Abstract
Aβ17-42 (so-called p3) amyloid is detected in vivo in the brains of individuals with Alzheimer's disease or Down's syndrome. We investigated the polymorphism of Aβ17-42 oligomers based on experimental data from steady-state NMR measurements, electron microscopy, two-dimensional hydrogen exchange, and mutational studies, using all-atom molecular-dynamics simulation with explicit solvent. We assessed the structural stability and the populations. Our results suggest that conformational differences in the U-turn of Aβ17-42 lead to polymorphism in β-sheet registration and retention of an ordered β-strand organization at the termini. Further, although the parallel Aβ17-42 oligomer organization is the most stable of the conformers investigated here, different antiparallel Aβ17-42 organizations are also stable and compete with the parallel architectures, presenting a polymorphic population. In this study we propose that 1), the U-turn conformation is the primary factor leading to polymorphism in the assembly of Aβ17-42 oligomers, and is also coupled to oligomer growth; and 2), both parallel Aβ17-42 oligomers and an assembly of Aβ17-42 oligomers that includes both parallel and antiparallel organizations contribute to amyloid fibril formation. Finally, since a U-turn motif generally appears in amyloids formed by full proteins or long fragments, and since to date these have been shown to exist only in parallel architectures, our results apply to a broad range of oligomers and fibrils.
Introduction
Amyloidogenic proteins are characterized by their tendency to aggregate into β-sheet-rich amyloid fibrils that are associated with a number of progressive degenerative diseases. Some of the most devastating diseases affect the central nervous system, including Alzheimer's disease (AD), Parkinson's disease, Huntington's disease, prion diseases (“mad cow” disease), amyotrophic lateral sclerosis, and Down's syndrome (DS) (1–5), in addition to type II diabetes mellitus (6).
AD is characterized by the presence of extracellular senile amyloid plaques and an intraneuronal neurofibrillary tangle in the brain. Biochemical analysis has revealed that the main constituent is a small polypeptide, Aβ1-40 or Aβ1-42, that is derived from endoproteolytic cleavage of the transmembrane amyloid precursor protein (APP) (7). The hydrophilic residue-rich N-terminus (residues 1–28) is exposed to the aqueous extracellular environment, whereas the hydrophobe-rich C-terminus (residues 29–42) is deeply embedded in the membrane (8,9). In the so-called “amyloidogenic cleavage” pathway leading to the production of Aβ1-40/Aβ1-42, APP is cleaved in the extracellular domain by β secretase, whereas γ secretase cleaves APP in the transmembrane region. In a second pathway, so-called “nonamyloidogenic cleavage” (2,10–13), α secretase and γ secretase cleave the APP and yield Aβ17-42 (14), the so-called p3. Thus, in addition to Aβ1-40/Aβ 1-42, the plaques contain Aβ17-42 fragments. Aβ17-42 is prevalent in the brains of patients with AD and in a subset of patients with dystrophic neuritis, but is absent or sparse in age-matched patients without AD (non-AD brain) (15). Aβ17-42 is also present in neurotic plaques in individuals with DS (5). The function and toxic properties of Aβ17-42, the importance of its accumulation in AD and DS, and the mechanism behind fibril formation are still unclear. Although the Aβ17-42 fragment was initially thought to be nonamyloidogenic, in vitro experiments showed that Aβ17-42 fragments can form amyloid fibrils (15–18) even though the intermediate soluble oligomeric state may not be stable, and can fibrillate directly (19).
Studies in humans, wild-type rodents, transgenic mice, cultured cells, and in vitro systems suggest that soluble, oligomeric assembly intermediates of amyloidogenic proteins are the primary pathogenic effectors (20–22), with most data obtained for Aβ1-42 (23–27). Abundant data from other proteins demonstrate that this is a common mechanism (28–32). Investigating the structures of oligomers is a challenging task because of their small size and short-lived nature compared with mature amyloid fibrils (33,34). On the other hand, mature fibrils can also be toxic, and the toxicity may also change with amyloid morphologies (35). This is particularly the case for brittle fibrils (36). The complexity of the underlying mechanisms of amyloid toxicity largely derives from the interplay of the stabilities, and oligomer and amyloid fibril variability.
Studies of polymorphism are important because understanding the range of potential conformations under various conditions and the self-assembly mechanism by which ordered fibrils assemble from the conformational ensemble may assist researchers in designing effective drugs. Experimentally, polymorphism is well documented in amyloid formation (18,36–51). Molecular-dynamics (MD) simulations are a useful computational tool to address amyloid conformational variability (52). In recent studies (53,54), MD simulations of Aβ17-42 were performed based on the Lührs model (35) and for Aβ1-40 fibrillar architecture (55,56). In this work, we constructed 14 oligomers with different backbone conformations due to the U-turn segment variability, and with a range of β-strand arrangements. We did not study polymorphism as it relates to side-chain conformations. We performed all-atom MD simulations in explicit solvent to investigate the polymorphism and conformational stability of the constructed Aβ17-42 oligomers. Analysis of the conformational stabilities and energies of Aβ17-42 octamer models based on experimental structures presented two dominant forms, thus indicating that competing polymorphic populations already exist at the oligomer level.
Materials and Methods
Single- and double-layer models of Aβ17-42
The initial coordinates of an Aβ17-42 octamer were extracted from the averaged NMR-based structure (PDB code: 2BEG) derived from quenched hydrogen/deuterium-exchange NMR (35). We extracted the third monomer conformation of the Aβ17-42 peptide from the β-amyloid pentamer and extended the number of monomers to eight. Initial conformations of the Aβ17-42 octamer were also extracted from the solid-state NMR (ssNMR)-based model of Aβ1-40 (57) (R. Tycko, National Institutes of Health, personal communication, 2009) by removing residues Gly9-Lys16 and adding residues Ile41 and Ala42 to the C-termini of each monomer. Finally, initial configurations were also extracted from the Ma-Nussinov computational model (58). All of the models were used to generate parallel, antiparallel, and a combination of parallel and antiparallel organizations of the Aβ17-42 oligomers. Double-layered structural models of Aβ17-42 oligomers were modeled using these organizations. The two β-sheets were associated together via the C-terminal/C-terminal interface and stacked in an antiparallel orientation, with maximum overlap of the hydrophobic region. A total of 14 octamer models were constructed, including single and double layers, three turn types, and three parallel/antiparallel organizations, and simulated.
