Abstract
TGF-β family members are highly pleiotropic cytokines with diverse regulatory functions. TGF-β is normally found in the latent form associated with latency-associated peptide (LAP). This latent complex can associate with latent TGFβ-binding protein (LTBP) to produce a large latent form. Latent TGF-β is also found on the surface of activated FOXP3+ regulatory T cells (Tregs), but it is unclear how it is anchored to the cell membrane. We show that GARP or LRRC32, a leucine-rich repeat molecule of unknown function, is critical for tethering TGF-β to the cell surface. We demonstrate that platelets and activated Tregs co-express latent TGF-β and GARP on their membranes. The knockdown of GARP mRNA with siRNA prevented surface latent TGF-β expression on activated Tregs and recombinant latent TGF-β1 is able to bind directly with GARP. Confocal microscopy and immunoprecipitation strongly support their interactions. The role of TGF-β on Tregs appears to have dual functions, both for Treg-mediated suppression and infectious tolerance mechanism.
Keywords: transforming growth factor beta, Tregs, latency-associated peptide
TGF-β family members (β1, β2, β3 isoforms) are highly pleiotropic cytokines that have critical functions in cell differentiation, tissue morphogenesis and modulation of cell growth, inflammation, matrix synthesis, and apoptosis. Dysregulations in TGF-β function are associated with multiple pathological conditions including tumor cell growth, fibrosis, emphysema, and autoimmunity (1). All 3 TGF-β isoforms are synthesized as homodimeric proproteins. The proproteins are cleaved in the Golgi apparatus by a furin-like convertase to produce the dimeric propeptides called latency-associated peptide (LAP) that noncovalently associates with the dimeric mature TGF-β to prevent its activity (2). There are multiple mechanisms of activating TGF-β from its latency by pathways that include protease plasmin, matrix metalloproteases, thrombospondin-1 (TSP1), and certain αV integrins (3). TGF-β can be secreted in a small latent form associated with LAP, or this complex can further associate with latent-TGF-β-binding protein (LTBP) to produce a large latent form for deposition onto the extracellular matrix. In addition, small latent TGF-β can be expressed on the membrane of many cell types, including megakaryocytes, platelets (4), immature dendritic cells (DCs) (5), and activated FOXP3+ regulatory T cells (Tregs) (6–8), and has important functions in tissue healing and immune regulation. However, it is unknown how this membrane small latent TGF-β is anchored to the cell surface.
It has been recently shown that megakaryocytes and activated Tregs expressed high levels of mRNA for a member of the leucine-rich repeat family of proteins that has been termed GARP or LRRC32 and that platelets express this molecule on their membrane (9, 10). The GARP or LRRC32 gene consists of 662 aa and encodes an 80-kDa transmembrane protein with an extracellular region composed primarily of 20 leucine-rich repeats (11, 12). As the Garp gene is expressed in multiple cell types in the mouse during embryogenesis, it has been proposed that Garp plays an important role in development, but its actual function is unknown (13). Since platelets and activated Tregs contain both GARP and latent TGF-β on their membranes, we hypothesized that GARP might bind and anchor latent TGF-β. Here, we show that GARP is critical for the surface expression of latent TGF-β by binding to the complex and functioning as its cell surface receptor.
Results
GARP or LRRC32 Is Selectively Expressed on Activated FOXP3+ Tregs.
