Abstract
Single-molecule techniques are developed to examine mechanistic features of individual E. coli replisomes during synthesis of long DNA molecules. We find that single replisomes exhibit constant rates of fork movement, but the rates of different replisomes vary over a surprisingly wide range. Interestingly, lagging strand synthesis decreases the rate of the leading strand, suggesting that lagging strand operations exert a drag on replication fork progression. The opposite is true for processivity. The lagging strand significantly increases the processivity of the replisome, possibly reflecting the increased grip to DNA provided by 2 DNA polymerases anchored to sliding clamps on both the leading and lagging strands.
Keywords: polymerase, clamp loader, replicase, sliding clamp, helicase
Replisome machines contain a helicase for DNA unwinding, 2 polymerases for replication of both strands of DNA, and a primase for initiation of lagging strand Okazaki fragments (1–3). Replisomes also contain ring shaped sliding clamp proteins that tether both polymerases to DNA for high processivity [reviewed in ref. 2]. Sliding clamp proteins require a clamp loader machine that couples ATP hydrolysis to open and close clamps onto primed sites. These several components are proposed to function together in a highly coordinated fashion during leading and lagging strand replication (1, 2, 4, 5).
The antiparallel structure of DNA requires that 1 strand (the lagging strand) is synthesized as fragments that are extended in the opposite direction of the continuous leading strand. This opposite directionality is resolved by formation of a DNA loop during extension of each Okazaki fragment [i.e., the “trombone model” of replication (5)]. Furthermore, each Okazaki fragment requires an RNA primer and assembly of a sliding clamp to initiate chain extension. The repeated initiation events and DNA looping during lagging strand replication may influence the rate and processivity of the replisome.
Replisome machines are highly processive entities that synthesize extremely long DNA molecules, making their rate and processivity quite difficult to study by ensemble methods because of the low resolving limit of most gel electrophoretic techniques. Thus, ensemble rate studies can only capture the first 10–20 s of a rapidly moving replisome, and establish a lower limit for processivity. Furthermore, individual replisomes may move forward in an uneven fashion, and this behavior will be masked in an ensemble analysis, which quickly loses synchronicity. Single-molecule studies circumvent these limitations by real-time observations of individual replication forks over the entire distance of a highly processive binding event. Long DNA products are imaged in the microscope for direct measurements of DNA length.
A further advantage to the single-molecule approach is the ability to apply a constant flow of buffer during replication. The use of a flow during replication enables a rigorous test of replisome processivity, as proteins that dissociate from the replisome will be quickly carried away from the reaction chamber, preventing them from reassociating with the DNA.
The current report examines the rate and processivity of the E. coli replisome. The E. coli replisome consists of the ring shaped hexameric DnaB helicase, DnaG primase, the DNA polymerase III* replicase (Pol III*), and β sliding clamps. Processivity estimates of the E. coli replisome from ensemble studies indicate a lower limit of 50 kb, and an average bulk rate of ≈500–700 nt/s at 37 °C (6, 7). Processivity measurements from other recent single-molecule studies differ over a wide range, from 3 kb to >80 kb (8, 9). In each of these previous studies replication was performed in the presence of excess proteins that could replace a replisome protein that dissociates from DNA, and thus the true processivity of the replisome has not been examined.
DnaB helicase, Pol III*, and the β clamp are tightly associated replisome components, while primase is loosely attached and comes on and off the replisome during synthesis (i.e., it acts distributively) (10). The Pol III* assembly contains 2–3 Pol III cores attached to 1 clamp loader apparatus (7). Two Pol III cores within Pol III* are tethered to the leading and lagging strands by β clamps, suggesting that Pol III* tightly adheres to the replication fork for high processivity. However, recent studies show that translesion DNA polymerases (i.e., Pol II and Pol IV) efficiently take over β clamps from Pol III* in a moving replisome, implying that Pol III* may dissociate and reassociate with β during replication (11). Indeed, the lagging strand Pol III core within Pol III* rapidly dissociates from β upon finishing an Okazaki fragment, and it repeatedly cycles to new β clamps on upstream RNA primers (12, 13). These findings bring into question the stability of Pol III* within the moving replisome.