MD simulation procedure
MD simulations of solvated Aβ17-42 oligomers were performed in the NPT ensemble using the NAMD program (59) with the CHARMM27 force field (60,61). The oligomers were energy-minimized and explicitly solvated in a TIP3P water box (62,63) with a minimum distance of 10 Å from any edge of the box to any Aβ atom. Any water molecule within 2.5 Å of the Aβ was removed. Counterions Na+ were added at random locations to neutralize the Aβ17-42 charge. The Langevin piston method (59,64,65) with a decay period of 100 fs and a damping time of 50 fs was used to maintain a constant pressure of 1 atm. A temperature of 330 K was controlled by a Langevin thermostat with a damping coefficient of 10 ps−1 (59). The short-range van der Waals interactions were calculated using the switching function, with a twin range cutoff of 10.0 and 12.0 Å. Long-range electrostatic interactions were calculated using the particle mesh Ewald method with a cutoff of 12.0 Å (66,67). The equations of motion were integrated using the leapfrog integrator with a step of 1 fs.
The solvated systems were energy-minimized for 2000 conjugated gradient steps, and the distance between the β-sheets in Aβ17-42 was fixed in the range of 2.2–2.5 Å. The counterions and water molecules were allowed to move. The hydrogen atoms were constrained to the equilibrium bond using the SHAKE algorithm (68). The minimized solvated systems were energy-minimized for an additional 5000 conjugate gradient steps at 250 K, with all atoms allowed to move. Then the systems were heated from 250 K to 300 K for 300 ps and equilibrated at 330 K for 300 ps. All simulations ran for 30 ns. These conditions were applied to test the stabilities of all Aβ17-42 oligomers.
Analysis details
To obtain the relative structural stability of the Aβ17-42 oligomers, the Aβ trajectories of the last 5 ns were first extracted from the explicit MD simulation, excluding water molecules. Fig. S1 and Fig. S2 in the Supporting Material present two examples of the structural stabilities of Aβ17-42 oligomers. The solvation energy of each system was calculated using the generalized Born method with molecular volume (GBMV) (69,70). Details on the GBMV method are provided in the Supporting Material.
We further examined the stability of the oligomers by following the changes in the number of hydrogen bonds between the β strands with the hydrogen bond cutoff set to 2.5 Å, and by monitoring the change in the intersheet distance (Cα backbone-backbone distance) in the core domain of all of the oligomers. The core domain is explained in detail in the Supporting Material.
Results and Discussion
Construction of the conformational ensemble of Aβ17-42 octamers
To compare potential oligomers that would be expected to relate to the ensuing amyloid fibril variants, we constructed a conformational ensemble of 14 initial arrangements and generated 500 conformations for each arrangement by MD simulation.
First, we chose three monomeric conformations based on experimental data and computational results. Two experiment-based models of the three-dimensional structures of Aβ oligomers were recently obtained: the first, called the Lührs model (35), presents a parallel single-layer structure of Aβ1-42 oligomers based on hydrogen/deuterium-exchange NMR data, combined with side-chain packing constraints from pairwise mutagenesis, ssNMR, and high-resolution cryo-electron microscopy (PDB code: 2BEG); the second, called the Tycko model (57), exhibits a parallel double-layer structure of Aβ1-40 fibrils based on ssNMR. A third computational model, proposed by Ma and Nussinov (58), predicts a parallel structure for Aβ10-35. According to all three of these models, the buried salt bridge (residues Asp23-Lys28) stabilizes the U-turn. In addition, they suggest that parallel oligomers are associated in a parallel orientation to form the Aβ fibrils. The main difference between these three models is the slight variation in the U-turn location and size in the Aβ peptide (Ser26-Ile31 for the Lührs model, Asp23-Gly29 for the Tycko model, and Val24-Asn27 for the Ma-Nussinov model), and therefore in the turn shape. The U-turn bent β-sheet of the Ma-Nussinov computational model is similar to that of the Tycko model.
To select conformations with the most likely arrangements and keep the number of arrangements manageable with respect to computational power and timescales, we consider three main types of variability in oligomer conformations: 1), β-strand backbone alignments in parallel and/or antiparallel orientations; 2), slight changes in backbone conformations due to variation in the turn; and 3), a change in association due to different matches of the β-sheet layers. Table 1 and Table S1 list the three arrangements of tested octamers and their overall features.
Table 1.
Constructed Aβ17-42 oligomers based on the Lührs (35), Tycko (57), and Ma-Nussinov (58) models, including single and double layers
| Model∗ number | Model | Conformational structure | Layer | Conformational energy† (kcal/mol) | Population‡ (%) | Figure |
|---|---|---|---|---|---|---|
| M1 | Tycko | Parallel | Single | −2287.88 (104.78) | 12 | 1 A |
| M2 | Lührs | Parallel | Single | −2244.66 (103.01) | 12 | 1 B |
| M3 | Tycko | Parallel | Double | −2170.56 (102.45) | 10 | 2 A |
| M4 | Lührs | Parallel 1 | Double | −2172.05 (106.88) | 10 | 2 C |
| M5 | Lührs | Parallel 2 | Double | −2135.91 (107.65) | 9 | 2 D |
| M6 | Lührs | Parallel 3 | Double | −2005.83 (100.95) | 5 | 2 B |
| M7 | Lührs | Parallel dimers associated in antiparallel | Single | −2074.97 (106.19) | 7 | 1 C and Fig. S3 |
| M8 | Lührs | Parallel dimers associated in antiparallel | Double | −2114.67 (103.83) | 5 | 2 E and Fig. S3 |
| M9 | Tycko | Parallel dimers associated in antiparallel | Single | −1961.70 (105.06) | 4 | Fig. S4 |
| M10 | Ma- Nussinov | Antiparallel | Single | −2112.22 (101.79) | 8 | 1 D and Fig. S5 |
| M11 | Ma- Nussinov | Antiparallel | Double | −2095.03 (99.54) | 7 | Fig. S5 |
| M12 | Lührs | Antiparallel | Single | −1966.88 (104.04) | 4 | 1 E and Fig. S3 |
| M13 | Tycko | Antiparallel 1 | Single | −1970.76 (93.14) | 4 | 1 F and Fig. S4 |
| M14 | Tycko | Antiparallel 2 | Single | −1909.77 (106.38) | 3 | Fig. S4 |
Model 1; see Table S1 for variation classification.
Conformational energies were computed using the GBMV calculations (69,70). Standard deviation values are presented in parenthesis.
Percentage of populations or the relative probabilities of the conformational structures were computed using MC simulations.
We start by comparing parallel and antiparallel alignments using single-layer octamers (Fig. 1). The first two models, M1 (Fig. 1 A) and M2 (Fig. 1 B), are parallel; here our goal is to reproduce the experimental conformations with the full-length Aβ peptides used by Tycko and Lührs, respectively. Six other arrangements with antiparallel β-strands in a single layer are constructed as reference states. M7 (Fig. 1 C and Fig. S3) and M9 (Fig. S4) use four parallel dimers associated in an antiparallel arrangement to form a β-sheet octamer based on Lührs and Tycko's monomers, respectively. M10 (Fig. 1 D and Fig. S5), M12 (Fig. 1 E and Fig. S3), M13 (Fig. 1 F and Fig. S4), and M14 (Fig. S4) are models with all β-strands organized in antiparallel alignments. Side-chain registration in the antiparallel alignments is selected to maximize the backbone hydrogen bonds and the hydrophobic core within the β-sheets (M10 and M11), although a model with less backbone hydrogen bonds but more compact turn interactions is also tested (M13).