Consistent with a previous publication (10), we found that GARP mRNA is selectively expressed in fresh human Tregs and rapidly up-regulated after activation of CD4+CD25hi Tregs with anti-CD3/CD28 and IL-2 (Fig. 1A). Only very low levels of mRNA were detected in CD4+CD25– T cells after activation for 5 days. While the addition of TGF-β1 resulted in the induction of FOXP3 mRNA in CD4+CD25– T cells (14), the level of GARP mRNA was not dramatically increased. Cell surface expression of either LAP or GARP was not significantly detected on freshly isolated Tregs (CD25hi), but the expression of both LAP and GARP was rapidly up-regulated after activation (Fig. 1B). GARP and LAP occasionally could be detected on <5% of activated CD4+CD127+CD25– T cells (Fig. S1). However, when we activated the CD4+CD25int population, which contains mostly CD45RO+FOXP3– T cells and some FOXP3+ Tregs, the vast majority of LAP and GARP could be detected on FOXP3+ Tregs. The rapid appearance and disappearance of LAP+/GARP+ FOXP3– T cells during the 36 h of activation of CD25hi and CD25int most likely represent Tregs that have down-regulated their FOXP3 and could be on their way to cell death. Platelets also co-expressed LAP and GARP on their membranes (Fig. 1C). In contrast to previous reports (5), we were unable to detect any significant surface expression of LAP or GARP on plasmacytoid or myeloid DCs from peripheral blood (Fig. 1D). Interestingly, the cell surface expression of both LAP and GARP requires the Golgi apparatus, since the addition of monensin or brefeldin A during the activation culture prevented their surface expression (Fig. 1E).
Fig. 1.
GARP and LAP are selectively expressed on activated FOXP3+ Tregs and platelets. (A) Level of FOXP3 and GARP mRNA on fresh (0 h) and activated CD4+CD25– T cells and Tregs (CD25hi) at 12, 24, 120 h, or 120 h with TGF-β1. Tregs were rested until day 14 (0 h) and restimulated for 18 h. (B) Flow cytometric analysis of surface LAP, GARP and intracellular FOXP3 on fresh (0 h) and activated Tregs (CD25hi) and CD4+CD25int T cells. (C) Surface staining of LAP and GARP on platelets based on FSC/SSC and CD61 expression. (D) LAP and GARP surface staining of plasmacytoid and myeloid DCs from PBMCs by gating on CD303+Lin-1– and CD1c+, respectively. (E) Surface LAP and GARP expression on Tregs after 12 h activation in the absence (none) or presence of monensin or brefeldin A for the last 8 h. Data are representative of 3 independent experiments. Numbers indicate percentage in each quadrant for B–E.
GARP Associates with Latent TGF-β Complex and Is Critical for Its Surface Expression.
Since the kinetics of induction and pattern of expression of LAP and GARP on activated Tregs appeared to be similar, we used siRNA technology to knockdown TGF-β1 or GARP mRNA to assess if the expression of these molecules is related. Freshly isolated Tregs transfected with TGF-β1 siRNA and activated for 48 h expressed GARP, but not LAP on their surface, while Tregs transfected with GARP siRNA failed to express either molecule. Expression of GARP or LAP was not affected when Tregs were transfected with a control nonspecific siRNA (Fig. 2A). This result suggested that GARP was required for the expression of latent TGF-β1 on the cell surface. To test whether latent TGF-β1 can associate with GARP, we incubated the 2 activated Treg populations (TGF-β1 siRNA or GARP siRNA) with recombinant human (rh) TGF-β1, LAP, LAP plus TGF-β1, or latent TGF-β1. LAP was only detected on the TGF-β1 siRNA transfected GARP+LAP– Tregs when they were incubated with either LAP mixed with TGF-β1 or latent TGF-β1, but not mature TGF-β1 or LAP alone, indicating that the TGF-β1/LAP complex was needed for interaction with GARP (Fig. 2B). We next tested whether the interaction of TGF-β2 or TGF-β3 with LAP of TGF-β1 results in binding to GARP. Interestingly, the mixture of TGF-β2 and LAP resulted in binding to GARP, while the mixture