The current study develops single-molecule techniques to visualize E. coli replisomes in real-time, allowing both their rate and processivity to be determined with precision. The results show that DnaB and Pol III*-β are indeed highly processive, and replicate an average of 86.5 kb of DNA in 1 binding event; some replisomes are processive for up to 300 kb. Pol III* appears to release from the fork before DnaB helicase because reactions that contain an excess of Pol III* result in continued extension of DNA to an average length of 140 kb. We also develop methods to study replisomes that only perform leading strand synthesis, and compare the results to replisomes that replicate both leading and lagging strands. Interestingly, we find that lagging strand synthesis has opposite effects on the rate and processivity of the replisome. However, lagging strand synthesis reduces the rate of fork progression by ≈23%, which may reflect priming or the strain of DNA looping during Okazaki fragment synthesis. Although, lagging strand replication enhances the processivity of the replisome by 61%. We propose that either primase or 2 DNA polymerases attached to sliding clamps on both the leading and lagging strands, stabilize the replisome, and enhance its processivity relative to a replisome that operates only on the leading strand.
Results and Discussion
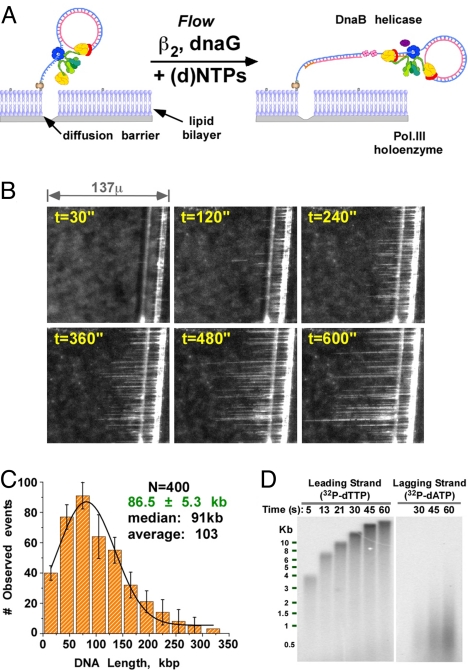
E. coli replisomes are highly processive and the leading and lagging strand polymerases are tightly coupled. The E. coli replisome was assembled onto a 5′ biotinylated 100mer minicircle substrate using DnaB, Pol III*, and β, and then immobilized on a lipid bilayer through the 5′ biotinylated minicircle tail. Upon including dNTPs and rNTPs in the buffer flow, the replisome advances around the circle multiple times (i.e., rolling circle replication). The leading strand product is a continuous single-strand (ss) DNA “tail” that becomes the template for lagging strand synthesis, forming a long double-strand (ds) DNA (see the scheme in Fig. 1A). The force of a hydrodynamic flow pushes the DNA-lipid complex to a diffusion barrier etched in the glass surface, concentrating at a defined position within the flow cell numerous DNA molecules that can be examined simultaneously in 1 visual field (14). Real-time rolling circle replication is detected using a fluorescent intercalating dye (Yo-Pro-1).
Fig. 1.
Single-molecule rolling circle replication on a lipid bilayer produces DNA curtains at a diffusion barrier. (A) Scheme of single-molecule rolling circle replication on a lipid bilayer. During leading strand synthesis (blue), DNA is displaced from the 100mer circle as the replisome ‘rolls’ around the template. The newly synthesized 5′ ssDNA “tail” is converted to dsDNA by lagging-strand synthesis (red). (B) Growth of a rolling circle DNA curtain in real-time as visualized by Yo-Pro-1 fluorescence. (C) Histogram of observed DNA lengths and 1-Gaussian fitting analysis (green). (D) Alkaline gel analysis of ensemble reaction leading and lagging strand products of minicircle replication.
A distinctive feature of the assay design is the use of a constant flow of buffer to deliver dNTPs and other reagents to the immobilized replisome. We leveraged this feature to determine the processivity of individual replisomes by omitting Pol III* and DnaB helicase from the buffer flow. Thus, when either Pol III* or DnaB dissociate from the replication fork, they cannot rebind DNA because they will be carried away in the flow, and growth of the DNA molecule will come to an abrupt halt. We used a flow of 100 μL/min, which exerts a force of only 1.45 pN (Fig. S1). Primase and the β clamp were included in the buffer flow because a new molecule of primase and β are needed to initiate each Okazaki fragment (10, 12).
A growing curtain of DNA molecules is observed at the diffusion barrier after replication is initiated (Fig. 1B and Movie S1). Most DNA molecules stop growing within 20 min, indicating loss of either DnaB or Pol III* (or both) from the DNA. The length of individual DNA molecules varies over a considerable range; some are <30 kb while others are >300 kb. The length distribution of 400 individual molecules, fit to a single Gaussian function, yields an average length of 86.5 ± 5.3 kb (Fig. 1C). DNA molecules that grow outside the visual field show the same length distribution, and therefore exposure to the laser does not affect the results. Ensemble experiments indicate that the dye used to stain DNA does not influence minicircle replication using similar conditions as the single molecule experiments (Fig. S2).