Figure 1.
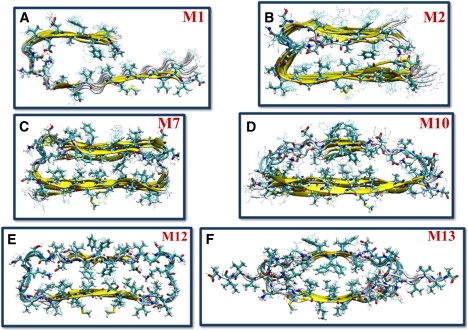
Initial structural models of single-layer Aβ17-42 oligomers. Parallel: (A) Tycko's model M1 and (B) Lührs' model M2. Antiparallel: (C) Lührs' model M7, (D) Ma-Nussinov's model M10, (E) Lührs' model M12, and (F) Tycko's model M13.
Six models with double-layer arrangements (dimer of tetramers; Fig. 2 and Fig. S3 and Fig. S5) are also tested to explore variations in the associations of oligomers, focusing on a comparison of different matches of β-sheet layers. M3 (Fig. 2 A) can be viewed as a double layer of M1 (Fig. 1 A). M4 (Fig. 2 C), M5 (Fig. 2 D), and M6 (Fig. 2 B) are stacked forms of M2 (Fig. 1 B). M3 and M6 have similar hydrophobic interactions between two layers. However, since the turns are different for M3 and M6, the M3 model has a larger contact surface between the two layers. The M4 and M5 models shift the side-chain registration between the two layers as compared with the M6 model. However, the difference between the M4 and M5 hydrophobic contacts is small. Finally, M8 (Fig. 2 E and Fig. S3) tests the double-layer form of M7 (Fig. 1 C and Fig. S3), and M11 (Fig. S5) is the double-layer form of M10 (Fig. 1 D and Fig. S5).
Figure 2.
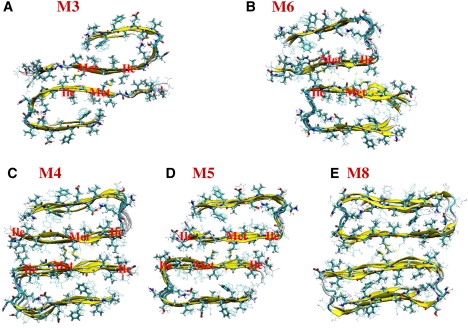
Initial structural models of double-layer Aβ17-42 oligomers: (A) Tycko's model M3 (B) Lührs' model M6, (C) Lührs' model M4, (D) Lührs' model M5, and (E) Lührs' model M8.
Using the models described above, we can examine the effects of the backbone turn conformation in a single layer by comparing M1 with M2; M3 with M4; and M10, M12, and M13 (Table 1 and Table S1).
Conformational stability of parallel Aβ17-42 oligomers: polymorphic U-turn shapes and hydrophobic association
For the single-layer structure, both Tycko's and Lührs' conformers associate into a stable β-sheet whereas the turn region remains flexible. Backbone and U-turn root mean-square deviations (RMSDs) indicate that the parallel conformers M1 and M2 are stable (Fig. S6). In addition, analysis of the averaged Asp23-Lys28 distance in M1 and M2 indicates that the salt bridge in these conformers is also stable (Fig. S7). M1 has a slightly better association among the β-strands assisted by backbone hydrogen bonds (Fig. 3 A). The β-sheet distances (Cα backbone–backbone distance; Fig. 3 C) within the U-turn expand to ∼14–15 Å, which is relatively large in comparison with the mostly packed association in the mature fibrils (∼10–11 Å). This expansion indicates that the turn region is still not locked in either the M1 or M2 model at the octamer level. However, M1 has a slightly tighter turn region, and thus has slightly more backbone hydrogen bonds than M2. The energy difference between M1 and M2 is very small (Table 1). Even though for the conformers sampled, M1 has slightly lower average energies than M2, the difference is within the SD. In conclusion, for a single layer, the two experimental models with different U-turn shapes show a similar behavior and are similar in energies, indicating polymorphic parallel single layers of Aβ17-42 oligomers.
Figure 3.
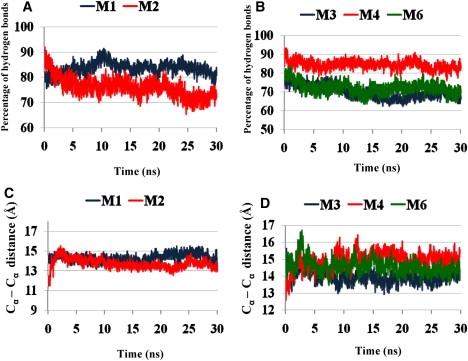
Fraction of the number of hydrogen bonds (in percentage) between all β-strands compared with the initial Aβ17-42 oligomer parallel conformation for (A) a single layer of Tycko's model M1 (blue) and Lührs' model M2 (red), and (B) a double layer of Tycko's model M3 (blue), Lührs' model M4 (red), and Lührs' model M6 (green). (C) Cα backbone–backbone distance of the single layer for Tycko's model M1 (blue) and Lührs' model M2 (red). (D) Cα backbone–backbone distance of the double layer for Tycko's model M3 (blue), Lührs' model M4 (red), and Lührs' model M6 (green).
In the M3 and M6 double-layer structures (Fig. 2, A and B), two β-sheets are stacked against each other in a twofold symmetry-like orientation with distinct C-terminal/C-terminal interfaces. In comparison with alternate associations (N-terminal to C-terminal, or N-terminal to N-terminal) this orientation was found to be the most stable because it provided maximized hydrophobic interactions (53). Apparently, the double-layer structures further rigidify the turn, as seen in Fig. 3 D, where the double layers have similar β-sheet distances (Cα backbone–backbone distance) within the U-turn in the single-layer model (Fig. 3 C), but with smaller overall RMSD (Fig. S6 and Fig. S8). Backbone and U-turn RMSDs exhibit stable structures (Fig. S8). In addition, analysis of the averaged Asp23-Lys28 distance in the parallel conformers M3 and M4 indicates the stability of the salt bridge in these studied conformers (Fig. S9). At the octamer level, the formation of double-layer structures sacrifices the backbone hydrogen bonds, as indicated by the smaller percentage of hydrogen bonds (Fig. 3 B) compared with the single-layer models (Fig. 3 A). The gain of hydrophobic interactions from the formation of the double-layer structure is still insufficient to offset the loss of backbone hydrogen bonds. Therefore, as seen in Table 1, the single-layer conformers of both models M1 and M2 have lower energy compared to the constructed models of the double layers M3 and M6 (as well as M4 and M5). For oligomers larger than those studied here, double layers should be more stable than single layers.