of TGF-β3 and LAP failed to bind to GARP (Fig. 2C). Although different isoforms of TGF-β naturally associate with their own distinct LAPs, it has been reported that LAP of TGF-β1 can bind and inactivate TGF-β2 and TGF-β3 with apparent Kd values of 1.9 and 0.4 nM, respectively (15). However, our result suggests that either TGF-β3 does not interact with LAP of TGF-β1 or their interactions produce a conformation that does not bind to GARP. Moreover, while TGF-β2 could interact with LAP to bind to GARP, it appears to be less efficient than with TGF-β1 based on the lower mean fluorescence intensity of LAP detection. The interaction of various molecules including αV integrins and TSP1 with the RGD sequence in LAP has been implicated in releasing and activating TGF-β (4, 16, 17). Using Jurkat cells expressing surface GARP, we tested whether TSP1, RGD, or RGDS peptides can block the binding of latent TGF-β1 to GARP. Preincubation of rhTSP1 or RGD/RGDS peptides did not block the binding of latent TGF-β1 to GARP (Fig. 2D). This result suggested that it is unlikely that the binding of GARP to latent TGF-β1 occurs via the RGD site on LAP or that latent TGF-β1 binds via the RGD site to another cell surface molecule that then interacts with GARP. To further support the association of GARP with LAP, we performed immunoprecipitation and colocalization confocal imaging studies. When LAP was immunoprecipitated from the surface of 48 h activated Tregs, GARP could be detected by immunoblot (Fig. 2E). Likewise, confocal imaging of surface-stained LAP and GARP on 48 h activated Tregs strongly demonstrated their colocalization (Fig. 3). Finally, to determine whether GARP and latent TGF-β1 can directly bind to each other, we performed a flow cytometric protein-protein interaction assay. Dynabeads were conjugated to either anti-LAP or anti-GARP mAbs and then incubated with a mixture of latent TGF-β1 and GARP-Fc fusion proteins, LAP and GARP-Fc, or TGF-β1 and GARP-Fc followed by flow cytometric detection of the anti-LAP Dynabead complex with fluorochrome conjugated anti-hFc or anti-GARP Dynabead complex with anti-LAP Abs. Beads conjugated with GARP bound latent TGF-β1, but not LAP and beads conjugated with latent TGF-β1, but not LAP, bound GARP (Fig. 2F). This result indicates that GARP can directly bind to latent TGF-β1.
Fig. 2.
Surface expression of latent TGF-β1 requires GARP association. (A) Tregs were transfected with nonspecific (siNS), TGF-β1 (siTGFβ1), or GARP (siGARP) siRNA and rested in IL-2 culture medium for 24 h before stimulation for 48 h to assess surface expression of GARP and LAP. (B) Tregs transfected with TGF-β1 or GARP siRNA were activated for 48 h then incubated for 30 min at 37 °C with 5 μg/mL mature rhTGF-β1, LAP, LAP + TGF-β1, or latent TGF-β1 and then surface stained for LAP. (C) GARP transfected Jurkat cells were incubated for 30 min at 37 °C without (NONE) or with 5 μg/mL LAP, TGF-β1, TGF-β2, or TGF-β3 alone or mixed with LAP and then stained for surface LAP. Jurkat cells transfected with the RFP vector were used as a control. (D) GARP-transfected Jurkat cells were incubated with 5 μg/mL latent TGF-β1 (LTGFβ1) or preincubated for 20 min at 37 °C with a 10-fold excess of TSP1, RGD, or RGDS peptides before latent TGF-β1 incubation and then stained for surface LAP. (E) Tregs were expanded in vitro for 14 days and then restimulated for 48 h before immunoprecipitation with anti-LAP (left lane) or isotype (right lane) followed by immunoblot with anti-GARP. (F) Dynabeads conjugated to αGARP (Left) or αLAP (Right) were incubated with a mixture of GARP-Fc + TGF-β1 (shaded histogram), GARP-Fc + LAP (dashed histogram), or GARP-Fc + latent TGF-β1 (solid histogram) followed by flow cytometric analysis with PE-labeled anti-hLAP (Left) or anti-hFc (Right). Data are representative of 3 independent experiments for A, B, C, D, and F and 2 for E. Numbers indicate percentage in each quadrant for A–D.
