRC. Biochim. Biophys. Acta. 2002;1564:412–420. doi: 10.1016/s0005-2736(02)00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Huang Y, Wang XB, Becker FF, Gascoyne PRC. Biophys. J. 2000;78:2680–2689. doi: 10.1016/S0006-3495(00)76812-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geier BM, Wendt B, Arnold WM, Zimmermann U. Biochim. Biophys. Acta. 1987;900:45–55. doi: 10.1016/0005-2736(87)90276-8. [DOI] [PubMed] [Google Scholar]
- 21.Labeed FH, Coley HM, Hughes MP. Biochim. Biophys. Acta. 2006;1760(6):922–929. doi: 10.1016/j.bbagen.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 22.Pethig R, Talary MS. IET Nanobiotechnol. 2007;1(1):2–9. doi: 10.1049/iet-nbt:20060018. [DOI] [PubMed] [Google Scholar]
- 23.Ratanachoo K, Gascoyne PRC, Ruchirawat M. Biochim. Biophys. Acta. 2002;1564:449–458. doi: 10.1016/s0005-2736(02)00494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao C, Luong JHT. Biotechnol. Prog. 2003;19:1000–1005. doi: 10.1021/bp025733x. [DOI] [PubMed] [Google Scholar]
- 25.Xiao C, Luong JHT. Toxicol. Appl. Pharmacol. 2005;206(2):102–112. doi: 10.1016/j.taap.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 26.van der Schalie WH, James RR, Gargan TP. Biosen. Bioelectron. 2006;22(1):18–27. doi: 10.1016/j.bios.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 27.Huang Y, Wang XB, Becker FF, Gascoyne PRC. Biophys. J. 1997;73:1118–1129. doi: 10.1016/S0006-3495(97)78144-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Huang Y, Wang XB, Becker FF, Gascoyne PRC. Anal. Chem. 1999;71:911–918. doi: 10.1021/ac981250p. [DOI] [PubMed] [Google Scholar]
- 29.Huang Y, Yang J, Wang XB, Becker FF, Gascoyne PRC. J. Hematother. Stem Cell Res. 1999;8:481–490. doi: 10.1089/152581699319939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cook JA, Mitchell IB. Anal. Biochem. 1989;179(1):1–7. doi: 10.1016/0003-2697(89)90191-7. [DOI] [PubMed] [Google Scholar]
- 31.Kannan K, Holcombe RF, Jain SK, Alvarez-Hernandez X, Chervenak R, Wolf RE, Glass J. Mol. Cell. Biochem. 2000;205:53–33. doi: 10.1023/a:1007080910396. [DOI] [PubMed] [Google Scholar]
- 32.Tuntawiroon J, Mahidol C, Navasumrit P, Autrup H, Ruchirawat M. Carcinogenesis. 2006;28(4):816–822. doi: 10.1093/carcin/bgl175. [DOI] [PubMed] [Google Scholar]
- 33.Nüβe O, Lindau M. J. Cell Biol. 1988;107(6 Pt 1):2117–2123. doi: 10.1083/jcb.107.6.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stark G. J. Membr. Biol. 2005;205:1–16. doi: 10.1007/s00232-005-0753-8. [DOI] [PubMed] [Google Scholar]
- 35.Malorni W, Paradisi S, Iosi F, Santini MT. Cell Biol. Toxicol. 1993;9(2):119–130. doi: 10.1007/BF00757574. [DOI] [PubMed] [Google Scholar]
- 36.Wang X-J, Yang J, Gascoyne PR. Biochim. Biophys. Acta. 1999;1426:53–68. doi: 10.1016/s0304-4165(98)00122-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gimsa J, Marszalek P, Loewe U, Tsong TY. Biophys. J. 1997;60:749–760. doi: 10.1016/S0006-3495(91)82109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y, Wang XB, Becker FF, Gascoyne PRC. Biochim. Biophys. Acta. 1996;1282:76–84. doi: 10.1016/0005-2736(96)00047-8. [DOI] [PubMed] [Google Scholar]
- 39.Chan KL, Gascoyne PRC, Becker FF, Pethig R. Biochim. Biophys. Acta. 1997;1349:182–196. doi: 10.1016/s0005-2760(97)00092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruckner JV, Ramanathan1 R, Monica, Lee KM, Muralidhara S. J. Pharm. Exp. Ther. 2002;300(1):273–281. doi: 10.1124/jpet.300.1.273. [DOI] [PubMed] [Google Scholar]
- 41.Kawai M, Chen J, Cheung CYS, Change TKH. Mol. Cell. Biochem. 2003;248(1–2):57–65. doi: 10.1023/a:1024101430363. [DOI] [PubMed] [Google Scholar]
