Abstract
Expression of T1ST2, the IL-33R, by Th2 cells requires GATA3. Resting Th2 cells express little GATA3, which is increased by IL-33 and a STAT5 activator, in turn increasing T1ST2 from its low-level expression on resting Th2 cells. Th2 cells that have upregulated T1ST2 produce IL-13, but not IL-4, in response to IL-33 plus a STAT5 activator in an antigen-independent, NF-κB-dependent, cyclosporin A (CsA)-resistant manner. Similarly, Th17 cells produce IL-17A in response to IL-1β and a STAT3 activator and Th1 cells produce IFNγ in response to IL-18 and a STAT4 inducer. Thus, each effector Th cell produces cytokines without antigenic stimulation in response to an IL-1 family member and a specific STAT activator, implying an innate mechanism through which memory CD4 T cells are recruited by an induced cytokine environment.
Keywords: IL-33, TSLP, IL-18, STAT5, T1ST2
IL-33, a member of the IL-1 family, is synthesized as a 31-kDa precursor and cleaved in vitro by caspase-1 to a mature 18-kDa form (1). T1ST2, originally identified as an orphan receptor expressed primarily on Th2 and mast cells (2–4), has now been shown to bind to IL-33 and to form a complex with IL-1RAcP (5, 6) that mediates IL-33 signaling (1). IL-33 transduces its effects through the classical IL-1 signaling machinery, including activation of NF-κB and MAPKs (1).
Administration of IL-33 to mice causes profound alterations in mucosal tissues, including lung, esophageal, and intestinal eosinophilia, and splenomegaly, and enhanced serum IgA and IgE (1). Inhibition of IL-33, either by anti-T1ST2 antibody or T1ST2 immunoglobin, blocks secretion of IL-4, IL-5, and IL-13 and markedly suppresses eosinophilic inflammation in recipients of ovalbumin-specific Th2 cells challenged with ovalbumin (2, 7). T1ST2-deficient mice show a defect in primary response to schistosoma egg antigens (8) and IL-33 treatment at the time of Trichuris muris infection allows susceptible mice to expel the parasite (9).
These results indicate an important role for IL-33 and T1ST2 in allergic/parasite-induced inflammatory responses. However, the molecular regulation of T1ST2 expression and how IL-33 mediates its function remain unclear. Here we show that T1ST2 expression by Th2 cells is regulated by GATA3 and STAT5. Further, IL-33 and STAT5 activators such as IL-2, IL-7, and TSLP act synergistically to induce and maintain GATA3 expression. IL-33 then acts on T1ST2-expressing Th2 cells to induce production of IL-13, but not IL-4, in a TCR-independent, NF-κB-, and p38-dependent manner. Such TCR-independent IL-13 production by Th2 cells is similar to the previous demonstration by others that treatment of Th1 cells with IL-18 and IL-12 induces IFNγ production (10). Consistent with these findings, we also show that Th17 cells (11–13) treated with IL-1β and a STAT3 activator (IL-6, IL-21, or IL-23) produce IL-17A in a TCR-independent manner. Thus, all 3 effector Th populations respond to an IL-1 family member and a STAT activator with the production of a key effector cytokine, providing a mechanism for innate activation of memory or effector CD4 T cells.
Results
T1ST2 Expression on Th2 Cells.
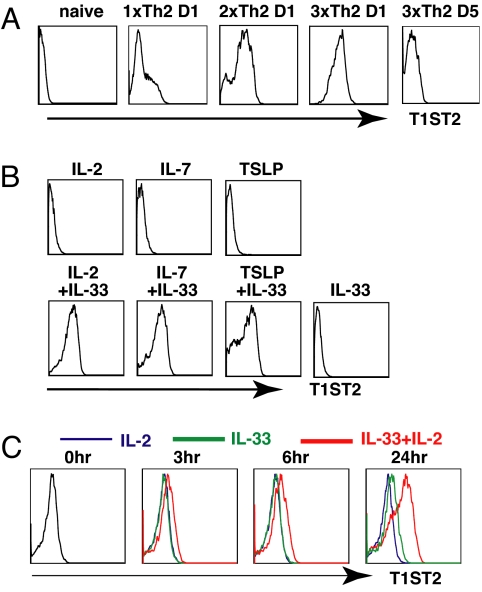
T1ST2 is under tight regulation while IL-1RAcP is broadly expressed. Naïve CD4 T cells do not express detectable T1ST2 (Fig. 1A). Culture under Th2 conditions for 4 days (1xTh2) results in a small fraction of the cells expressing high levels of T1ST2. More Th2 cells express T1ST2 after a second round of priming (2xTh2) and essentially all 3-round primed Th2 cells (3xTh2) express T1ST2 (Fig. 1A). T1ST2 expression falls strikingly when 3xTh2 cells are “rested” in IL-2- (Fig. 1 A and B), IL-7-, or TSLP-containing medium (Fig. 1B). Rested Th2 cells expressing low levels of T1ST2 were cultured in IL-2, IL-7, or TSLP alone, IL-33 alone, or IL-33 plus IL-2, IL-7, or TSLP for 2 days. T1ST2 expression was significantly upregulated in the cells cultured in IL-33 combined with IL-2, IL-7, or TSLP, but not in the cells cultured in IL-2, IL-7, or TSLP alone or IL-33 alone (Fig. 1B). T1ST2 upregulation was first detected at 24 h of culture in IL-2 plus IL-33 (Fig. 1C).
