Abstract
We present a general method called dynamic single-molecule colocalization for quantitating the associations of single cell surface molecules labeled with distinct autofluorescent proteins. The chief advantages of the new quantitative approach are that, in addition to stable interactions, it is capable of measuring nonconstitutive associations, such as those induced by the cytoskeleton, and it is applicable to situations where the number of molecules is small.
Main Text
Tracking single molecules on the cell membrane, at subdiffraction resolution, has great potential for elucidating the organization of proteins on the cell surface and how this changes during signal transduction (1). Recently, multicolor imaging has allowed the trajectories of individual molecules to be related to other larger structures on the cell surface, such as supramolecular clusters of proteins, which has provided new insights into the structure of the cell surface and the role of the cytoskeleton in its organization (2, 3). A natural extension of these studies is the multicolor imaging of individual molecules to identify proteins that form constitutively-associated oligomers or are more weakly associated because, for example, they form transiently associated oligomers or because their diffusion is constrained to the same part of the cell membrane. Kusumi and co-workers have previously shown the feasibility of tracking molecules labeled with two distinct fluorophores and of detecting constitutive, artificial dimers via colocalization (4). We now extend this general approach by quantifying the extent of association. This allows us to identify, along with constitutive dimers and monomers, functionally important but much weaker associations occurring at the cell surface.
Previously we developed Two Color Coincidence Detection (TCCD) in which two overlapped confocal beams independently excited single diffusing membrane proteins labeled with a specific red- or blue-tagged antibody fragment (5). Association was quantified by calculating the number of red and blue coincident fluorescence events that occurred above the level expected by chance. Here we extend the TCCD concept to two dimensions, using a method we call Dynamic Single-Molecule Colocalization, or DySCo, which relies on quantitative video analysis of the fluorescence from two distinct fluorophores diffusing at the cell surface and imaged using total-internal reflection fluorescence microscopy (Fig. 1 A). The advantages of tracking molecules is that there is no need to correct the data for chance coincidence events, since the probability of two nonassociated molecules tracking within a short distance of one another for a number of frames is very low (Fig. 1 B). In addition, we can detect immobile or slowly diffusing molecules that are unobservable using TCCD. Finally, very weakly expressed proteins can be analyzed, whereas TCCD requires the event rate to be reasonably high to make data collection feasible. We apply this method to the same proteins used to validate TCCD (5), i.e., the monomer CD86, the dimer CD28, and the T-cell receptor (TCR), expressed on unactivated Jurkat T cells (or derivatives thereof; see the Supporting Material) and investigate the role of the cytoskeleton in their organization.
Figure 1.
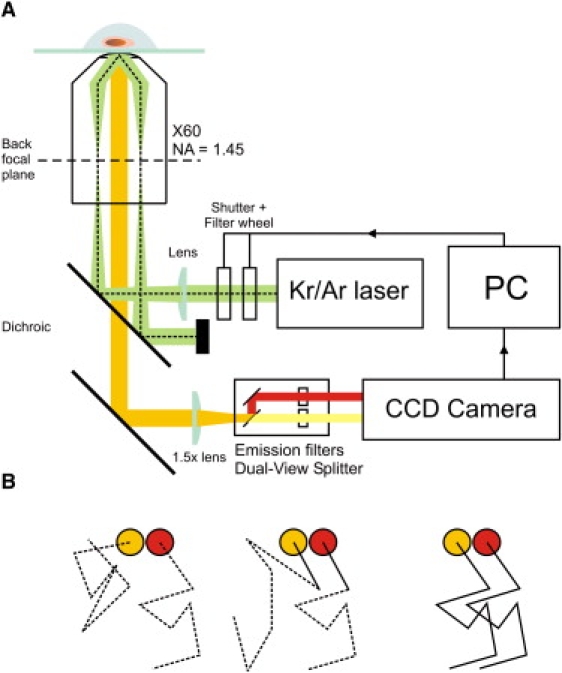
(A) Experimental setup – full details of which can be found in the Supporting Material. T cells expressing proteins labeled with mCherry and Citrine are attached to a glass coverslip coated with nonactivating donkey anti-mouse antibody and the diffusion of the individual molecules on the basal surface of the cell is imaged after exciting simultaneously with overlapped 488 and 568 nm laser beams in total internal reflection geometry. Example raw data can be found as Movie S1. (B) Principle of the DySCo method. Associated molecules track within a short distance of one another for multiple frames (right) whereas unassociated molecules show no correlated motion (left). Unassociated molecules may track together by chance over a short distance (center), but the probability of this occurring for multiple frames is small.
Initially we used Citrine and mCherry, monomeric YFP and RFP variants respectively, which allowed the attachment of the labels to the cytoplasmic domains of the proteins, thus decreasing the chance of disrupting any extracellular interaction. However, due to their convoluted surface, the observable area and signal/noise ratio achievable with unactivated T cells in TIRF imaging is very poor relative to activated T cells, which spread (3). To ameliorate this loss in signal, three copies in tandem of Citrine (3cit) and three mCherry (3ch) FPs are used as fluorescent tags. An optimized Bayesian-based tracking algorithm, following on from our previous work (6) and that of Oh and co-workers (7), was used to identify tracks from individual molecules (see the Supporting Material and Movie S1, Movie S2, Movie S3, and Movie S4).