At first glance, a comparison between the double layers of the two conformers M3 and M6 shows that Tycko's conformer M3 is preferred over Lührs' M6 (Table 1), due to the strong hydrophobic interactions between the two β-sheets of the two layers (Fig. 2, A and B). The sheet-to-sheet packing is stabilized by a nonpolar zipper through van der Waals and hydrophobic interactions. The double-layer conformer constructed based on Lührs' model M6 (Fig. 2 B) has hydrophobic interactions between the C-termini of the two layers (Ile31 interacting with Met35) similar to those observed in the conformer constructed using Tycko's model M3 (Fig. 2 A). However, in M6 the overlap of the hydrophobic region (Ile31-Ile41) at the C-terminal/C-terminal interface is small. This indicates that a different U-turn shape can affect the hydrophobic associations between two layers. Two different double-layer models based on the Lührs turn structure (M4 and M5) attempt to maximize the overlap of this hydrophobic region, involving hydrophobic interactions between Ile31-Ile41 and Met35-Met35 at the C-terminal/C-terminal interface (Fig. 2, C and D) (53). As a result, both M4 and M5 are significantly lower in energy than M6, and the M4 conformer is the most stable conformer among these three conformers. The GBMV energy of the M4 conformer is fairly similar to Tycko's double-layered conformer M3 (Fig. 2 A, Table 1). This similarity indicates that two double-layer Aβ17-42 structures based on the different U-turn shapes also have similar stabilities, and consequently further confirms a likely polymorphic scenario in amyloid formation.
Conformational stability of nonparallel Aβ17-42 oligomers
Although parallel Aβ1-40 and Aβ1-42 oligomers are stable in experiment and simulations, there are no experimental data with a similar resolution for the p3 (Aβ17-42); only low-resolution electron microscopy images are available (18). ssNMR suggests a stable Aβ16-22 antiparallel β-sheet organization (71) and a mixed picture for Aβ25-35 (72). Focusing on stability and populations, we tested other architectures based on the experimental Tycko and Lührs oligomers.
We modeled two octamers, each consisting of four parallel dimers associated in an antiparallel orientation. The first octamer is based on the Lührs model (M7; Fig. 1 C and Fig. S3), and the second on Tycko's model (M9; Fig. S4). We obtained the conformer populations by means of GBMV calculations and Monte Carlo (MC) simulations, and the results show that the Lührs conformer is the preferred dimer model for the antiparallel orientation (Table 1). The octamer based on Tycko's model (M9) is disfavored due to the small number of hydrogen bonds between the β-strands of the monomers as compared to the Lührs model. Fig. 4 A describes the change in the number of hydrogen bonds between the β strands in the single layer of the Lührs model (M7) during the simulations. For the octamer modeled as four dimers associated in antiparallel orientation, >50% of the hydrogen bonds are retained during the 30 ns simulations. The change in the intersheet distance across the cavity is shown in Fig. 4 B for this conformer. The intersheet distance is in the range of 12–13 Å, indicating that the turn region is more rigid in the antiparallel alignment.
Figure 4.
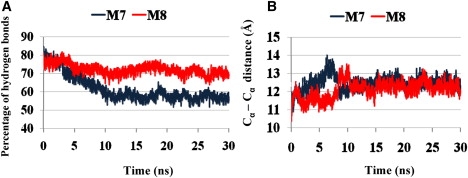
(A) Fraction of the number of hydrogen bonds (in percentage) between all β-strands compared with the initial single-layer M7 (blue) Aβ17-42 oligomer shown in Fig. 1C, and double-layer M8 (red) Aβ17-42 oligomer shown in Fig. 2E. (B) Cα backbone–backbone distance of the single-layer M7 (blue) Aβ17-42 oligomer shown in Fig. 1C, and double-layer M8 (red) Aβ17-42 oligomer shown in Fig. 2E.
A second set of antiparallel octamers was built by alternating single monomers (M10, M12, M13, and M14). The antiparallel β-sheets organization of the monomers was modeled using both the experimental models and the Ma-Nussinov computational model. Fig. 1, D–F, show single layers of three octamer models of the Aβ17-42 antiparallel organization. Of interest, both the Tycko (M13 and M14) and Lührs (M12) models show similar energy values obtained in the GBMV calculations (Table 1). The antiparallel model based on the Ma-Nussinov computational model (M10) shows a higher stability and higher population compared to the experimental models (Table 1). This is the outcome of more hydrogen bonds and hydrophobic interactions as compared to the experimental models (Fig. 1, D–F). Yet, all the antiparallel models tested have higher energies than the parallel models.
The constructed double layer of the preferred octamer for dimers associated in an antiparallel orientation based on the Lührs model (M8; Fig. 2 E). This double-layer model is more stable than the single-layer model M7, despite the small oligomer size. Apart from the hydrogen bonds between the β-strands of the monomers as in the single layer, the double layer also possesses hydrophobic interactions between the two layers, which increases the oligomers' stability. Another double-layer model (M11) with antiparallel β-strand alignment is not more stable than the corresponding single-layer model (M10). However, the energy difference between the single layer and the double layer is small. Taken together with the M8 conformer, the preference for single-layer oligomers holds only for small parallel β-sheet conformers.
Polymorphic of Aβ17-42 oligomers: energy and populations
Using all 14 models and 7000 conformations (500 for each model) generated through MD simulations, we estimate the overall populations for each model based on MC simulations with the energy landscape computed with GBMV for all conformers. For the complex kinetics of amyloid formation, this group is likely to represent only a very small percentage of the ensemble. Nevertheless, the carefully selected models cover the most likely organizations. The population results are reported in Table 1.
The most striking observation from these calculations is the equal populations of the parallel models constructed with Tycko and Lührs' structures at both the single-layer (M1 and M2) and double-layer (M3 and M4) level, despite the small energy difference found when we examine the average energies for each model. The fact that the two parallel models that differ in the U-turn conformation exhibit similar populations for both single and double layers strongly supports the notion that the type of U-turn may be a primary characteristic of the polymorphism of mature Aβ17-42 fibrils.