Formation of long leading/lagging strand duplex DNA products, within a flow cell that continuously removes dissociated proteins, provides a rigorous test that the helicase, leading/lagging strand DNA polymerases and clamp loader stay associated at the moving replication fork. These studies therefore strongly support the DNA looping “trombone” model of replication. Although this hypothesis is widely accepted, it has been difficult to demonstrate unequivocally that the lagging strand DNA polymerase truly remains bound to the replisome during replication. In particular, the lagging strand polymerase must cycle on and off DNA during synthesis of multiple Okazaki fragments. Elegant electron microscopy studies demonstrate DNA loops in the T4 and T7 systems, but similar studies have not been performed in the E. coli system (15–18). The current study demonstrates that the lagging strand polymerase remains continuously associated with the moving replisome in the E. coli system, implying that DNA loops are formed during extension of each Okazaki fragment.
To estimate the number of Okazaki fragments, and thus number of DNA loops produced by the replisome during synthesis of an 86 kb duplex, we determined the frequency of Okazaki fragment synthesis in an ensemble assay under conditions similar to the single-molecule assays (Fig. 1D). The synthetic minicircle contains no dT residues on the inner circle, which allows the lagging strand to be uniquely labeled using α32P-dATP; the leading strand can be labeled using α32P-dTTP. The alkaline agarose gel analysis gives an average Okazaki fragment length of 0.65 kb. Therefore, the lagging strand polymerase remains continuously attached to the replisome during production of approximately 130 Okazaki fragments to form an 86 kb duplex. Because the polymerase remains attached to the replisome, yet comes on and off DNA for each fragment, ≈130 DNA loops must be formed during an average processive binding event. It should be noted that Okazaki fragment size is dictated by fork speed and primase concentration, and is typically 1–2 kb under conditions of this report when measured at 37 °C (10). The shorter length observed here likely results from the slower rate of fork progression at the 23 °C temperature used in this report.
Pol III* Often Dissociates from the Replisome Before DnaB.
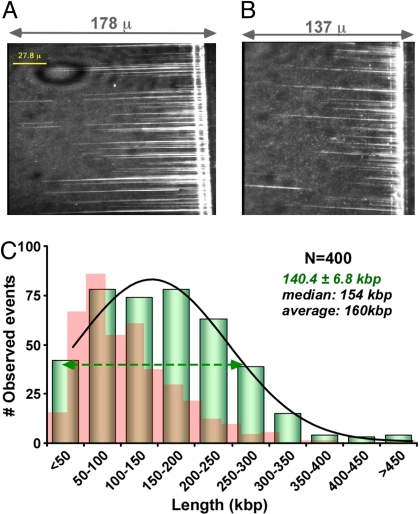
The replisome will be brought to a sudden halt upon dissociation of either Pol III* or DnaB helicase. To determine the processivity of DnaB helicase, we included Pol III* in the buffer flow, along with β and primase. Thus, if Pol III* dissociates from the replisome, it should quickly be replaced by Pol III* in the buffer flow, and fork propagation should continue. Comparison of DNA curtains produced when Pol III* is present or absent from the buffer flow shows distinctly longer DNA molecules when Pol III* is present (compare Fig. 2 A and B). Analysis of 400 molecules, fit to a single Gaussian function, yields an average length of 140.4 ± 6.8 kb (Fig. 2C). This length of DNA is 62% longer than observed when Pol III* is omitted from the flow (compare red and green histograms in Fig. 2C). A simple explanation for this result is that Pol III* often dissociates from the replisome before DnaB falls off DNA, and when another Pol III* is available to associate with DnaB, it reforms the replisome and the replication fork continues. This observation underscores the importance of continuously removing dissociated proteins from a reaction to make accurate measurements of replisome processivity.
Fig. 2.
DNA length is enhanced by excess Pol III* in the buffer flow. DNA curtains produced when (A) Pol III* is present in the flow, (B) Pol III* is absent from the flow during replication. (C) Histograms of product length when Pol III* is present in the flow (green), or absent from the buffer flow (red).
Individual Replisomes Exhibit Different Intrinsic Rates of Fork Movement.