Fig. 1.
STAT5 is required for T1ST2 expression. (A) Naïve CD4 T cells purified from 5C.C7 transgenic Rag2−/− mice were cultured under Th2 conditions for indicated periods. D1 indicates cells that had just completed Th2 priming. D5 indicates cells cultured in IL-2-containing cRPMI for 5 days after completion of Th2 priming. (B) 3xTh2 cells that had been maintained in IL-2 medium for 2 weeks were cultured in medium containing indicated cytokines for 48 h and then stained for T1ST2. (C) 3xD14 Th2 cells were cultured in the cytokines for the indicated periods and then stained for T1ST2. The experiments were performed 3 times.
T1ST2 Expression Is GATA3 Dependent.
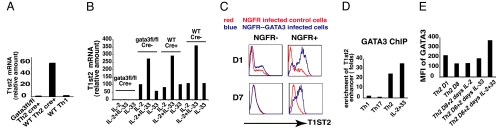
T1ST2 is expressed on Th2 cells but not on Th1 or naïve CD4 T cells, suggesting that GATA3, the master transcription factor for Th2 differentiation, has a role in regulating T1ST2. Two-round primed Th2 (2xTh2) cells derived from Gata3-fl/fl mice or wild-type mice were infected with a GFP-Cre retrovirus. Cre+ cells were separated by cell sorting, cultured under Th2 conditions for an additional round to reduce the amount of residual GATA3 protein in the Cre-expressing cells, and then collected to measure T1st2 mRNA (Fig. 2A). T1st2 mRNA in wild-type Th2 Cre+ cells was 60-fold higher than in Gata3-fl/fl Th2 Cre+ cells. Indeed, the amount of T1st2 mRNA in the GATA3-deficient Th2 cells was no greater than in Th1 cells.
Fig. 2.
T1ST2 expression is GATA3 dependent. (A) 2xTh2 cells derived from Gata3“floxed” or C57BL/6 mice were retrovirally infected with GFP-Cre during the third round of Th2 priming. GFP-Cre positive cells were sorted and cultured under Th2 conditions for an additional round and cells were collected to measure T1st2 mRNA by QPCR. Four-round primed Th1 (4xTh1) cells derived from C57BL/6 mice were cultured and examined in parallel. (B) 3xTh2 cells from Gata3“floxed” or C57BL/6 mice were retrovirally infected with GFP-Cre. GFP-Cre positive and negative cells were sorted and cultured under Th2 conditions for an additional round and then maintained in IL-2-containing medium. Two weeks later, the cells were divided into 3 portions, cultured in medium containing IL-2 alone, IL-33 alone, or IL-2 plus IL-33. Two days later, cells were collected to measure T1st2 mRNA. (C) CD4 T cells were cultured under Th2 conditions and 2 days later the cells were retrovirally infected with Gata3-NGFR or NGFR. After an additional 2 days, cells were stained to measure NGFR and T1ST2 expression immediately (D1) or after 7 days of culture in IL-2-containing medium (D7). (D) 1xTh1, 1xTh17, and 1xTh2 cells were fixed immediately after completion of priming and immunoprecipitated with anti-GATA3. D7 1xTh2 cells were cultured in IL-2 plus IL-33 for 2 days and ChIP with anti-GATA3 or control mouse IgG was performed. (E) MFI of GATA3 staining for D1 2xTh2 cells; D8 2xTh2 cells; D8 cells cultured in IL-2, IL-33, or IL-2 plus IL-33 for an additional 48 h (D8 + 2 days IL-2, D8 + 2 days IL-33, D8 + 2 days IL-2 + 33). The experiments were performed two times.
We next asked whether the IL-33/STAT5-mediated upregulation of T1ST2 in resting Th2 cells is also GATA3 dependent. Three-round primed Th2 (3xTh2) cells from Gata3-fl/fl or wild-type mice were infected with a GFP-Cre retrovirus. Cre+ and Cre− cells were cultured under Th2 conditions for an additional round and then maintained in IL-2-containing medium for 2 weeks. Cells were then placed in medium containing IL-2 alone, IL-33 alone, or IL-2 plus IL-33 for 2 days. T1st2 mRNA, which was already very low in Gata3-fl/fl/Cre+ cells, was not induced in these cells by any treatment. By contrast, T1st2 mRNA was induced, to a similar degree, in wild-type/Cre+, wild type/Cre−, and Gata3-fl/fl/Cre− cells and was best in response to stimulation with IL-2 plus IL-33 (Fig. 2B). Thus, T1st2 induction by IL-2/IL-33 is GATA3 dependent.
To directly link GATA3 to T1ST2 expression, CD4 T cells were cultured under Th2 conditions for 2 days and infected with a Gata3-nerve growth factor receptor (NGFR) retrovirus or a control NGFR retrovirus. After a 4-day-Th2 priming period, a small fraction of both the NGFR− and the NGFR+ cells from control retroviral infection expressed T1ST2, representing endogenous early Th2 T1ST2 induction (Fig. 2C). In the cells expressing viral Gata3 (NGFR+ cells from the NGFR-Gata3-infected culture), the majority of the cells expressed high levels of T1ST2. After being cultured in IL-2-containing medium for 7 days, T1ST2 on the NGFR− cells strikingly declined while the majority of viral GATA3+ cells continued to express T1ST2, although at a somewhat lower level, implying that continued high-level GATA3 expression is important for maintaining T1ST2 levels.