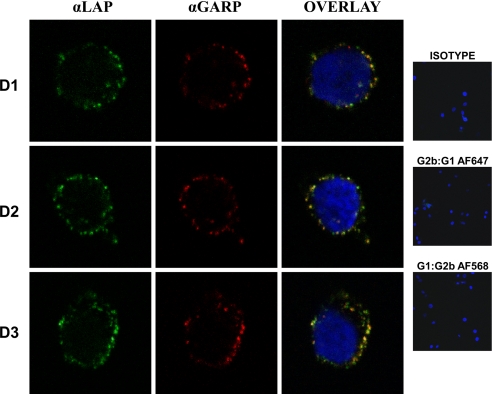
Fig. 3.
Colocalization of GARP and latent TGF-β1 on the surface of activated Tregs. Tregs activated for 48 h from 3 different donors (D1, D2, D3) were surface-stained with anti-GARP and anti-LAP or isotype controls. The cells were imaged with a Leica SP2-AOBS confocal microscope.
Regulation of GARP Is Independent of FOXP3.
Since GARP and LAP expression was observed in FOXP3+ Tregs, we next evaluated whether the de novo induction of FOXP3 in CD4+FOXP3– T cells was sufficient to induce GARP and LAP expression on the cell surface. Naïve CD45RA+ and memory CD45RO+ T cells activated with anti-CD3/CD28 in the presence of TGF-β1 can be induced to express FOXP3 (Fig. 4A) although such cells lack regulatory function (18). In contrast to Tregs, activation of naïve or memory T cells in the presence or absence of TGF-β1 failed to express significant levels of surface LAP or GARP on primary stimulation (Fig. S1A) or following multiple rounds of restimulation without TGF-β1 (Fig. 4A and Fig. S1B). It has been shown that high level and prolonged expression of FOXP3 following lentiviral-mediated transfection can result in the complete acquisition of the Treg phenotype in CD4+FOXP3– T cells (19). Although the TGFβ-induced FOXP3+ cells expressed similar level of FOXP3 as the Tregs upon restimulation, they fail to express surface latent TGF-β or GARP (Fig. S1B) and continue to lack Treg phenotype. Therefore, it does not appear that expression of FOXP3 in non-Tregs is sufficient to drive the surface expression of GARP and LAP.
Fig. 4.
GARP is required for the surface expression of latent TGF-β1. (A) FACS-sorted Tregs (CD25hi), naïve (CD45RA+), and memory (CD45RO+) T cells were activated for 5 days in the absence or presence of TGF-β1 then rested for 7 days and restimulated for 48 h without TGF-β1 before analysis of FOXP3 with surface GARP and LAP expressions. (B) Tregs were transfected with nonspecific (siNS) or FOXP3 (siFOXP3) siRNA and cultured for 5 days before activation for 48 h and analysis of FOXP3 with surface GARP and LAP expressions. (C) Naïve T cells were transduced with control (vector) or GARP encoded RFP-expressing lentiviral vectors and expanded for 7 days with anti-CD3/CD28 Dynabeads before restimulation for 48 h to evaluate for surface GARP and LAP (Left) and intracellular FOXP3 expressions among RFP+ and RFP− cells (Right). Data are representative of 3 independent experiments. Numbers indicate percentage in each quadrant.
To evaluate whether FOXP3 was essential for the expression of GARP and LAP on Tregs, we first knocked down the level of FOXP3 on CD25hi cells with siRNA for 5 days and determined the induction of GARP and LAP after 48 h of restimulation. Although FOXP3 expression was completely suppressed with the siRNA compared to the nonspecific control, the expression of GARP and LAP was not affected (Fig. 4B). This result indicates that the rapid expression of GARP and LAP was not controlled by an immediate downstream effect of FOXP3. However, it remains possible that FOXP3 might have an indirect effect on GARP expression after a more prolonged period of suppression of FOXP3 expression. Since virtually all T cells express TGF-β1 mRNA and are capable of secreting TGF-β1, we evaluated whether forced expression of GARP in CD4+FOXP3− T cells was sufficient to permit surface expression of latent TGF-β. Indeed, transduction of GARP into CD4+FOXP3− T cells allows for the surface expression of LAP, but not an increase in the percentage of FOXP3+ cells (Fig. 4C). Therefore it appears that GARP associates with latent TGF-β intracellularly and transports it to the cell surface via the Golgi apparatus. Furthermore, this result demonstrates that the failure to detect latent TGF-β on activated non-Tregs was due to the lack of GARP expression in these cells.