For the Aβ17-42 oligomers, the parallel conformers had the highest population of all the conformers studied here. Furthermore, apart from the parallel organization, antiparallel Aβ17-42 oligomers are also populated. Here, the most stable conformer was obtained based on the Ma-Nussinov computational model. The energy difference between this conformer and both experimental models for the single layer is ∼140 kcal/mol (Table 1). The computed model is more stable due to the tightly packed β-sheets, compared with the experimental models in which such tightly packed conformations are not observed. As shown in Fig. 1 D and Fig. S5, the antiparallel Aβ17-42 oligomer based on the computational model involves strong hydrophobic interactions (e.g., between Leu and Phe) that are not observed in the two experimental models (Figs. 1, E and F, and Fig. S3 and Fig. S4). The population size of the computational model is also higher. Other studied organizations consist of parallel dimers associated in an antiparallel orientation. The Lührs conformer exhibits the largest stability and the highest population (Table 1), due to the tightly packed β-sheets (Figs. 1 C and 2 E).
Conformational differences in the U-turns between the Tycko and Lührs models can lead to polymorphism as reflected in sheet–sheet registration and the extent of ordered/disordered β-strand organization at the termini for full-length Aβ1-40/Aβ1-42. In Tycko's model, residues 1–9 are structurally disordered, residues 10–22 and 30–40 form β-strands, and residues 23–29 form a turn. In Lührs' model, residues 1–16 are disordered, and residues 17–26 and 31–42 form β-strands that are connected by a four-residue (27–30) U-turn. Both Aβ1-40/Aβ1-42 models present parallel sheet organization; however, the different turn shape is reflected in their polymorphic behavior. Our results regarding the relative conformational stabilities and populations demonstrate that the major characteristics of the full-length Aβ1-40/Aβ1-42 models have been preserved in the Aβ17-42 amyloid formation.
Thus, it appears that more than one parallel conformer of the Aβ17-42 oligomer is responsible for p3 amyloid fibril formation. Previous ssNMR measurements (43) of the amylin fibril responsible for type II diabetes suggest a similar scenario. In summary, it is not only one conformation that fibrillates; rather, there exists a conformational variability in a range of different populations, including amorphous aggregates in dynamics equilibrium. A broad repertoire of turn conformations and side-chain orientations may lead to polymorphic oligomers and fibrils. Which ones prevail is likely to depend on the conditions, e.g., sequence such as mutational variants, length, concentration, pH, ionic strength, and temperature, presence of various ligands and solutes, and different surface properties. Using MD simulations, Takeda and Klimov (73) investigated the binding of Aβ peptides to amyloid fibril and found that Aβ peptides may associate with fibrils, forming elements of parallel and antiparallel organizations with similar preferences.
Conclusions
Using all-atom MD simulations in explicit solvent, we investigated polymorphic architectures for Aβ17-42, which is cleaved by α secretase from Aβ1-42 in vivo and is associated with AD and DS. Several monomers and oligomer organizations were studied. These were based on experimental models developed by Tycko and co-workers (57) and Lührs et al. (35). Although both presented a parallel assembly, this does not rule out other potential assemblies whose populations are too low to be detected under our particular experimental conditions. Indeed, the Lührs and Tycko structures differed in their turn conformations, pointing to fibril polymorphism. Furthermore, these experimental models were for Aβ1-40/Aβ1-42, whereas the truncated Aβ17-42 monomers can potentially display different preferred conformational states. No relatively high-resolution experimental structure is available for Aβ17-42, and no simulations of its potential architectures have been performed. Since oligomers are a toxic species and constitute the fibril seeds, we focused here on their stabilities and populations. Although we did not investigate such factors in this study, it behooves us to emphasize that amyloid fibril formation depends not only on nucleation and kinetics, but also on conditions such as temperature, concentration, and pH.
The major conclusion drawn from this study is that the U-turn conformation is the primary factor in amyloid polymorphism for Aβ17-42. Previous studies have confirmed that the Asp23-Lys28 salt bridge plays an important role in turn stability. Melquiond et al. (74) studied the role of the stability of Asp23-Lys28 in Aβ fibril formation. The Ma-Nussinov model (58) similarly suggests that the salt bridge stabilizes the oligomer and can play a role in Aβ oligomer formation. In the Aβ1-40 (57) oligomer structure, the Asp23-Lys28 salt bridge is intramolecular, whereas in Aβ1-42 (35) the Asp23-Lys28 salt bridge is intermolecular. The stability of the Asp23-Lys28 in Aβ fragments has been studied previously (75,76,77). Thus, in this work we did not focus on the stabilizing effect of the salt bridge; rather, we examined the difference in the U-turn shapes of these two experimental models and its contribution to the oligomer variation. These distinct turn shapes (and probably many others for which no experiment-based coordinates are currently available) lead to oligomer polymorphism as the positions of the sheets, the sheet–sheet interactions, and the consequent lengths of the disordered N-terminals are altered.
We also found out that the difference in the U-turn shape is coupled with the way two layers of β-sheet oligomers associate into a larger double-layer oligomer. This issue is well illustrated by the double-layer models M3 and M4, which have different U-turn shapes and different hydrophobic patterns of association. Thus, for a given U-turn type, different hydrophobic association patterns can lead to polymorphism with large differences in energy and populations (e.g., M4 and M6), or with small differences in energy and populations (e.g., M4 and M5).
Although the population of an antiparallel β-strand organization is not negligible, our results demonstrate that the parallel Aβ17-42 oligomer is the most stable organization, and the conformer that was modeled using Tycko's model (57) is slightly preferred over Lührs' model for the single layer (35). On the other hand, for the double layer, both models have similar stabilities (a difference of ∼2 kcal/mol). The main difference between these two experimental models is in the U-turn shape. The turn determines the intersheet registration. Therefore, the U-turn conformation plays a key role in the stability and polymorphic conformational ensemble of parallel Aβ17-42 oligomers. Furthermore, in vivo the amyloid fibril is a supramolecular structure composed of several protofibrils that wind around each other, mostly with a left-handed twist along their main axis (78–87). Different U-turn conformations can affect the twist angles and thus lead to different morphologies in electron microscopy measurements. We therefore conclude that Aβ17-42 conformers with different U-turn shapes and hence different morphologies can be detected by experiment. It is important to note here that these may only present the prevailing organization under experimental conditions, and the probability of detecting the various morphologies will depend on the size of the population. In general, for longer sequences, a parallel organization is favored because identical residues stack on top of each other and the identical charge repulsion at the terminals is overcome (88). We further emphasize that it remains to be determined which organizations preferentially interact with and penetrate the bilayer; however, the question is relevant because interaction with the bilayer is currently believed to underlie toxicity.
Insights into polymorphic behavior at the molecular level should assist investigators in understanding the mechanism of aggregation and toxicity, and may pave the way to novel therapeutic approaches and more effective drug design.
Supporting Material
Additional text, nine figures, and a table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)01051-0.