GATA3 binds strongly to the −12K T1st2 enhancer as indicated by chromatin immunoprecipitation (ChIP) sequencing (J.Z., G.W., unpublished observations). ChIP with an anti-GATA3 antibody revealed a remarkable enrichment of the −12K T1st2 enhancer sequences in TCR-stimulated Th2 cells, but not in Th1 or Th17 cells (Fig. 2D). Using mouse IgG as a control for anti-GATA3 resulted in no enrichment.
GATA3 levels decline when Th2 cells are rested in IL-2 medium. Culturing such rested cells in medium containing IL-33 plus IL-2 for 2 days substantially increased GATA3 protein levels compared to cells cultured in IL-33 alone or in IL-2 alone (Fig. 2E). In “2-day IL-33 plus IL-2” cells, GATA3 binds to the T1st2 gene as shown by the striking enrichment of the −12K enhancer sequences by anti-GATA3 precipitation. The capacity of GATA3 to bind to specific sites in the T1st2 locus implies its role in regulating T1ST2 expression is direct.
STAT5 Plays Direct Roles in GATA3 Upregulation and T1ST2 Reexpression.
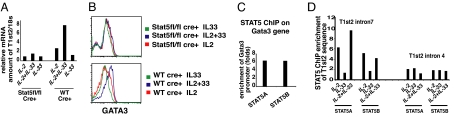
IL-2, IL-7, and TSLP, in combination with IL-33, each led to upregulation of T1ST2 (Fig. 1B) and each activates STAT5. To determine whether STAT5 mediated the effects of these cytokines in T1ST2 upregulation, STAT5 was depleted from the responding cells (Fig. 3A). Two-round primed Th2 (2xTh2) cells from Stat5a/Stat5b-fl/fl or wild-type mice were infected with a GFP-Cre retrovirus. Cre+ cells were sorted, on the basis of GFP expression and cultured for 6 days in IL-4-containing medium to maintain viability. These cells were then cultured in IL-4-containing medium supplemented with IL-2, IL-33, or IL-2 plus IL-33. T1st2 mRNA induction, measured 48 h later, occurred only in WT Cre+ cells cultured with IL-2 plus IL-33. Stat5a/Stat5b-fl/fl/Cre+ cells, in which the Stat5a and Stat5b genes were deleted, had markedly decreased T1st2 (Fig. 3A).
Fig. 3.
STAT5 is directly involved in GATA3 and T1ST2 upregulation. (A and B) 2xTh2 cells from Stat5“floxed” or C57BL/6 mice were retrovirally infected with GFP-Cre. GFP-Cre positive cells were sorted and cultured in IL-4-containing-cRPMI for 6 days. Cells were then cultured in IL-2, IL-33, or IL-2 plus IL-33 for 48 h, with IL-4 in the medium, and then collected to measure T1st2 mRNA (A) or stained for GATA3 expression (B). (C) D7 1xTh2 cells were cultured in IL-2 plus IL-33 for an additional 2 days, and ChIP with anti-STAT5A, anti-STAT5B was performed. (D) D7 1xTh2 cells were cultured in IL-2, IL-33, or IL-2 plus IL-33 for 24 h and ChIP with anti-STAT5A, anti-STAT5B, or control mouse IgG was performed. The experiments were performed twice.
Because T1ST2 expression requires both STAT5 and GATA3 and GATA3 appears to act directly on T1st2, we asked whether STAT5 is required for GATA3 induction or STAT5 functions together with GATA3 to induce T1ST2 expression. Induction of GATA3 expression by IL-33 plus IL-2 was observed in wild-type/Cre+ cells but not in the Stat5a/Stat5b-fl/fl/Cre+ cells (Fig. 3B), implying that STAT5, and IL-33, is required for GATA3 induction and thus has an “upstream” role in T1ST2 induction. ChIP using anti-STAT5A and anti-STAT5B antibodies resulted in a significant enrichment of a −800-bp Gata3 promoter sequence, indicating that STAT5 binds to the Gata3 gene, consistent with its playing a direct role in upregulating GATA3 expression (Fig. 3C). ChIP revealed that STAT5 binds to T1st2 intron 7, but not intron 4, sequences (Fig. 3D). From these experiments, it can be concluded that GATA3 expression is dependent on both STAT5 activation and IL-33 and that GATA3 and STAT5 likely jointly regulate T1ST2 expression in Th2 cells.
Culture in IL-33 Enhances STAT5 Phosphorylation.
We asked whether IL-33 reciprocally regulates STAT5 activation. Three-round primed Th2 (3xTh2) cells expressing T1ST2 were cultured in medium with or without IL-33 for 20 h, and then washed to remove IL-33, and challenged with IL-2 for 15 min. Twenty-hour culture in IL-33 resulted in a greater degree of STAT5 phosphorylation in response to IL-2 than seen in Th2 cells cultured without IL-33 (supporting information (SI) Fig. S1A). Consistent with this, IL-2 induced more Socs3 mRNA in IL-33-cultured cells (Fig. S1B).
T1ST2-Expressing Cells Produce IL-13 in Response to IL-33.