Dual Role of TGF-β on Tregs for Mediating Suppression and Infectious Tolerance.
As the kinetics for induction of cell surface expression of GARP and LAP closely resembled the kinetics for activation of Treg suppressor function (20), we next evaluated the role of these molecules in Treg-mediated suppression. Tregs that lacked the surface expression of LAP following treatment with TGF-β1 siRNA or the expression of GARP and LAP following treatment with GARP siRNA were significantly less suppressive than Tregs treated with the control siRNA in an in vitro suppression assay (Fig. 5A and B). Although the role of cell surface or secreted TGF-β1 as a major effector molecule of Treg suppression in vitro has remained controversial (6, 21), these results indicate that TGF-β contributes moderately to Treg-mediated suppression in vitro, but TGF-β does not appear to be the dominant mechanism since suppression was not completely abrogated. We have shown previously that activated mouse Tregs can induce Foxp3 expression in CD4+FOXP3− T cells during a 4-day co-culture (7). Similarly, co-culture of activated human Tregs treated with nonspecific, but not TGF-β1 or GARP siRNA, induced FOXP3 in CD4+FOXP3− T cells (Fig. 5C). We have not tested whether these induced FOXP3+ T cells have regulatory functions, as we are unable to separate them from the large population of FOXP3− T cells in the culture.
Fig. 5.
Dual functions of TGF-β in Treg-mediated suppression and infectious tolerance mechanism. (A) Tregs were transfected with siNS, siGARP, or siTGFβ1 and preactivated for 24 h before assessing their suppression of the proliferation of CD4+CD25– T cells stimulated with HLA-DR+ APCs and soluble anti-CD3 for 3 days and pulsed with 3H-TdR. The top panel is the FOXP3 expression, and the middle panel is the surface GARP and LAP expression of the 3 Treg populations at the end of the 3-day suppression assay. *, P < 0.05 between the siNS and siGARP or siTGFβ1 Treg suppression. (B) Similarly, CFSE-labeled CD4+CD25− responders were activated with HLA-DR+ APCs and soluble anti-CD3 for 3 days alone (top row) or in the presence of Tregs transfected with siNS, siGARP, or siTGFβ1 at a ratio of 2:1, 4:1, and 8:1 responder:Treg (bottom rows). (C) CFSE-labeled CD4+CD25−CD127+CD45RA+ T cells were stimulated with anti-CD3/CD28 Dynabeads for 5 days alone (top panel) or with 24-h preactivated Tregs transfected with siNS, siGARP, or siTGFβ1 and analyzed for FOXP3 induction in the CFSE-labeled cells. Data are representative of 3 independent experiments. Numbers indicate percentage in each quadrant.
Discussion
Our results clearly demonstrate that GARP or LRRC32 functions as a carrier and cell surface receptor for latent TGF-β1. Using siRNA technology, we have shown in vitro that GARP+LAP− cells can bind latent TGF-β1, but we have not been able to determine whether in vivo the binding of latent TGF-β1 to GARP occurs exclusively intracellularly or whether GARP can also bind secreted latent TGF-β1 or latent TGF-β associated with LTBP. Although the GARP/LAP complex is expressed by platelets, within the immune system GARP/LAP expression is mostly observed on activated functional FOXP3+ Tregs (9). We did not observe significant surface expression of GARP or LAP in CD19+/CD20+ B cells, CD14+ monocytes, CD8+ T cells, natural killer (NK) cells, NK T cells, and immature/mature monocyte-derived DCs. However, it is possible that under certain conditions and with activation, these cells might express GARP. Thus far, the predominant function of GARP/LAP on human Tregs remains elusive. Our functional studies indicate that the GARP/LAP complex does contribute to Treg-mediated suppression in vitro but whether this result also holds true in vivo is unknown and remains controversial. We have recently proposed (7) that a major role of latent TGF-β on the surface of murine Tregs is to convert responder T cells into FOXP3+ Tregs through a mechanism of infectious tolerance when both populations are activated in concert via their TCRs. This TGFβ-mediated infectious tolerance mechanism appears to also be involved in human Tregs. The regulation of expression of GARP at the mRNA level (10) in the mouse and the regulation of the expression of cell surface LAP are similar to that seen on human Tregs. GARP expression at the protein level in the mouse has not been evaluated due to the lack of anti-murine GARP antibodies. As GARP and LAP are only expressed on TCR-activated FOXP3+ Tregs, 1 major function of this complex is to target delivery of TGF-β1 to sites of ongoing immune responses where Tregs can be activated by recognition of their cognate antigens on APCs. Following release of active TGF-β1, it might act locally on FOXP3– T cells to convert them to FOXP3+ Tregs (7), to generate Th17 effectors if an inflammatory milieu is present (22), to act directly on the DCs to modulate their functions, or to signal in an autocrine manner to maintain Treg functions. A major question that remains to be addressed is the mechanism by which active TGF-β1 is generated from the latent GARP/LAP complex either by cell-associated molecules such as the αV integrins or by soluble factors.