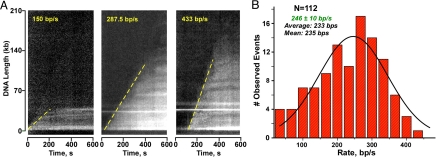
The replisome may move in a continuous uniform motion, or it may progress at an uneven rate because of periodic pausing. Real-time observations of individual replisomes show a uniform rate of fork movement (Fig. 3). However, examination of single molecules reveals a remarkable finding. Although a given replisome moves at a uniform rate, individual replisomes show surprisingly different rates that vary as much as 5-fold or more, as illustrated by the kymographs in Fig. 3A. The fact that individual replisomes move at different rates may explain why ensemble reactions of growing leading strand products give a broad smear in an alkaline gel (e.g., Fig. 1D). One possible explanation for the different rates of replisome movement is that a given replisome may develop a unique Okazaki fragment cycle (e.g., Okazaki fragment size) that is set stochastically by the first few fragments and may result in different rates of fork movement. Other possible explanations are discussed later in this report.
Fig. 3.
Individual replisomes move at different rates. (A) Representative kymographs of DNA molecules from real-time observation of DNA synthesis. The endpoint trajectories (DNA lengths) plotted versus time defines a curve with a slope that represents the rate of DNA synthesis. (B) Histogram of the rates of individual replisomes.
Analysis of 112 replisomes, fit to a single Gaussian function, yields a rate of 246 ± 10 nt/s (Fig. 3B). The assays of this report are performed at 23 °C. E. coli replisomes move ≈2-fold faster at 37 °C (6, 7, 9). We would like to note that the level of resolution of the EM-CCD camera is approximately 760 bp/pixel; therefore, brief pausing events that may occur during replisome translocation would not be detected under our present working conditions.
The Lagging Strand Enhances Replisome Processivity.
Lagging strand specific operations, such as priming or DNA extension in the opposite direction from fork progression, may exert influence the rate and/or processivity of the replisome [reviewed in ref. 19]. Indeed, a single-molecule study of T7 replication demonstrates that primase action causes the replication fork to pause (20). Additionally, a single-molecule study of the E. coli replisome on a λ DNA substrate showed that the presence of primase lowers the processivity of the replisome by ≈3-fold, from 10.5 kb to 3 kb (8). The decrease in processivity appeared to be because of the presence of primase, and not actual priming activity. However, the previous study used Pol III cores that were not preattached to the clamp loader, and the χ and ψ subunits were missing in most of the experiments. One or more of these differences may explain the low overall processivity observed in that study. Indeed, the χ subunit is known to enhance the processivity of Pol III*-β (21). The χ subunit is also known, to facilitate transfer of an RNA primer from primase to the clamp loader (22). Furthermore, the ψ subunit is required to stabilize the oligomeric structure of the clamp loader upon which the architecture of the replisome is built (23–25).
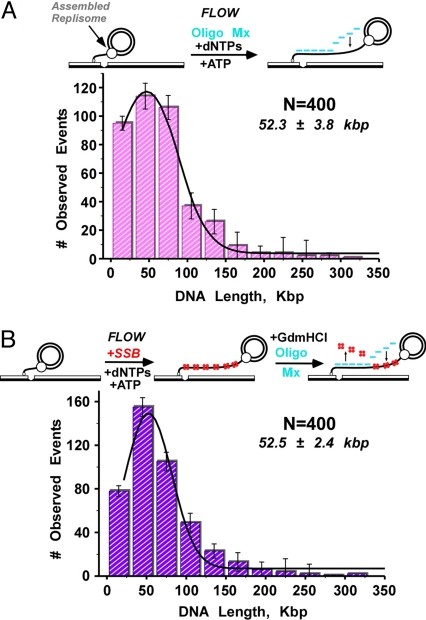
The experiments of Fig. 4 reexamine the influence of lagging strand synthesis on processivity using replisomes that are reconstituted using a fully assembled Pol III*. To study replisomes in the absence of lagging strand synthesis, we omitted primase from the buffer flow. The leading strand product of rolling circle replication is a long ssDNA composed of 100mer direct repeats of the minicircle sequence. The Yo-Pro-1 intercalating dye does not detect ssDNA, and therefore we converted the ssDNA to dsDNA by including in the flow a mixture of 400 nM each of 5 DNA 20mers that anneal end-to-end over the 100mer sequence (see scheme in Fig. 4A). This allows the observation of leading strand replication in real-time (see Movie S2). Ensemble reactions demonstrate that the oligonucleotides do not inhibit replication or slow fork movement (Fig. S3). Analysis of the final length of 400 molecules yields an average processivity, when fit to a single Gaussian function, of only 52.3 kb ± 3.8 kb (Fig. 4A). The experiments of Fig. 1 demonstrate that the leading/lagging strand replisome has a significantly greater processivity (86 kb) than observed for the leading strand replisome (52 kb). Taken together, these results indicate that lagging strand synthesis enhances replisome processivity by ≈61%.