After tracks were identified, we quantitated association by applying a nearest-neighbor distance approach similar to that used by others (8). Distances between red and yellow tracks were calculated for each video frame, and a track was taken to be associated if it remained within a defined separation for a chosen number of frames. For this work, we required associated tracks to stay within 300 nm for three or more frames. The degree of association was determined by calculating the total number of coincident tracks for a given color divided by the total number of tracks for that color, expressing this value as a percentage. We make our measurements relative to the red tracks, since these have lower detection efficiency. Using the method we can measure weak associations where only a fraction of the red tracks are coincident with yellow tracks using the above criterion. Our results are not strongly dependent on the distance used to identify associated molecules (see Fig. S1 in the Supporting Material).
The red and yellow single-molecule trajectories were analyzed to determine the diffusion coefficients of the proteins. These data gave similar values to those we have obtained using fluorescence correlation spectroscopy (Table S3 and Table S4). This shows that neither the tandem FPs used for labeling, nor the close proximity of the surface significantly affected protein diffusion. The results of DySCo analysis of the proteins are presented in Fig. 2. Our dimer control, CD28, gave a coincidence level of 22.7 ± 5.9%. We only detect yellow-red dimers, which would ideally give a maximum coincidence of 50% for homodimers. The deviation from this value of the coincidence measured for CD28 is assigned to a combination of FP photophysics and slow fluorophore maturation rate (see the Supporting Material). For CD86 we observe a significantly lower percentage of associated red molecules, 10.2 ± 3.0%, but this is still significantly above zero. We then analyzed the TCR, after confirming that the FP-tagged forms of the receptor triggered normally (see the Supporting Material). We found that the labeled TCR gave an association level of 4.8 ± 2.3%, lower than that obtained for CD86.
Figure 2.
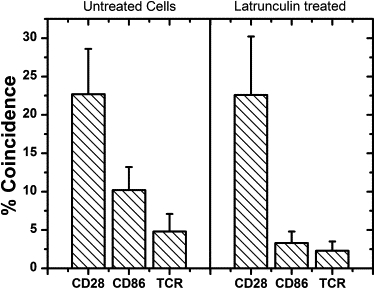
Molecule colocalization for cells with and without latrunculin treatment. See text for details. Data points are mean ± SE.
Control experiments on cells without FP-tagged proteins showed that noise in the data very occasionally produced apparent tracks in our analysis, but the level of coincidence was minimal compared to our FP-data: only 1 red apparent track in 21 was coincident with an apparent yellow track for data from 15 cells. We also estimated the expected fraction of tracks that would appear to be due to the random association of molecules at a given protein density. We did this by shuffling the red and yellow videos relative to one another, and rerunning the data analysis for nonmatched file pairs. This gave a value for apparent association of only 0.6%, indicating that we are observing nonrandom organization for CD86 and the TCR, in keeping with other studies (8, 9). This result is also in close agreement with our previous TCCD-based analysis for which nonrandom organization was observed (5), possibly due to constrained or partially constrained motion.
To investigate the role of the cytoskeleton in nonrandom organization, the experiment was repeated for T cells treated with latrunculin, which disrupts the cytoskeleton by inhibiting actin polymerization. This had little effect on the diffusion coefficients (Table S3) insofar as there was a small reduction for CD28, and ∼70–80% increases in the median for CD86 and the TCR, in keeping with other studies (10). However, significant reductions in association close to background levels were observed for both CD86 and TCR (Fig. 2), strongly suggesting that the apparent confinement is cytoskeleton-dependent. These results are in agreement with the actin-dependent micron-scale confinement recently observed for FcɛR1 (9). The confinement or partial confinement may be due to nonspecific interactions of the fluorescent proteins with the cytoskeleton or could be due to other proteins bound by the actin cytoskeleton acting as barriers to diffusion (10). The confinement by the cytoskeleton also appears greater for CD86 than TCR, which might reflect differences in the distribution of these proteins on the cell surface. Since changes in the cytoskeleton occur very soon after T-cell activation this partial confinement by the actin cytoskeleton may have functional significance (11).
The number of TCRs in the present experiments is ∼10-fold less than that of a normal Jurkat T cell (see the Supporting Material) which would reduce the concentration of dynamically associated molecules. However, we still find that under these conditions the T cell is activated normally. Our data therefore strongly suggests that the TCR is predominantly monovalent. Furthermore, since we detected 22.7% coincidence for CD28 and the coincidence level for the TCR expressed in latrunculin-treated cells was 2.3%, at most only ∼10% of the TCR in these experiments is associated, without the influence of the cytoskeleton. We cannot rule out that this is due to the presence of a low level of TCR dimers, but it seems more likely to be due to corralling effects (12), given our observations that the well-defined monomer CD86 exhibits nonzero coincidence.