Supporting Material
Acknowledgments
We thank the members of the Nussinov group (Drs. Yongping Pan, Chung-Jung Tsai, and Jin Liu) at the Center for Cancer Research Nanobiology Program, NCI-Frederick, for helpful discussions, and in particular we thank Dr. Hyunbum Jang for his helpful comments. All simulations were performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov). We thank Rob Tycko for providing the atomic coordinates of the model obtained in his laboratory.
This project was funded in whole or in part by federal funds from the National Cancer Institute, National Institutes of Health, under contract number NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
References
- 1.Sipe J.D. Amyloidosis. Annu. Rev. Biochem. 1992;61:947–975. doi: 10.1146/annurev.bi.61.070192.004503. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D.J. Cell biology of protein misfolding: the examples of Alzheimer's and Parkinson's diseases. Nat. Cell Biol. 2004;6:1054–1061. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- 3.Sunde M., Blake C.C. From the globular to the fibrous state: protein structure and structural conversion in amyloid formation. Q. Rev. Biophys. 1998;31:1–39. doi: 10.1017/s0033583598003400. [DOI] [PubMed] [Google Scholar]
- 4.Caughey B., Lansbury P.T. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 5.Lalowski M., Golabek A., Lemere C.A., Selkoe D.J., Wisniewski H.M. The “nonamyloidogenic” p3 fragment (amyloid β17–42) is a major constituent of Down's syndrome cerebellar preamyloid. J. Biol. Chem. 1996;271:33623–33631. doi: 10.1074/jbc.271.52.33623. [DOI] [PubMed] [Google Scholar]
- 6.Ahmad A., Millett I.S., Doniach S., Uversky V.N., Fink A.L. Stimulation of insulin fibrillation by urea-induced intermediates. J. Biol. Chem. 2004;279:14999–15013. doi: 10.1074/jbc.M313134200. [DOI] [PubMed] [Google Scholar]
- 7.Fraser P.E., Levesque L., McLachlan D.R. Biochemistry of Alzheimer's disease amyloid plaques. Clin. Biochem. 1993;26:339–349. doi: 10.1016/0009-9120(93)90110-r. [DOI] [PubMed] [Google Scholar]
- 8.Wertkin A.M., Turner R.S., Pleasure S.J., Golde T.E., Younkin S.G. Human neurons derived from a teratocarcinoma cell line express solely the 695-amino acid amyloid precursor protein and produce intracellular β-amyloid or A4 peptides. Proc. Natl. Acad. Sci. USA. 1993;90:9513–9517. doi: 10.1073/pnas.90.20.9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamour Y. Alzheimer's disease: a review of recent findings. Biomed. Pharmacother. 1994;48:312–318. doi: 10.1016/0753-3322(94)90177-5. [DOI] [PubMed] [Google Scholar]
- 10.Annaert W., De Strooper B. A cell biological perspective on Alzheimer's disease. Annu. Rev. Cell Dev. Biol. 2002;18:25–51. doi: 10.1146/annurev.cellbio.18.020402.142302. [DOI] [PubMed] [Google Scholar]
- 11.Wisniewski T., Ghiso J., Frangione B. Alzheimer's disease and soluble A β. Neurobiol. Aging. 1994;15:143–152. doi: 10.1016/0197-4580(94)90105-8. [DOI] [PubMed] [Google Scholar]
- 12.Checler F. Processing of the β-amyloid precursor protein and its regulation in Alzheimer's disease. J. Neurochem. 1995;65:1431–1444. doi: 10.1046/j.1471-4159.1995.65041431.x. [DOI] [PubMed] [Google Scholar]
- 13.Selkoe D.J. Cell biology of the amyloid β-protein precursor and the mechanism of Alzheimer's disease. Annu. Rev. Cell Biol. 1994;10:373–403. doi: 10.1146/annurev.cb.10.110194.002105. [DOI] [PubMed] [Google Scholar]
- 14.Wei W., Norton D.D., Wang X., Kusiak J.W. Aβ 17–42 in Alzheimer's disease activates JNK and caspase-8 leading to neuronal apoptosis. Brain. 2002;125:2036–2043. doi: 10.1093/brain/awf205. [DOI] [PubMed] [Google Scholar]
- 15.Higgins L.S., Murphy G.M., Forno L.S., Catalano R., Cordell B. P3 β-amyloid peptide has a unique and potentially pathogenic immunohistochemical profile in Alzheimer's disease brain. Am. J. Pathol. 1996;149:585–596. [PMC free article] [PubMed] [Google Scholar]
- 16.Dickson D.W. The pathogenesis of senile plaques. J. Neuropathol. Exp. Neurol. 1997;56:321–339. doi: 10.1097/00005072-199704000-00001. [DOI] [PubMed] [Google Scholar]
- 17.Pike C.J., Overman M.J., Cotman C.W. Amino-terminal deletions enhance aggregation of β-amyloid peptides in vitro. J. Biol. Chem. 1995;270:23895–23898. doi: 10.1074/jbc.270.41.23895. [DOI] [PubMed] [Google Scholar]
- 18.Schmechel A., Zentgraf H., Scheuermann S., Fritz G., Pipkorn R. Alzheimer β-amyloid homodimers facilitate A β fibrillization and the generation of conformational antibodies. J. Biol. Chem. 2003;278:35317–35324. doi: 10.1074/jbc.M303547200. [DOI] [PubMed] [Google Scholar]
- 19.Dulin F., Leveille F., Ortega J.B., Mornon J.P., Buisson A. P3 peptide, a truncated form of A β devoid of synaptotoxic effect, does not assemble into soluble oligomers. FEBS Lett. 2008;582:1865–1870. doi: 10.1016/j.febslet.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Kirkitadze M.D., Bitan G., Teplow D.B. Paradigm shifts in Alzheimer's disease and other neurodegenerative disorders: the emerging role of oligomeric assemblies. J. Neurosci. Res. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- 21.Thirumalai D., Klimov D.K., Dima R.I. Emerging ideas on the molecular basis of protein and peptide aggregation. Curr. Opin. Struct. Biol. 2003;13:146–159. doi: 10.1016/s0959-440x(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 22.Walsh D.M., Selkoe D.J. Oligomers on the brain: the emerging role of soluble protein aggregates in neurodegeneration. Protein Pept. Lett. 2004;11:213–228. doi: 10.2174/0929866043407174. [DOI] [PubMed] [Google Scholar]
- 23.Mattson M.P. Pathways towards and away from Alzheimer's disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh D.M., Selkoe D.J. Deciphering the molecular basis of memory failure in Alzheimer's disease. Neuron. 2004;44:181–193. doi: 10.1016/j.neuron.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Conway K.A., Lee S.J., Rochet J.C., Ding T.T., Williamson R.E. Acceleration of oligomerization, not fibrillization, is a shared property of both α-synuclein mutations linked to early-onset Parkinson's disease: implications for pathogenesis and therapy. Proc. Natl. Acad. Sci. USA. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Agnaf O.M., Nagala S., Patel B.P., Austen B.M. Non-fibrillar oligomeric species of the amyloid ABri peptide, implicated in familial British dementia, are more potent at inducing apoptotic cell death than protofibrils or mature fibrils. J. Mol. Biol. 2001;310:157–168. doi: 10.1006/jmbi.2001.4743. [DOI] [PubMed] [Google Scholar]