Fig. 4.
Processivity of the leading strand replisome in the absence of lagging strand synthesis. (A) (Top) Scheme for observation of leading strand replication. Five DNA 20mers anneal to the 100mer repeat sequence of the long leading strand ssDNA product, converting it to dsDNA for visualization using Yo-Pro-1. (Bottom) Length distribution of replication products. (B) (Top) Scheme of leading strand replication in presence of E. coli SSB, followed by 4 M Guanidine-HCl to remove SSB, and then addition of the 5 DNA 20mers to convert the ssDNA to dsDNA. (Bottom) Length distribution of replication products. The Gaussian fitting analysis is shown in green.
It is possible that the leading strand replisome is less processive because of the presence of the ssDNA 20mers. Therefore, we analyzed the processivity of the leading strand replisome by a multistep method that allowed addition of the DNA 20mers after replication (see scheme in Fig. 4B). In the first step, rolling circle replication was allowed to proceed for 30 min in the absence of primase and DNA 20mers. To ensure that leading strand replication had occurred, we used fluorescently labeled SSB (SSB labeled using either Texas Red or Oregon Green 488 retains full activity, as illustrated in Fig. S4). After replication, SSB was removed from the DNA by a buffer flow containing 4 M guanidine-HCl, and SSB removal was confirmed by monitoring loss of the fluorescent signal. Then we included the 5 DNA 20mers in the buffer flow (without guanidine-HCl) to visualize the dsDNA product. Measurement of 400 molecules yields an average length of 52.5 ± 2.4 kb (Fig. 4B). This length compares well with the reactions performed in the presence of the ssDNA 20mers.
In summary, the experiments of Fig. 4 demonstrate that lagging strand synthesis enhances processivity compared with a replisome that extends only the leading strand. Primase is not an integral part of the replisome, and thus does not seem a likely candidate as a stabilizing factor for increased replisome processivity, although this possibility cannot be excluded. However, a coupled leading/lagging strand replisome has more contacts to the replication fork compared with a leading strand-only replisome. Specifically, the leading strand-only replisome has only 1 Pol III core-to-β connection, whereas the coupled leading and lagging strand replisome has 2 Pol III core-to-β connections, 1 on each strand. We propose that these extra connections may underlie the higher processivity of the coupled leading/lagging replisome.
Lagging Strand Synthesis Decreases the Rate of Replisome Progression.
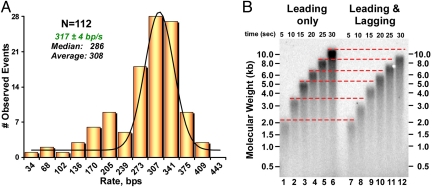
Real-time observations of the rate of 112 individual leading strand replisomes gives a rate, fit to a single Gaussian function, of 317 ± 4 nt/s (Fig. 5A). Interestingly this rate is approximately 23% faster than the leading/lagging strand replisome. It seems possible that the DNA 20mers may require some time for hybridization, and that the rate measurements could be an underestimate. Therefore, we also examined the rate of fork movement by ensemble assays. As explained earlier, a limitation of ensemble studies is the resolving limit of agarose gels, and therefore only the earliest times during a replication reaction can be examined before the DNA becomes too long. In the ensemble experiment of Fig. 5B, replication reactions were performed in the presence or absence of primase, and aliquots were quenched at 5 s, 10 s, 15 s, 20 s, 25 s, and 30 s. The result confirms that the leading strand-only replisome (321 nt/s) moves at a faster rate than the coupled leading/lagging strand replisome (278 nt/s).
Fig. 5.
The rate of leading strand replisomes. (A) Alkaline gel analysis of ensemble reaction products in which leading strand synthesis is monitored during minicircle leading-only replication (lanes 1–6) and leading/lagging replication (lanes 7–12). Red dotted lines mark the front of the migration bands in leading-only replication. (B) Histogram of the rates of individual replisomes. The Gaussian fitting analysis is shown in green.
In summary, the experiments of Fig. 5 demonstrate that lagging strand synthesis decreases the rate of replication fork progression. The effect of lagging strand synthesis on the rate of the replisome could be because of primase, or to DNA polymerase acting on the lagging strand.