The strength of the DySCo method is its ability to quantify homo- and hetero-interactions including both constitutive dimers and the much weaker interactions of transient dimers or of proteins whose diffusion is constrained by cytoskeleton-mediated corralling effects, as shown here. These weaker interactions are likely to underpin signal transduction at the cell surface and are thus of great importance (13, 14). An additional advantage of the method is that it is minimally perturbative; it is not, for example, reliant on the prior cross-linking of molecules (15) and, like TCCD, it provides an opportunity to analyze natively expressed proteins tagged only with labeled Fab fragments (5). Finally, experiments at very low surface densities are possible, which will allow the investigation of weakly expressed membrane proteins, such as G-protein coupled receptors.
Acknowledgments
The authors thank Mary Collins (University College London, UK) for kindly providing some of the expression vectors used in this work and Roger Tsien for Citrine and mCherry sequences.
This project was funded by the Engineering and Physical Sciences Research Council, Biotechnology and Biological Sciences Research Council, Medical Research Council and The Wellcome Trust, UK, and by Fundação para a Ciência e a Tecnologia, Portugal.
Editor: Lukas K. Tamm.
Footnotes
Paul D. Dunne, Ricardo A. Fernandes, and James McColl contributed equally to this work.
John R. James' present address is University of California at San Francisco, Department of Cellular and Molecular Pharmacology, San Francisco, California.
Tables, figures, and movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)01056-X.
Contributor Information
Simon J. Davis, Email: simon.davis@ndm.ox.ac.uk.
David Klenerman, Email: dk10012@cam.ac.uk.
Supporting Material
References and Footnotes
- 1.Sako Y., Minoghchi S., Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat. Cell Biol. 2000;2:168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- 2.Yokosuka T., Sakata-Sogawa K., Kobayashi W., Hiroshima M., Hashimoto-Tane A. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat. Immunol. 2005;6:1253–1262. doi: 10.1038/ni1272. [DOI] [PubMed] [Google Scholar]
- 3.Douglass A.D., Vale R.D. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koyama-Honda I., Ritchie K., Fujiwara T., Iino R., Murakoshi H. Fluorescence imaging for monitoring the colocalization of two single molecules in living cells. Biophys. J. 2005;88:2126–2136. doi: 10.1529/biophysj.104.048967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.James J.R., White S.S., Clarke R.W., Johansen A.M., Dunne P.D. Single-molecule level analysis of the subunit composition of the T cell receptor on live T cells. Proc. Natl. Acad. Sci. USA. 2007;104:17662–17667. doi: 10.1073/pnas.0700411104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon J.W., Bruckbauer A., Fitzgerald W.J., Klenerman D. Bayesian inference for improved single molecule fluorescence tracking. Biophys. J. 2008;94:4932–4947. doi: 10.1529/biophysj.107.116285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh S., Schenato L., Chen P., Sastry S. Tracking and coordination of multiple agents using sensor networks: System design, algorithms and experiments. Proc. IEEE. 2007;95:234–254. [Google Scholar]
- 8.Lachmanovich E., Shvartsman D.E., Malka Y., Botvin C., Henis Y.I. Co-localization analysis of complex formation among membrane proteins by computerized fluorescence microscopy: application to immunofluorescence co-patching studies. J. Microsc. 2003;212:122–131. doi: 10.1046/j.1365-2818.2003.01239.x. [DOI] [PubMed] [Google Scholar]
- 9.Andrews N.L., Lidke K.A., Pfeiffer J.R., Burns A.R., Wilson B.S. Actin restricts Fc epsilon RI diffusion and facilitates antigen-induced receptor immobilization. Nat. Cell Biol. 2008;10:955–963. doi: 10.1038/ncb1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Umemura Y.M., Vrljic M., Nishimura S.Y., Fujiwara T.K., Suzuki K.G.N. Both MHC class II and its GPI-anchored form undergo hop diffusion as observed by single-molecule tracking. Biophys. J. 2008;95:435–450. doi: 10.1529/biophysj.107.123018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Merwe P.A., Davis S.J., Shaw A.S., Dustin M.L. Cytoskeletal polarization and redistribution of cell-surface molecules during T cell antigen recognition. Semin. Immunol. 2000;12:5–21. doi: 10.1006/smim.2000.0203. [DOI] [PubMed] [Google Scholar]
- 12.Vereb G., Szollosi J., Matko J., Nagy P., Farkas T. Dynamic, yet structured: The cell membrane three decades after the Singer-Nicolson model. Proc. Natl. Acad. Sci. USA. 2003;100:8053–8058. doi: 10.1073/pnas.1332550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burbach B.J., Medeiros R.B., Mueller K.L., Shimizu Y. T-cell receptor signaling to integrins. Immunol. Rev. 2007;218:65–81. doi: 10.1111/j.1600-065X.2007.00527.x. [DOI] [PubMed] [Google Scholar]
- 14.Dustin M.L. Cell adhesion molecules and actin cytoskeleton at immune synapses and kinapses. Curr. Opin. Cell Biol. 2007;19:529–533. doi: 10.1016/j.ceb.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorsch S., Klotz K.-N., Engelhardt S., Lohse M.J., Bunemann M. Analysis of receptor oligomerization by FRAP microscopy. Nat. Methods. 2009;6:225–230. doi: 10.1038/nmeth.1304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