- 27.Bucciantini M., Giannoni E., Chiti F., Baroni F., Formigli L. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 28.Kayed R., Head E., Thompson J.L., McIntire T.M., Milton S.C. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 29.Cleary J.P., Walsh D.M., Hofmeister J.J., Shankar G.M., Kuskowski M.A. Natural oligomers of the amyloid-β protein specifically disrupt cognitive function. Nat. Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 30.Reixach N., Deechongkit S., Jiang X., Kelly J.W., Buxbaum J.N. Tissue damage in the amyloidoses: Transthyretin monomers and nonnative oligomers are the major cytotoxic species in tissue culture. Proc. Natl. Acad. Sci. USA. 2004;101:2817–2822. doi: 10.1073/pnas.0400062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demuro A., Mina E., Kayed R., Milton S.C., Parker I. Calcium dysregulation and membrane disruption as a ubiquitous neurotoxic mechanism of soluble amyloid oligomers. J. Biol. Chem. 2005;280:17294–17300. doi: 10.1074/jbc.M500997200. [DOI] [PubMed] [Google Scholar]
- 32.Malisauskas M., Ostman J., Darinskas A., Zamotin V., Liutkevicius E. Does the cytotoxic effect of transient amyloid oligomers from common equine lysozyme in vitro imply innate amyloid toxicity? J. Biol. Chem. 2005;280:6269–6275. doi: 10.1074/jbc.M407273200. [DOI] [PubMed] [Google Scholar]
- 33.Mousseau N., Derreumaux P. Exploring the early steps of amyloid peptide aggregation by computers. Acc. Chem. Res. 2005;38:885–891. doi: 10.1021/ar050045a. [DOI] [PubMed] [Google Scholar]
- 34.Makabe K., McElheny D., Tereshko V., Hilyard A., Gawlak G. Atomic structures of peptide self-assembly mimics. Proc. Natl. Acad. Sci. USA. 2006;103:17753–17758. doi: 10.1073/pnas.0606690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lührs T., Ritter C., Adrian M., Riek-Loher D., Bohrmann B. 3D structure of Alzheimer's amyloid-β (1–42) fibrils. Proc. Natl. Acad. Sci. USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka M., Chien P., Naber N., Cooke R., Weissman J.S. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 37.Petkova A.T., Leapman R.D., Guo Z., Yau W.M., Mattson M.P. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 38.Paravastu A.K., Petkova A.T., Tycko R. Polymorphic fibril formation by residues 10–40 of the Alzheimer's β-amyloid peptide. Biophys. J. 2006;90:4618–4629. doi: 10.1529/biophysj.105.076927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bradley M.E., Edskes H.K., Hong J.Y., Wickner R.B., Liebman S.W. Interactions among prions and prion “strains” in yeast. Proc. Natl. Acad. Sci. USA. 2002;99:16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paravastu A.K., Leapman R.D., Yau W.M., Tycko R. Molecular structural basis for polymorphism in Alzheimer's {β}-amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petkova A.T., Buntkowsky G., Dyda F., Leapman R.D., Yau W.M. Solid state NMR reveals a pH-dependent antiparallel β-sheet registry in fibrils formed by a β-amyloid peptide. J. Mol. Biol. 2004;335:247–260. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 42.Meinhardt J., Sachse C., Hortschansky P., Grigorieff N., Fandrich M. Aβ (1–40) fibril polymorphism implies diverse interaction patterns in amyloid fibrils. J. Mol. Biol. 2008;386:869–877. doi: 10.1016/j.jmb.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madine J., Jack E., Stockley P.G., Radford S.E., Serpell L.C. Structural insights into the polymorphism of amyloid-like fibrils formed by region 20–29 of amylin revealed by solid-state NMR and X-ray fiber diffraction. J. Am. Chem. Soc. 2008;130:14990–15001. doi: 10.1021/ja802483d. [DOI] [PubMed] [Google Scholar]
- 44.Derkatch I.L., Chernoff Y.O., Kushnirov V.V., Inge-Vechtomov S.G., Liebman S.W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brachmann A., Baxa U., Wickner R.B. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King C.Y. Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J. Mol. Biol. 2001;307:1247–1260. doi: 10.1006/jmbi.2001.4542. [DOI] [PubMed] [Google Scholar]
- 47.King C.Y., Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka M., Chien P., Yonekura K., Weissman J.S. Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell. 2005;121:49–62. doi: 10.1016/j.cell.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 49.Baxa U., Cassese T., Kajava A.V., Steven A.C. Structure, function, and amyloidogenesis of fungal prions: filament polymorphism and prion variants. Adv. Protein Chem. 2006;73:125–180. doi: 10.1016/S0065-3233(06)73005-4. [DOI] [PubMed] [Google Scholar]
- 50.Yamaguchi K., Takahashi S., Kawai T., Naiki H., Goto Y. Seeding-dependent propagation and maturation of amyloid fibril conformation. J. Mol. Biol. 2005;352:952–960. doi: 10.1016/j.jmb.2005.07.061. [DOI] [PubMed] [Google Scholar]
- 51.Verel R., Tomka I.T., Bertozzi C., Cadalbert R., Kammerer R.A. Polymorphism in an amyloid-like fibril-forming model peptide. Angew. Chem. Int. Ed. Engl. 2008;47:5842–5845. doi: 10.1002/anie.200800021. [DOI] [PubMed] [Google Scholar]
- 52.Ma B.Y., Nussinov R. Simulations as analytical tools to understand protein aggregation and predict amyloid conformation. Curr. Opin. Chem. Biol. 2006;10:445–452. doi: 10.1016/j.cbpa.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 53.Zheng J., Jang H., Ma B., Tsai C.J., Nussinov R. Modeling the Alzheimer Aβ17–42 fibril architecture: tight intermolecular sheet-sheet association and intramolecular hydrated cavities. Biophys. J. 2007;93:3046–3057. doi: 10.1529/biophysj.107.110700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng J., Jang H., Ma B., Nussinov R. Annular structures as intermediates in fibril formation of Alzheimer A β (17–42) J. Phys. Chem. B. 2008;112:6856–6865. doi: 10.1021/jp711335b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchete N.V., Tycko R., Hummer G. Molecular dynamics simulations of Alzheimer's β-amyloid protofilaments. J. Mol. Biol. 2005;353:804–821. doi: 10.1016/j.jmb.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 56.Buchete N.V., Hummer G. Structure and dynamics of parallel β-sheets, hydrophobic core, and loops in Alzheimer's A β fibrils. Biophys. J. 2007;92:3032–3039. doi: 10.1529/biophysj.106.100404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petkova A.T., Yau W.M., Tycko R. Experimental constraints on quaternary structure in Alzheimer's β-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma B., Nussinov R. Stabilities and conformations of Alzheimer's β-amyloid peptide oligomers (Aβ 16–22, Aβ 16–35, and Aβ 10–35): sequence effects. Proc. Natl. Acad. Sci. USA. 2002;99:14126–14131. doi: 10.1073/pnas.212206899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kale L., Skeel R., Bhandarkar M., Brunner R., Gursoy A. NAMD2: greater scalability for parallel molecular dynamics. J. Comput. Phys. 1999;151:283–312. [Google Scholar]