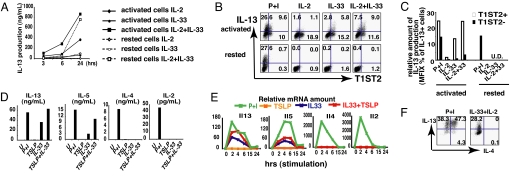
One-round primed Th2 (1xTh2) activated cells, of which ≈1/5 express high levels of T1ST2, and rested Th2 cells, which uniformly express low-level T1ST2, were cultured in medium containing IL-2 alone, IL-33 alone, or IL-2 plus IL-33. IL-13 production was detected at 3 h and 6 h in activated Th2 cells cultured in IL-2 plus IL-33 (Fig. 4 A, B, and C), largely in the T1ST2+ cells (Fig. 4 B and C). IL-33 alone also induced T1ST2+ activated cells to produce IL-13, to a level approximately half that induced by IL-33 plus IL-2 (Fig. 4 A, B, and C).
Fig. 4.
IL-33, together with activated STAT5, induces IL-13 and IL-5 but not IL-4 or IL-2. (A) D1 1xTh2 cells (activated) and D10 1xTh2 cells (rested) were cultured in medium containing IL-2, IL-33, or IL-2 plus IL-33 for the indicated periods and supernatants collected to measure IL-13 production by ELISA. (B) Cells in A were stimulated for 4 h to measure IL-13 production by intracellular staining. (C) Relative amount of IL-13 production was calculated by multiplying the MFI and the percentage of IL-13-producing cells. (D) 3xTh2 cells were stimulated with P+I, TSLP, IL-33, or TSLP+IL-33 for 48 h. The supernatants were collected. (E) 3xTh2 cells were stimulated with P+I, TSLP, IL-33, or TSLP+IL-33 for indicated periods and collected to measure Il13, Il5, Il4, and Il2 mRNA. (F) 3xTh2 cells were stimulated with P+I or IL-2 plus IL-33 for 4 h to test IL-4 and IL-13 production. The experiments were performed 3 times.
IL-13 production by rested Th2 cells was not detectable until 24 h and then only in cells cultured in both IL-33 and IL-2 (Fig. 4A). Rested Th2 cells require 24 h culture in IL-33 plus IL-2 to achieve high levels of T1ST2 expression (Fig. 1C); it is only in the T1ST2-expressing cells that IL-13 production is induced.
IL-13 production in response to PMA plus ionomycin (P+I) was similar in activated and rested Th2 cells (Fig. 4 B and C) and not limited to T1ST2+ cells, implying that the pathways through which IL-33 and P+I induce IL-13 are different.
IL-33 Plus a STAT5 Activator Can Induce IL-13 and IL-5 but Not IL-4 or IL-2 Production from Th2 Cells.
To examine which Th2 cytokine(s) IL-33 can induce, activated 3xTh2 cells were stimulated with IL-33, TSLP, IL-33 plus TSLP, or P+I for 48 h. IL-33 plus TSLP induced amounts of IL-13 similar to those induced by P+I and somewhat greater than those induced by IL-33 alone (Fig. 4D). No IL-13 was induced by TSLP alone. IL-5 production was induced by IL-33 plus TSLP and to a substantially smaller degree by IL-33. Neither IL-4 nor IL-2 was induced by IL-33 or IL-33 plus TSLP, although both were induced by P+I. Similar induction of IL-13 and IL-5 by Th2 cells was observed in response to IL-33 plus IL-2 or IL-33 plus IL-7, but no IL-4 or IL-2 was observed (Fig. S2).
Il13 and Il5 mRNA were induced by IL-33 or IL-33 plus TSLP at ≈30–70% of the levels induced by P+I (Fig. 4E). The kinetics of induction by P+I and by IL-33 were similar. Il4 and Il2 mRNAs were not induced in response to IL-33 plus TSLP or to IL-33 alone.
Cytokine production was also assessed by intracellular staining (Fig. 4F). P+I induced ≈85% of 3xTh2 to produce IL-13 and ≈50% to produce IL-4; IL-33 plus IL-2 induced 25–30% of the cells to produce IL-13, but none produced IL-4 (Fig. 4F). We conclude that without TCR stimulation, IL-33 plus IL-2 can induce Th2 cells to produce substantial amount of IL-13 but not IL-4.
IL-33/STAT5-Induced IL-13 Production Requires NF-κB and p38 but Not Nuclear Factor of Activated T Cells.
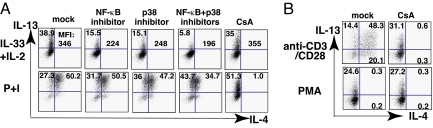
IL-33 has been reported to trigger the IL-1 signaling pathway, leading to activation of NF-κB, and the ERK, p38 and JNK MAPKs (1). To test whether IL-13 production was mediated through such pathways, 3xTh2 cells were pretreated for 30 min with an NF-κB inhibitor, Bay11–7082, or with MAPK inhibitors, and then cultured for 4 h in IL-33 plus IL-2. IL-13 production was decreased by pretreatment with the NF-κB inhibitor or the p38 inhibitor, SB203580, but not by pretreatment with inhibitors of ERK or JNK signaling, although these MAP kinases were activated in IL-33-stimulated Th2 cells (Fig. S3). The NF-κB inhibitor caused ≈70% inhibition, taking into account the diminution in the frequency of IL-13-producing cells and mean fluorescence intensity (MFI) of those cells (Fig. 5A). The p38 inhibitor caused a similar degree of inhibition (≈70%). When the p38 inhibitor and the NF-κB inhibitor were used together, the degree of inhibition was ≈90% (Fig. 5A).