Mutations in both GARP and LAP have been reported and result in complex clinical conditions. A base substitution from arginine to tryptophan in the coding region of the GARP gene has been identified in a large Samaritan kindred with Usher syndrome type 1, an autosomal recessive disease characterized by profound congenital sensorineural deafness, vestibular dysfunction, and progressive visual loss (23). It is unclear whether this mutation would result in a defective GARP protein that would fail to associate with LAP or potentially lead to enhanced release of active TGF-β. Mutations in the TGF-β1 signal peptide or the LAP coding region result in enhanced TGF-β activity as seen in Camurati-Engelmann disease, a rare, autosomal dominant condition characterized by sclerosing bone dysplasia and neurological deficiencies (24). Detailed studies of lymphocyte function, platelet function, or studies of tissue healing, remodeling, and fibrosis have not been performed in these patients. Mutations or deletions in GARP would only affect membrane bound TGF-β, and not the secreted pool, and therefore offer an opportunity to understand the unique contribution of membrane TGF-β to the regulation of immunity and inflammation. A recent study using antisense morpholino oligonucleotide to knockdown GARP in zebrafish demonstrated that GARP might be important in thrombus initiation on platelets (25). GARP also appears to be associated with infertility since up-regulation of GARP transcripts were observed in infertile human endometrium compared with fertile controls (26). Lastly, GARP mRNA is highly amplified in different tumors (27–34), but surface expression of GARP and its association with TGF-β in tumors has not been studied. Tumor cells may use GARP to express TGF-β or to capture TGF-β from their surroundings resulting in local suppression of anti-tumor immune responses or the induction of Tregs. Further studies of the regulation of GARP expression may lead to the development of drugs that can enhance or suppress the expression of GARP and membrane TGF-β and might be useful to treat autoimmunity or cancers and fibrotic diseases, respectively. Therefore, our discovery of the critical role of GARP in controlling surface expression of latent TGF-β will provide insights into another importance pathway of TGF-β regulation of morphogenesis, immune homeostasis, inflammation, and tissue remodeling.
Materials and Methods
Cell Purification.
Peripheral blood was obtained from healthy adult donors through the Department of Transfusion Medicine at the National Institutes of Health and approved by the NIAID institutional review board. The study was conducted in accordance with the Declaration of Helsinki. PBMCs were prepared over Ficoll-Paque Plus gradients (GE Healthcare). The CD4+ cells were enriched over the AutoMACS with CD4 microbead (Miltenyi). The cells were FACS-sorted with the FACSVantage DiVa or FACSAria for CD4+CD127+CD25−, CD4+CD25int (intermediate), and CD4+CD127–CD25hi (high). In some experiments, the cells were FACS-sorted for CD4+CD127+CD25–CD45RA+ or CD45RO+. Human APCs for in vitro suppression assay were obtained by depleting T cells from PBMCs with CD3 microbead (Miltenyi) using the AutoMACS followed by positive selection with HLA-DR microbead.