It is interesting to note that the distribution of rates for individual leading strand replisomes appears more uniform than the distribution observed for individual leading/lagging strand replisomes. This result suggests that lagging strand events may underlie the wide dispersion of rates observed for the leading/lagging replisome (i.e. Fig. 3). However, there appears to be a group of leading strand replisomes that move ≈100 nt/s slower than the average (Fig. 5A). Other possibilities that may explain the persistent difference in the rate of replication fork movement among individual replisomes include: (i) loss of a nonessential but important subunit (i.e., χ, Ψ, θ, ε); (ii) the leading strand polymerase may bind a τ subunit that occupies any of 3 different positions in the clamp loader, and different positions may impart functional consequences; (iii) different replisomes may have subunits that are partially unfolded, affecting their intrinsic rate; (iv) some subunits may have undergone oxidative damage during preparation, and this damage may affect their intrinsic rate.
Single molecule studies on a variety of enzymes have noted significant differences in the intrinsic catalytic rate of otherwise identical molecules (26–30). Although the physical basis for these differences is not understood, some proposals suggest that enzymes exist in different stable conformers that have distinct catalytic rates. However, single molecule studies of enzymes that demonstrate distinct rates of catalysis are typically performed over much shorter time scales than those of the current report. Additional studies will be needed to address this fascinating issue.
Conclusions
The current study outlines single-molecule methods to analyze the processivity and rate of replisome action in real-time, both for a coupled leading/lagging strand replisome and for a replisome that only operates on the leading strand. Growth of a curtain of DNA molecules is performed in a flow of buffer that sweeps dissociated Pol III* and DnaB from the reaction, preventing their reassociation with DNA and providing firm measurements of processivity. Previous ensemble and single-molecule studies of replisome processivity contained an excess of unassociated replisome proteins, which may replace a protein that dissociates from DNA and thereby mask a true measure of processivity. Indeed, we find here that including excess Pol III* in the buffer flow results in significantly longer DNA products. We presume that when Pol III* dissociates from the replisome, another Pol III* in the flow replaces it and continues fork movement with DnaB. Interestingly, replisomes that perform only leading strand synthesis move 23% faster than replisomes that perform both leading and lagging strand synthesis, indicating that lagging strand synthesis slows the rate of fork progression. In contrast, replisome processivity is enhanced 61% by lagging strand synthesis. It seems likely that the additional connection to the replication fork, provided by a lagging strand polymerase held to DNA by the sliding clamp, lends additional stability to the replisome for enhanced processivity.
Experimental Procedures
Materials.
Pol III subunits (α, ε, θ, τ, δ, δ′, χ, ψ, β), primase, SSB, and DnaB helicase were purified as described in ref. 13. Pol III* (Pol III core3τ3δδ′χψ) was reconstituted and purified as described in ref. 7. Oligonucleotides were synthesized by IDT and gel purified. Glucose oxidase and catalase were from Sigma. Yo-Pro-1 was from Invitrogen (Molecular Probes). Photo clear silicone-based elastomer (Sylgard 184) was from Dow-Corning. Lipids were from Avanti Polar Lipids Inc.
Flow Cell Construction.
Flow cells were prepared using photo-clear silicone-based elastomer poured into a negative lithography mold; the bottom contained a 2.5 mm × 25 mm strip of 50 μm thick 3M tape (i.e., forming a channel). After solidification, a rectangular block containing the channel was cut out; entry and exit ports were punched into either end of the channel. A #1 coverslip was scratched with a diamond-tipped scribe to create diffusion barriers. The elastomer block and the coverslip were treated in a plasma oven, then welded together. A lipid bilayer was formed on the glass surface inside the flow cell using a mixture of freshly sonicated liposomes (DOPC supplemented with 8% DOPE-mPEG550 and 0.5% DOPE-biotin) in buffer L1 (10 mM Tris·HCl, pH 7.8, 100 mM NaCl) as described in ref. 14.
Total Internal Reflectance Fluorescence (TIRF) Microscopy.
Microscopy was performed using an Olympus IX70 inverted microscope fitted with a 60× TIRF objective (N.A. = 1.45), a Prior motorized stage, and a motorized shutter. Yo-Pro-1 was excited using a solid state 488-nm laser (Coherent) at 1.5 mW. Fluorescence emission was collected back through the objective and filtered for residual laser light before being captured by a 512 × 512 back thinned EM CCD camera (Hamamatsu). The motorized shutter permitted a 100-ms exposure every 1 s. Buffer was driven through the flow cell using a syringe pump (KD Scientific). Image collection and data work-up were facilitated using the Slidebook Software suite (Intelligent Imaging Inc.).
Leading and Lagging Strand Replication.