Fig. 5.
IL-33 plus IL-2-induced IL-13 production is NF-κB and p38 dependent but NFAT independent. (A) 3xTh2 cells were pretreated with the indicated inhibitors for 30 min and then stimulated with IL-33 plus IL-2 or P+I for 4 h to test IL-4 and IL-13 production. (B) 3xTh2 cells were pretreated with DMSO (mock) or CsA for 30 min and then stimulated with PMA or PB-anti-CD3/CD28 for 6 h to determine IL-4 and IL-13 production. The experiments were performed 3 times.
Nuclear factor of activated T cells (NFAT), activated by TCR through calcineurin, regulates transcription of multiple cytokine genes, including Il2, Il4, and Il13 (14). Cyclosporin A (CsA), an inhibitor of calcineurin-mediated activation of NFAT, failed to block IL-33 plus IL-2-induced IL-13 production by 3xTh2 cells (Fig. 5A). By contrast, CsA completely abolished IL-4 production induced by P+I or by anti-CD3/CD28 (Fig. 5 A and B). CsA only partially inhibited IL-13 production in response to P+I or anti-CD3/CD28 (Fig. 5 A and B). Consistent with this, PMA alone induced IL-13 but not IL-4 production; PMA-induced IL-13 production was CsA resistant (Fig. 5B).
IL-33 combined with IL-2, IL-7, or TSLP also led to cell proliferation. Rested Th2 cells that were cultured in the medium containing IL-33 combined with IL-2, IL-7, or TSLP showed CFSE dilution indicating several cell divisions (Fig. S4). By contrast, cells that were cultured without any cytokine, IL-33 alone or STAT5 activators alone, showed no or only a limited number of cell divisions.
Upregulation of Receptors for IL-1 Family Members and Master Regulators in Th1 and Th17 Cells.
The capacity of IL-33 plus a STAT5 activator to regulate expression of GATA3 and T1ST2 led us to ask whether the master regulators and IL-1R family members expressed by Th1 and Th17 cells were similarly regulated.
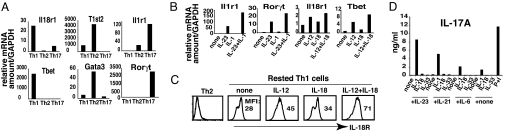
Selective expression of receptors for an IL-1 family member was shown in CD4 T cells primed under Th1, Th2, or Th17 conditions for 4 days. IL-18Rα showed heightened expression on Th1 cells, IL-33R on Th2 cells, and IL-1R1 on Th17 cells (Fig. 6A). As a control, we show the selective expression of the master regulator genes for the 3 cell types, Tbet, Gata3, and Rorc.
Fig. 6.
IL-1, together with IL-23, IL-21, or IL-6 induces IL-17A production. (A) 1xTh1, 1xTh2, and 1xTh17 cells were collected after 4-day priming and mRNA for Tbet, Gata3, Rorc, Il18r1, T1st2, and Il1r1 measured. (B) Rested 1xTh17 and 1xTh1 cells were cultured in cRPMI with indicated cytokines for 24 h and then collected to test expression of indicated mRNAs. (C) Rested 1xTh1 cells were cultured in cRPMI with the indicated cytokines for 24 h and then stained for IL-18Rα expression. (D) 1xTh17 cells were cultured in cRPMI for 2 days and then stimulated with the indicated combination of cytokines for 48 h. Supernatants were collected and ELISA was performed to test IL-17A production. The experiments were performed 3 times.
Because T1ST2 and GATA3 are regulated by IL-33 and STAT5 activators in Th2 cells, we asked whether expression of IL-18Rα and T-bet in Th1 cells and of IL-1R1 and RORγt in Th17 cells are similarly regulated by the appropriate IL-1 family member and the STAT known to be important in commitment to their lineage. Th1 cells cultured in IL-18 plus IL-12 showed increased Il18r and Tbet mRNA levels (Fig. 6B) and elevated IL-18R1 expression (Fig. 6C). Markedly elevated Il1r1 and Rorc mRNA levels are found in Th17 cells cultured in IL-23 plus IL-1 (Fig. 6B). IL-12 activates STAT4, which is important for Th1 differentiation (15). IL-23 activates STAT3, a critical factor for Th17 differentiation (11–13). Thus, the IL-1 receptor family members expressed in Th1 and Th17 cells appear to be regulated in a way parallel to the regulation of T1ST2 by GATA3 and STAT5 in Th2 cells.
IL-1 and STAT3 Activators Cause TCR-Independent IL-17A Production.
It would be interesting to establish whether an IL-1 receptor family member and a STAT activator collaborate in cytokine-dependent cytokine production in all 3 Th lineages. IL-1 and STAT3 activators (IL-6, IL-21, and IL-23) have been reported to be important for Th17 development and maintenance (11–13, 16). On the basis of these reports, we tested Th17 cells for IL-17 production in response to stimulation with IL-1 and a STAT3 activator, independent of TCR engagement. One-round primed Th17 (1xTh17) cells were cultured in cRPMI for 2 days and then stimulated with IL-1 family cytokines alone or with IL-23, IL-21, or IL-6 (Fig. 6D). None of the STAT3 activators alone caused detectable IL-17A production; IL-1β alone induced IL-17A but the combination of IL-1β with IL-23, IL-21, or IL-6 caused considerably greater production. Such IL-17A production is only slightly inhibited by CsA (Fig. S5). Neither IL-18 nor IL-33 alone or in combination with the STAT3 activators had any effect on IL-17A production.