- 42.Choi SH, Nishikawa M, Sakoda A, Sakai Y. Toxicol. in Vitro. 2004;18(3):393–402. doi: 10.1016/j.tiv.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 43.Søderlund EJ, Brunborg G, Holme JA, Hongslo JK, Nelson1 SD, Dybing E. Mutagenesis. 1991;6(1):25–30. doi: 10.1093/mutage/6.1.25. [DOI] [PubMed] [Google Scholar]
- 44.International Programme on Chemical Safety (IPCS) Finland: WHO; Environmental health criteria 28 Acrylonitrile. 1983
- 45.Whysner J, Steward RE, Chen D, Conaway CC, Verna LK, Richie JP, Ali N, Williams GM. Arch. Toxicol. 1998;72:429–438. doi: 10.1007/s002040050523. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Wang Y, Jing J, Xu Y, Klauig JE. Toxicologist. 1998;42(1S):76. [Google Scholar]
- 47.Zhang H, Kamendulis LM, Jiang J, Xu Y, Klaunig JE. Carcinogensis. 2000;21(4):727–733. doi: 10.1093/carcin/21.4.727. [DOI] [PubMed] [Google Scholar]
- 48.Zhang H, Kamendulis LM, Klaunig JE. Toxicol. Sci. 2002;67:247–255. doi: 10.1093/toxsci/67.2.247. [DOI] [PubMed] [Google Scholar]
- 49.Fraschini A, Bottone MG, Scovassi AI, Denegri M, Risueño MC, Testillano PS, Martin TE, Biggiogera M, Pellicciari C. Histol. Histopathol. 2005;20:107–117. doi: 10.14670/HH-20.107. [DOI] [PubMed] [Google Scholar]
- 50.Sobell HM. Proc. Natl. Acad. Sci. U. S. A. 1985;82:5328–5331. doi: 10.1073/pnas.82.16.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.International Programme on Chemical Safety (IPCS) Finland: WHO; Environmental health criteria 208 Carbon Tetrachloride. 1999
- 52.Plaa GL. Annu. Rev. Pharmacol. Toxicol. 2000;40:43–65. doi: 10.1146/annurev.pharmtox.40.1.43. [DOI] [PubMed] [Google Scholar]
- 53.International Programme on Chemical Safety (IPCS) Finland: WHO; Environmental health criteria 40 Endusulfan. 1984 doi: 10.1007/BF02783154. [DOI] [PubMed]
- 54.Huggins GB, Ueda N, Wiessler M. Proc. Natl. Acad. Sci. U. S. A. 1981;78(2):1185–1188. doi: 10.1073/pnas.78.2.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.IARC monographs on the evaluation of carcinogenic risks to humans. [Retrieved November 2007];Some N-Nitroso compounds. 1998 Vol. 17 Available online http://monographs.iarc.fr/ENG/Monographs/vol17/volume17.pdf.
- 56.Tominaga Y, Tsuzuki T, Shiraishi A, Kawate H, Sekiguchi M. Carcinogenesis. 1997;18(5):889–896. doi: 10.1093/carcin/18.5.889. [DOI] [PubMed] [Google Scholar]
- 57.Peter B, Wartena M, Kampinga HH, Konings AW. Biochem. Pharmacol. 1992;43(4):705–715. doi: 10.1016/0006-2952(92)90234-a. [DOI] [PubMed] [Google Scholar]
- 58.WHO. [Retrieved October 2007];International Programme on Chemical Safety, Environmental Health Criteria 3: Paraquat and Diquat. 1984 Available online http://www.inchem.org/documents/ehc/ehc/ehc39.htm.
- 59.Darken MA. Pharmacol. Rev. 1964;16:223–243. [PubMed] [Google Scholar]
- 60.Lehinger AL, Nelson DL, Cox MM. Principles of Biochemistry. 2nd ed. New York: Worth Publishers; 1993. [Google Scholar]
- 61.Bastlová T, Vodièka P, Peterková K, Hemminki K, Lambert B. Carcinogenesis. 1995;16(10):2357–2362. doi: 10.1093/carcin/16.10.2357. [DOI] [PubMed] [Google Scholar]
- 62.Dypbukt JM, Costa LG, Manzo L, Orrenius S, Nicotera P. Carcinogensis. 1992;13(3):47–424. doi: 10.1093/carcin/13.3.417. [DOI] [PubMed] [Google Scholar]
- 63.Vodicka P, Stetina R, Kumar R, Plna K, Hemminki K. Carcinogensis. 1996;17(4):801–808. doi: 10.1093/carcin/17.4.801. [DOI] [PubMed] [Google Scholar]
- 64.Williams KR, Lee I, Giddings JC. Separation and characterization of 0.01–50 µm particles using FIFFF. Chapter 15. In: Provder T, editor. Particle Size Distribution II; ACS Symposium Series 472; American Chemical Society; Washington, DC. 1991. [Google Scholar]