A 100mer synthetic DNA minicircle substrate containing a 40 dT 5′ tail was prepared as described in ref. 13. DnaB helicase (18.2 pmol, 365 nM) was assembled onto the minicircle DNA (655 fmol, 13.1 nM) in 50 μL Buffer A (20 mM Tris·HCl, pH 7.5, 5 mM DTT, 40 μg/mL BSA, and 4% glycerol) containing 8 mM MgOAc2 and 1.25 mM ATP, followed by incubation for 30s at 37 °C. To this solution was added a 25 μL reaction containing Pol III* (675 fmol, 27 nM), β2 (1.85 pmol, 74 nM as dimer), 60 μM each dCTP and dGTP, and 8 mM Mg(OAc)2 in Buffer A. After 6 min at 37 °C, 1 μL of the reaction was added to 1 mL Buffer B (8 mM MgOAc2, 60 μM each of dCTP and dGTP, and 50 nm Yo-Pro-1 in Buffer A). The reaction was passed through the flow cell at 500 μL/min for 30 s, then at 10 μL/min for 30 s. DNA replication was initiated upon flowing (100 μL/min) Buffer A containing 60 μM of each dNTP, 250 μM each of CTP, TTP, and UTP, 1 mM ATP, 462 nM SSB4, 300 nM primase, 50 nM β2, 50 nM Yo-Pro-1, 0.8% glucose, 0.01% β-mercaptoethanol, 0.57 U glucose oxidase, and 2.1 U catalase. Control experiments using biotinylated λ DNA show that a flow rate of 100 μL/min exerts a force of 1.45 pN and stretches dsDNA to 88% of its full contour length (Fig. S1). All DNA lengths were corrected using this value. To detect dsDNA we used 50 nM Yo-Pro-1, a single intercalating moiety that binds and dissociates from DNA much more rapidly than Yo-Yo1.
Ensemble reactions were performed similarly except that after incubation with DnaB (30 s) and Pol III* (6 min), replication was initiated upon adding SSB, primase, remaining dNTPs and rNTPs, along with either 32P-dATP or 32P-dTTP (primase was not present for leading strand only replication).
Leading Strand Replication Reactions.
Reactions were performed as described above except primase, SSB, β2, CTP, GTP, and UTP were omitted from the flow to prevent lagging strand synthesis. The leading ssDNA product was visualized by including in the buffer flow 400 nM each of 5 DNA 20mers that hybridize to the 100mer sequence. We also performed leading strand synthesis by including SSB, and omitting the DNA 20mers, primase and β2 from the flow. SSB was labeled with either Texas Red or OregonGreen488 maleimide to follow synthesis. Buffer B containing 4M guanidine-HCl was passed over the flow cell to remove SSB (confirmed visually by loss of SSB fluorescence), and then the ssDNA was converted to dsDNA by annealing the 5 DNA 20mers for visualization by Yo-Pro-1 as described above.
Supplementary Material
Acknowledgments.
We thank Eric Greene's group at Columbia University for help in microscopy set-up and advice on formation of lipid bilayers, Steven Kowalczykowski (U. C. Davis) for encouragement and advice in the microscopy set-up, Albert Libchaber (Rockefeller University) and Dr. Axel Buguin (Institut Curie) for assistance in flow cell construction, Martin Eber and Kathy Lindsay of Olympus Inc. for their expert assistance, Vadim Sherman and Joe Golja of the Rockefeller University Instrument Shop for the construction of various types of flow cell platforms, and members of the laboratory for advice and critical reading of the manuscript. This work supported by Howard Hughes Medical Institute and National Institutes of Health Grant GM38839.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906157106/DCSupplemental.