Discussion
IL-1 family cytokines play critical roles in immune regulation and inflammation (17, 18). IL-33, the most recently identified member of the IL-1 family, has been shown to be involved in allergic and parasitic inflammatory responses. IL-33 binding to T1ST2, together with IL-1RAcP, leads to activation of NF-κB, and the ERK, p38, and JNK MAPKs, causing enhanced TCR-stimulated IL-13 and IL-5, but not IL-4, production from Th2 cells (1). Here, we showed that activated Th2 cells produce IL-13 in response to IL-33 independent of TCR stimulation but only in cells expressing high levels of T1ST2.
Naïve CD4 and Th1 cells do not express T1ST2. As cells are repetitively stimulated under Th2 conditions, the proportion of the activated cells that express high levels of T1ST2 rises. When Th2 cells were allowed to “rest” for several days in medium containing IL-2, IL-7, or TSLP, T1ST2 expression declines strikingly. Culture of resting Th2 cells in medium containing IL-33 combined with IL-2, IL-7, or TSLP, but not IL-33 alone, causes upregulation of T1ST2 expression within 24 h.
We demonstrated that T1ST2 expression is dependent on GATA3, the critical Th2 transcription factor. Th2 cells from which the Gata3 gene was deleted showed a severe impairment in both TCR-induced and IL-33 plus STAT5-induced T1ST2 reexpression. Introduction of retroviral GATA3 during the first round of Th2 priming strikingly increases the proportion of T1ST2+ cells. ChIP studies showed direct binding of GATA3 to the −12K enhancer in the T1st2 locus in recently primed Th2 cells and in IL-33 plus IL-2 cultured Th2 cells. These lines of evidence indicate that GATA3 plays an important role in T1ST2 expression.
The importance of STAT5 in GATA3 reexpression in resting Th2 cells is based on experiments showing that in established Th2 cells from which STAT5 was deleted, IL-2 plus IL-33 failed to induce GATA3 upregulation and the subsequent enhancement of T1ST2 expression was abolished. Furthermore, ChIP analysis revealed direct binding of STAT5 to the Gata3 promoter. Thus, GATA3 is upregulated through the concerted action of IL-33 and IL-2. IL-7 or TSLP, which activate NF-κB and STAT5, respectively, and GATA3 in turn, directly regulates T1ST2 expression. STAT5 also binds to the T1st2 locus, consistent with its cooperation with GATA3 in T1ST2 upregulation. Interestingly, while IL-33 and a STAT5 activator regulate sensitivity to IL-33, IL-33 enhances STAT5 phosphorylation in response to IL-2, indicating a reciprocal regulation of receptor expression and activity.
Not only is the expression of T1ST2 in Th2 cells regulated by GATA3, an association of Tbet expression with Il18r1 levels in Th1 cells and of Rorc expression with Il1r1 mRNA levels in Th17 cells was observed. It had been reported that IL-18Rα is upregulated in established Th1 cells by TCR stimulation and IL-12, a STAT4 activator (19), which might question a role for T-bet in regulation of IL-18Rα expression. However, TCR/IL-12-dependent upregulation of IL-18Rα does not occur in Th1 cells prepared from IFNγ-deficient CD4 T cells and is restored by addition of exogenous IFNγ (20). Because IFNγ activates T-bet, these reports are consistent with a role for T-bet in Th1-cell IL-18Rα induction. Further, we showed that T-bet and IL-18Rα expression in Th1 cells are regulated by the joint action of IL-18 and IL-12, parallel to the upregulation of GATA3 and T1ST2 by IL-33 and IL-2, IL-7, or TSLP in Th2 cells. A similar regulation of Rorc and Il1r1 mRNA levels was observed in Th17 cells. An increase in Rorc expression is associated with elevation of Il1r1 expression in rested Th17 cells, as described above, and such elevation is observed in cells cultured in both IL-1β and ΙL-23. Consistent with our results, it has recently been reported that IL-1R is selectively expressed on Th17 cells and Th17 cells produced IL-17A in response to IL-1 and IL-23 (16).
In T1ST2-expressing Th2 cells, IL-33 alone or IL-33 plus a STAT5 activator induce IL-13 and IL-5 production within 4 h but neither IL-4 nor IL-2 is produced. IL-13 production in response to IL-33 plus a STAT5 activator is CsA resistant but NF-κB dependent; when stimulated through TCR or P+I, IL-13 production is only partly inhibited by CsA. Further, PMA alone induces IL-13 production, which is completely CsA resistant. By contrast, TCR- or P+I-stimulated IL-4 production is completely calcineurin/NFAT dependent and PMA alone fails to induce IL-4. Thus, IL-4 production appears totally dependent on the calcium/calmodulin/calcineurin/NFAT pathway whereas IL-13 production is stimulated through this pathway but also can respond to NF-κB. Indeed, 2 putative NF-κB binding sites have been reported in the Il13 5′ flanking region (21). It has also been reported that the affinity of NF-κB for the murine Il4 promoter is too weak to be detected (22). In keeping with the requirements for cytokine-stimulated induction of IL-13 and IL-17 in Th2 and Th17 cells, respectively, in Th1 cells, IL-18 plus IL-12-induced IFNγ production is NF-κB and p38 dependent and NFAT independent (10, 23, 24).