- 60.MacKerell A.D., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 61.Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S. CHARMM—a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 62.Mahoney M.W., Jorgensen W.L. A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J. Chem. Phys. 2000;112:8910–8922. [Google Scholar]
- 63.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 64.Martyna G.J., Tobias D.J., Klein M.L. Constant-pressure molecular-dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 65.Feller S.E., Zhang Y.H., Pastor R.W., Brooks B.R. Constant-pressure molecular-dynamics simulation—the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 66.Darden T., York D., Pedersen L. Particle mesh Ewald—an N.Log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 67.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 68.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of Cartesian equations of motion of a system with constraints—molecular-dynamics of N-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 69.Lee M.S., Feig M., Salsbury F.R., Brooks C.L. New analytic approximation to the standard molecular volume definition and its application to generalized born calculations. J. Comput. Chem. 2003;24:1348–1356. doi: 10.1002/jcc.10272. [DOI] [PubMed] [Google Scholar]
- 70.Lee M.S., Salsbury F.R., Brooks C.L. Novel generalized Born methods. J. Chem. Phys. 2002;116:10606–10614. [Google Scholar]
- 71.Balbach J.J., Ishii Y., Antzutkin O.N., Leapman R.D., Rizzo N.W. Amyloid fibril formation by A β (16–22), a seven-residue fragment of the Alzheimer's β-amyloid peptide, and structural characterization by solid state NMR. Biochemistry. 2000;39:13748–13759. doi: 10.1021/bi0011330. [DOI] [PubMed] [Google Scholar]
- 72.Ma B., Nussinov R. The stability of monomeric intermediates controls amyloid formation: Aβ 25–35 and its N27Q mutant. Biophys. J. 2006;90:3365–3374. doi: 10.1529/biophysj.105.075309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takeda T., Klimov D.K. Replica exchange simulations of the thermodynamics of Aβ fibril growth. Biophys. J. 2009;96:442–452. doi: 10.1016/j.bpj.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Melquiond A., Dong X., Mousseau N., Derreumaux P. Role of the region 23–28 in Aβ fibril formation: insights from simulations of the monomers and dimers of Alzheimer's peptides Aβ40 and Aβ42. Curr. Alzheimer Res. 2008;5:244–250. doi: 10.2174/156720508784533330. [DOI] [PubMed] [Google Scholar]
- 75.Baumketner A., Bernstein S.L., Wyttenbach T., Lazo N.D., Teplow D.B. Structure of the 21–30 fragment of amyloid β-protein. Protein Sci. 2006;15:1239–1247. doi: 10.1110/ps.062076806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baumketner A., Shea J.E. The structure of the Alzheimer amyloid β10–35 peptide probed through replica-exchange molecular dynamics simulations in explicit solvent. J. Mol. Biol. 2007;366:275–285. doi: 10.1016/j.jmb.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 77.Tarus B., Straub J.E., Thirumalai D. Dynamics of Asp23-Lys28 salt-bridge formation in Aβ10–35 monomers. J. Am. Chem. Soc. 2006;128:16159–16168. doi: 10.1021/ja064872y. [DOI] [PubMed] [Google Scholar]
- 78.Bauer H.H., Aebi U., Haner M., Hermann R., Muller M. Architecture and polymorphism of fibrillar supramolecular assemblies produced by in vitro aggregation of human calcitonin. J. Struct. Biol. 1995;115:1–15. doi: 10.1006/jsbi.1995.1024. [DOI] [PubMed] [Google Scholar]
- 79.Goldsbury C.S., Cooper G.J., Goldie K.N., Muller S.A., Saafi E.L. Polymorphic fibrillar assembly of human amylin. J. Struct. Biol. 1997;119:17–27. doi: 10.1006/jsbi.1997.3858. [DOI] [PubMed] [Google Scholar]
- 80.Ionescu-Zanetti C., Khurana R., Gillespie J.R., Petrick J.S., Trabachino L.C. Monitoring the assembly of Ig light-chain amyloid fibrils by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 1999;96:13175–13179. doi: 10.1073/pnas.96.23.13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harper J.D., Lieber C.M., Lansbury P.T. Atomic force microscopic imaging of seeded fibril formation and fibril branching by the Alzheimer's disease amyloid-β protein. Chem. Biol. 1997;4:951–959. doi: 10.1016/s1074-5521(97)90303-3. [DOI] [PubMed] [Google Scholar]
- 82.Chamberlain A.K., MacPhee C.E., Zurdo J., Morozova-Roche L.A., Hill H.A. Ultrastructural organization of amyloid fibrils by atomic force microscopy. Biophys. J. 2000;79:3282–3293. doi: 10.1016/S0006-3495(00)76560-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jimenez J.L., Nettleton E.J., Bouchard M., Robinson C.V., Dobson C.M. The protofilament structure of insulin amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2002;99:9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rochet J.C., Lansbury P.T. Amyloid fibrillogenesis: themes and variations. Curr. Opin. Struct. Biol. 2000;10:60–68. doi: 10.1016/s0959-440x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 85.Dobson C.M. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 86.Uversky V.N., Fink A.L. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim. Biophys. Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 87.Kanno T., Yamaguchi K., Naiki H., Goto Y., Kawai T. Association of thin filaments into thick filaments revealing the structural hierarchy of amyloid fibrils. J. Struct. Biol. 2005;149:213–218. doi: 10.1016/j.jsb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 88.Tsai H.H., Gunasekaran K., Nussinov R. Sequence and structure analysis of parallel β helices: implication for constructing amyloid structural models. Structure. 2006;14:1059–1072. doi: 10.1016/j.str.2006.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