References
- 1.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 2.Johnson A, O'Donnell M. Cellular DNA replicases: Components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg A, Baker TA. DNA Replication. New York: W.H. Freeman; 1992. [Google Scholar]
- 4.McHenry CS. Chromosomal replicases as asymmetric dimers: Studies of subunit arrangement and functional consequences. Mol Microbiol. 2003;49:1157–1165. doi: 10.1046/j.1365-2958.2003.03645.x. [DOI] [PubMed] [Google Scholar]
- 5.Sinha NK, Morris CF, Alberts BM. Efficient in vitro replication of double-stranded DNA templates by a purified T4 bacteriophage replication system. J Biol Chem. 1980;255:4290–4293. [PubMed] [Google Scholar]
- 6.Mok M, Marians KJ. The Escherichia coli preprimosome and DNA B helicase can form replication forks that move at the same rate. J Biol Chem. 1987;262:16644–16654. [PubMed] [Google Scholar]
- 7.McInerney P, Johnson A, Katz F, O'Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 8.Tanner NA, et al. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998a. [DOI] [PubMed] [Google Scholar]
- 9.Tanner NA, Loparo JJ, Hamdan SM, Jergic S, Dixon NE, van Oijen AM. Real-time single-molecule observation of rolling-circle DNA replication. Nucleic Acids Res. 2009;37:e27. doi: 10.1093/nar/gkp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zechner EL, Wu CA, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. II. Frequency of primer synthesis and efficiency of primer utilization control Okazaki fragment size. J Biol Chem. 1992;267:4045–4053. [PubMed] [Google Scholar]
- 11.Indiani C, Langston LD, Yurieva O, Goodman MF, O'Donnell M. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc Natl Acad Sci USA. 2009;106:6031–6038. doi: 10.1073/pnas.0901403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stukenberg PT, Turner J, O'Donnell M. An explanation for lagging strand replication: Polymerase hopping among DNA sliding clamps. Cell. 1994;78:877–887. doi: 10.1016/s0092-8674(94)90662-9. [DOI] [PubMed] [Google Scholar]
- 13.McInerney P, O'Donnell M. Functional uncoupling of twin polymerases: Mechanism of polymerase dissociation from a lagging-strand block. J Biol Chem. 2004;279:21543–21551. doi: 10.1074/jbc.M401649200. [DOI] [PubMed] [Google Scholar]
- 14.Graneli A, Yeykal CC, Prasad TK, Greene EC. Organized arrays of individual DNA molecules tethered to supported lipid bilayers. Langmuir. 2006;22:292–299. doi: 10.1021/la051944a. [DOI] [PubMed] [Google Scholar]
- 15.Lee J, Chastain PD, 2nd, Griffith JD, Richardson CC. Lagging strand synthesis in coordinated DNA synthesis by bacteriophage t7 replication proteins. J Mol Biol. 2002;316:19–34. doi: 10.1006/jmbi.2001.5325. [DOI] [PubMed] [Google Scholar]
- 16.Lee J, Chastain PD, 2nd, Kusakabe T, Griffith JD, Richardson CC. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol cell. 1998;1:1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 17.Nossal NG, Makhov AM, Chastain PD, 2nd, Jones CE, Griffith JD. Architecture of the bacteriophage T4 replication complex revealed with nanoscale biopointers. J Biol Chem. 2007;282:1098–1108. doi: 10.1074/jbc.M606772200. [DOI] [PubMed] [Google Scholar]
- 18.Park K, Debyser Z, Tabor S, Richardson CC, Griffith JD. Formation of a DNA loop at the replication fork generated by bacteriophage T7 replication proteins. J Biol Chem. 1998;273:5260–5270. doi: 10.1074/jbc.273.9.5260. [DOI] [PubMed] [Google Scholar]
- 19.Yao NY, O'Donnell M. Replisome dynamics and use of DNA trombone loops to bypass replication blocks. Mol Biosyst. 2008;4:1075–1084. doi: 10.1039/b811097b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 21.Kelman Z, Yuzhakov A, Andjelkovic J, O'Donnell M. Devoted to the lagging strand-the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 1998;17:2436–2449. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuzhakov A, Kelman Z, O'Donnell M. Trading places on DNA–a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–163. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
- 23.Glover BP, McHenry CS. The chi psi subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J Biol Chem. 1998;273:23476–23484. doi: 10.1074/jbc.273.36.23476. [DOI] [PubMed] [Google Scholar]
- 24.Onrust R, Finkelstein J, Naktinis V, Turner J, Fang L, O'Donnell M. Assembly of a chromosomal replication machine: Two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. I. Organization of the clamp loader. J Biol Chem. 1995;270:13348–13357. doi: 10.1074/jbc.270.22.13348. [DOI] [PubMed] [Google Scholar]
- 25.Anderson SG, Williams CR, O'Donnell M, Bloom LB. A function for the psi subunit in loading the Escherichia coli DNA polymerase sliding clamp. J Biol Chem. 2007;282:7035–7045. doi: 10.1074/jbc.M610136200. [DOI] [PubMed] [Google Scholar]
- 26.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 27.Lu HP, Xun L, Xie XS. Single-molecule enzymatic dynamics. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 28.van Oijen AM, Blainey PC, Crampton DJ, Richardson CC, Ellenberger T, Xie XS. Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science. 2003;301:1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 29.Wuite GJ, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404:103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 30.Xie XS. Single-molecule approach to dispersed kinetics and dynamic disorder: Probing conformational fluctuation and enzymatic dynamics. J Chem Phys. 2002;117:11024–11032. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.