Flow Cytometric Analysis.
FOXP3 expression was detected with anti-FOXP3 mAbs after fixing and permeabilizing the cells with a Fixation/Permeabilization kit (eBioscience). LAP and GARP expression was surface stained with anti-LAP and anti-GARP mAbs before fixation/permeabilizing for FOXP3 detection. For GARP, a secondary detection with fluorochrome conjugated anti-mouse IgG2b was needed.
Antibodies and Reagents.
CD4, CD25, CD45RA, CD45RO, anti-mouse IgG1, anti-mouse IgG2b, goat (Fab′)2 anti-hFc, and carboxyfluorescein succinimidyl ester (CFSE) were from Invitrogen. Linage-1 mixture, CD127, streptavidin, monensin, and brefeldin A were from BD. CD1c and CD303 were from Miltenyi. Anti-LAP unconjugated and PE-conjugated IgG1 mAbs (clone 27232), biotinylated goat anti-hLAP, recombinant human (rh) LAP, latent TGF-β1, and thrombospondin-1 were from R&D Systems. Unconjugated anti-GARP IgG2b mAbs (clone Plato-1) and rhGARP (amino acid 20–627) fused to the Fc portion of human IgG1 were from Alexis Biochemicals. rhTGF-β1, -β2, -β3, and rhIL-2 were from Peprotech. RGD and RGDS peptides were from Sigma-Aldrich. Anti-CD3 (UCHT1), -CD28, and -FOXP3 (clone 236A/E7) mAbs were from eBioscience. FACSCalibur was used for data acquisition and the data were analyzed with FlowJo software (Tree Star).
Cell Culture.
For cell stimulation, 24-well culture plates (Corning) were coated with anti-CD3/CD28 at 5 μg/mL each for 2–4 h at 37 °C. The cells were cultured in complete RPMI 1640 supplemented with 2% heat-inactivated autologous human serum and 100 U/mL rhIL-2 (Peprotech). For induction of FOXP3, naïve CD4+CD127+CD25–CD45RA+ and memory CD45RO+ T cells were stimulated for 5 d with plate-bound anti-CD3/CD28 (5 μg/mL) ± 5 ng/mL rhTGF-β1 (Peprotech). For expansion, Tregs were stimulated with anti-CD3/CD28 Treg Dynabead (Invitrogen) and 100 U/mL IL-2. For in vitro suppression assay, 50,000 FACS-sorted allogeneic CD4+CD25– T cells were unlabeled or labeled with 2 μM CFSE and stimulated with 25,000 nonirradiated autologous CD3-depleted HLA-DR+ APCs and 0.25 μg/mL OKT3 (Ortho Biotech) alone or with varying numbers of preactivated (24 h) Tregs. The cells were cultured for 3 days in 96-well flat bottom plates (Corning) and pulsed with 3H-TdR (1 μCi/well) for the last 6–8 h or FACS analysis for CFSE labeled experiment. For the infectious tolerance experiments, 50,000 CFSE-labeled CD4+CD25–CD127+CD45RA+ T cells were stimulated with anti-CD3/CD28 Dynabeads at 2:1 cell-to-bead ratio for 5 days alone (top panel) or with 25,000 preactivated (24 h) Tregs transfected with siNS, siGARP, or siTGFβ1 and analyzed for FOXP3 induction in the CFSE-labeled cells. The cells were placed in 96-well flat bottom plates and cultured in X-VIVO 15 (Lonza) serum-free medium with 100 U/mL IL-2. For both the suppression and infectious tolerance experiments, the Tregs were first transfected by electroporation then rested for 24 h in 100 U/mL IL-2 before preactivation with plate-bound anti-CD3/CD28 for 24 h before testing their function.
Quantitative Real-Time PCR Analysis.