As is well known, IL-4 and IL-13 are closely related Th2 cytokines encoded by adjacent genes on the same chromosome, ≈12 kb apart. Among the type I cytokines, they are most similar to one another in amino acid sequence. They have a similar structure, share a receptor (the type II IL-4 receptor), and have a common ability to activate STAT6. However, they have distinct physiologic functions. IL-4 plays a critical role in Th2 differentiation and immunoglobulin class switching (25) whereas IL-13 is a major effector of allergic inflammation (26). Anti-IL-13, but not anti-IL-4, inhibits elicitation of airway hypersensitivity responses in mice whereas anti-IL-4 can block induction of hypersensitivity (27, 28). IL-13, but not IL-4, is essential for expulsion of parasites in Nocardia brasiliensis infection (29, 30). Consistent with the above, serum IL-13 concentrations during allergic inflammatory responses are measurable, while IL-4 is barely detectable. The cytokine-mediated, NF-κB-dependent pathway may provide a partial explanation for this large amount of IL-13 in vivo.
What is the physiological significance of cytokine-induced effector cytokine production? Cytokine-induced cytokine production provides a means through which differentiated Th cells can be stimulated to produce their effector cytokines in response to innate stimuli that cause the production of IL-1, IL-18, or IL-33 and of activators of STAT3, STAT4, or STAT5. For each of the involved STATs, a non-T cell-dependent activator exists. For STAT3, it is IL-6, or IL-23, for STAT4, IL-12, and for STAT5, IL-7 or TSLP. Thus, innate stimuli can act on non-T cells inducing them to produce cytokines that, in turn, mobilize potent effector cytokine production by memory CD4 T cells. Indeed, TSLP has been reported to cause Th2 cytokine production in allergic inflammatory skin reactions (31). Thus, in acute or chronic inflammatory settings in which there is heightened and/or dysregulated production of an IL-1 family member and of a STAT3, STAT4, or STAT5 producer, an opportunity for production of key effector cytokines exists from differentiated CD4 T cells independent of the presence of the cognate antigens of those T cells and thus for inducing or propagating the inflammatory state.
Materials and Methods
Mice and Cell Culture.
Taconic supplied the 5C.C7 transgenic Rag2−/−, C57BL/6, and BALB/c mice. Mice homozygous for the floxed Gata3 gene (32) or for floxed Stat5a and Stat5b genes (33) were previously described. CD4 T cells and antigen-presenting cell (APC) purification and Th1, Th2, and Th17 cell priming are described in SI Text.
Flow Cytometry.
To stain phospho-STAT5, cells were fixed with 4% paraformaldehyde and permeabilized with 90% ice-cold methanol at −20 °C overnight. To stain GATA3, cells were fixed with the fixation and permeabilization kit from eBioscience. Fluorescein-anti-IL-18Rα was from R&D, FITC-anti-T1ST2 from MD Bioscience, PE-streptavidin, from Jackson ImmunoResearch, biotin-anti-NGFR from Alexis, and 647-anti-GATA3 and PE-anti-Phospho-STAT5 from BD-PharMingen. Other cell surface marker and intracellular staining are described in SI Materials and Methods.
Retroviral Infection.
GFP-Cre, NGFR, and Gata3-NGFR retroviral supernatants were prepared as described previously (34). Th2 cells retroviral infection was performed as in SI Materials and Methods.
ELISA.
ELISA was performed according to the ELISA kit instruction. IL-4, IL-13, IL-5, IL-2, IL-17A ELISA kits were purchased from eBioscience.
Quantitative PCR (QPCR).
See SI Materials and Methods for details.
ChIP.
ChIP assay was performed as previous described (35). For details see SI Materials and Methods.
Supplementary Material
Acknowledgments.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID). We thank John O'Shea and Arian Laurence for providing us with Stat5a/Stat5b fl/fl mice prepared in Lothar Hennighausen's laboratory in the National Institute of Diabetes and Digestive and Kidney Diseases, Sarah Tanksley (Laboratory of Immunology, NIAID) for cell sorting, and Shirley Starnes for editorial assistance.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906988106/DCSupplemental.
References
- 1.Schmitz J, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 2.Lohning M, et al. T1/ST2 is preferentially expressed on murine Th2 cells, independent of interleukin 4, interleukin 5, and interleukin 10, and important for Th2 effector function. Proc Natl Acad Sci USA. 1998;95:6930–6935. doi: 10.1073/pnas.95.12.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moritz DR, Rodewald HR, Gheyselinck J, Klemenz R. The IL-1 receptor-related T1 antigen is expressed on immature and mature mast cells and on fetal blood mast cell progenitors. J Immunol. 1998;161:4866–4874. [PubMed] [Google Scholar]
- 4.Xu D, et al. Selective expression of a stable cell surface molecule on type 2 but not type 1 helper T cells. J Exp Med. 1998;187:787–794. doi: 10.1084/jem.187.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ali S, et al. IL-1 receptor accessory protein is essential for IL-33-induced activation of T lymphocytes and mast cells. Proc Natl Acad Sci USA. 2007;104:8660–18665. doi: 10.1073/pnas.0705939104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chackerian AA, et al. IL-1 receptor accessory protein and ST2 comprise the IL-33 receptor complex. J Immunol. 2007;179:2551–2555. doi: 10.4049/jimmunol.179.4.2551. [DOI] [PubMed] [Google Scholar]
- 7.Coyle AJ, et al. Crucial role of the interleukin 1 receptor family member T1/ST2 in T helper cell type 2-mediated lung mucosal immune responses. J Exp Med. 1999;190:895–902. doi: 10.1084/jem.190.7.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Townsend MJ, Fallon PG, Matthews DJ, Jolin HE, McKenzie AN. T1/ST2-deficient mice demonstrate the importance of T1/ST2 in developing primary T helper cell type 2 responses. J Exp Med. 2000;191:1069–1076. doi: 10.1084/jem.191.6.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humphreys NE, Xu D, Hepworth MR, Liew FY, Grencis RK. IL-33, a potent inducer of adaptive immunity to intestinal nematodes. J Immunol. 2008;180:2443–2449. doi: 10.4049/jimmunol.180.4.2443. [DOI] [PubMed] [Google Scholar]
- 10.Robinson D, et al. IGIF does not drive Th1 development but synergizes with IL-12 for interferon-gamma production and activates IRAK and NFkappaB. Immunity. 1997;7:571–581. doi: 10.1016/s1074-7613(00)80378-7. [DOI] [PubMed] [Google Scholar]
- 11.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 12.Dong C. TH17 cells in development: An updated view of their molecular identity and genetic programming. Nat Rev Immunol. 2008;8:337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- 13.Thomas K, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 14.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: Regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 15.Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu Rev Immunol. 2003;21:713–758. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- 16.Chung Y, et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009;30:576–587. doi: 10.1016/j.immuni.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev. 2008;223:20–38. doi: 10.1111/j.1600-065X.2008.00624.x. [DOI] [PubMed] [Google Scholar]
- 18.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 19.Yoshimoto T, et al. IL-12 upregulates IL-18 receptor expression on T cells, Th1 cells, and B cells: Synergism with IL-18 for IFN-gamma production. J Immunol. 1998;161:3400–3407. [PubMed] [Google Scholar]
- 20.Smeltz RB, Chen J, Hu-Li J, Shevach EM. Regulation of interleukin (IL)-18 receptor alpha chain expression on CD4(+) T cells during T helper (Th)1/Th2 differentiation. Critical downregulatory role of IL-4. J Exp Med. 2001;194:143–153. doi: 10.1084/jem.194.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das J, et al. A critical role for NF-kappa B in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nat Immunol. 2001;2:45–50. doi: 10.1038/83158. [DOI] [PubMed] [Google Scholar]
- 22.Casolaro V, et al. Inhibition of NF-AT-dependent transcription by NF-kappa B: Implications for differential gene expression in T helper cell subsets. Proc Natl Acad Sci USA. 1995;92:11623–11627. doi: 10.1073/pnas.92.25.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J, Murphy TL, Ouyang W, Murphy KM. Induction of interferon-gamma production in Th1 CD4+ T cells: Evidence for two distinct pathways for promoter activation. Eur J Immunol. 1999;29:548–555. doi: 10.1002/(SICI)1521-4141(199902)29:02<548::AID-IMMU548>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 24.Berenson LS, Yang J, Sleckman BP, Murphy TL, Murphy KM. Selective requirement of p38alpha MAPK in cytokine-dependent, but not antigen receptor-dependent, Th1 responses. J Immunol. 2006;176:4616–4621. doi: 10.4049/jimmunol.176.8.4616. [DOI] [PubMed] [Google Scholar]
- 25.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: Signaling mechanisms and biologic functions. Annu Rev Immunol. 1999;17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 26.Wynn TA. IL-13 effector functions. Annu Rev Immunol. 2003;21:425–456. doi: 10.1146/annurev.immunol.21.120601.141142. [DOI] [PubMed] [Google Scholar]
- 27.Grunig G, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–2263. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wills-Karp M, et al. Interleukin-13: Central mediator of allergic asthma. Science. 1998;282:2258–2261. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 29.Ramalingam TR, et al. Unique functions of the type II interleukin 4 receptor identified in mice lacking the interleukin 13 receptor alpha1 chain. Nat Immunol. 2008;9:25–33. doi: 10.1038/ni1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenzie GJ, Bancroft A, Grencis RK, McKenzie AN. A distinct role for interleukin-13 in Th2-cell-mediated immune responses. Curr Biol. 1998;8:339–342. doi: 10.1016/s0960-9822(98)70134-4. [DOI] [PubMed] [Google Scholar]
- 31.He R, et al. TSLP acts on infiltrating effector T cells to drive allergic skin inflammation. Proc Natl Acad Sci USA. 2008;105:11875–11880. doi: 10.1073/pnas.0801532105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu J, et al. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- 33.Cui Y, et al. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu J, Guo L, Watson CJ, Hu-Li J, Paul WE. Stat6 is necessary and sufficient for IL-4's role in Th2 differentiation and cell expansion. J Immunol. 2001;166:7276–7281. doi: 10.4049/jimmunol.166.12.7276. [DOI] [PubMed] [Google Scholar]
- 35.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.