Total RNA was extracted from cells with an RNeasy Plus Kit (Qiagen). RT-PCR was performed with ≈1 μg of isolated RNA for cDNA synthesis using SuperScript II RNase H- Reverse Transcriptase (Invitrogen). Real-time PCR was performed in triplicate according to the Taqman Universal 2× master mix and run on the ABI/PRISM 7900 Sequence Detector System (Applied Biosystems). The amount of FOXP3 and GARP mRNA expression was normalized to the 18S rRNA and calculated according to the comparative Ct method as described by Applied Biosystems. The TagMan Gene Expression Assays from FOXP3 (Hs01085834_m1) and GARP (Hs00194136_m1) were from Applied Biosystems.
siRNA Experiments.
GARP, TGF-β1, FOXP3, and nonsilencing control siRNAs were from Invitrogen (Stealth Select RNAi). To transfect the Tregs, 200 pmols siRNA were mixed with 100 μL human T cell Nucleofector solution (Amaxa Biosystems), and 5 × 106 cells were resuspended in this mixture. The cell suspension was immediately electroporated by the Nucleofector II instrument (Amaxa Biosystems) and placed in 37 °C prewarmed 100 IU/mL IL-2 culture medium. For the GARP and TGF-β1 knockdown experiments, the transfected Tregs were rested in 100 U/mL IL-2 for 24 h before stimulation with plate-bound anti-CD3/CD28. For the FOXP3 knockdown experiments, the transfected Tregs were rested for 5 d before stimulation with anti-CD3/CD28.
Immunoprecipitation.
Day 14 expanded Tregs (20 × 106) were preactivated for 48 h with anti-CD3/CD28, then incubated with anti-LAP or IgG (Jackson Immunoresearch) for 1 h at 4 °C and then washed to remove unbound Abs. The cells were detergent solubilized in 1% Brij 96 lysis buffer containing protease inhibitors. LAP complexes were precipitated with anti-mouse IgG Dynabeads. Immunoprecipitates were resolved on 10%–14% acrylamide gels (Invitrogen) under reducing conditions and transferred to nitrocellulose membranes (Amersham). Blots were incubated with the anti-GARP and then horseradish peroxidase-conjugated anti-IgG2b. Reactivity was revealed by enhanced chemiluminescence. For flow cytometric protein-protein interaction experiment, 4.5 μm (1 × 106) anti-mouse IgG Dynabeads (Invitrogen) were incubated in PBS with either 20 μg/mL anti-LAP or anti-GARP mAbs at room temperature (RT) for 20 min and placed on magnet to wash away the supernatant. The anti-LAP or anti-GARP conjugated Dynabeads were incubated with a mixture of 20 μg/mL GARP-Fc + 20 μg/mL LAP, GARP-Fc + latent TGF-β1 or GARP-Fc + TGF-β1 recombinant proteins for 20 min at RT in PBS and then placed on magnet to wash away the supernatant. Finally the anti-LAP Dynabead samples were stained with goat anti-hFc PE and the anti-GARP Dynabead samples were stained with biotinylated goat anti-hLAP followed by streptavidin PE and then analyzed by flow cytometry.
Confocal Microscopy.
Tregs were activated for 48 h then surface-stained with anti-GARP IgG2b and anti-LAP IgG1 mAbs followed by anti-mouse IgG2b AF 568 and IgG1 AF 647. For background controls, isotype staining and anti-GARP IgG2b with anti-IgG1 AF 647 or anti-LAP IgG1 with anti-IgG2b AF 568 were used. Tregs were then fixed with 4% paraformaldehyde in PBS for 30 min at 4 °C and cytospun onto slides, permeabilized with 0.05% Triton X-100 for 5 min at RT. After 3 PBS washes, the nuclei were stained with 40 ng/mL Hoechst 33342 (Invitrogen) for 5 min. Slides were rinsed and mounted with a coverslip using Fluoromount-G (Southern Biotechnology). Images were collected on a SP2-AOBS confocal microscope (Leica Microsystems) by the NIAID Biological Imaging Facility.
GARP Transduction.
CD4+ T cells were purified using magnetic Dynabeads. CD45RO−CD25− population was sorted via FACSAria (BD). The naive cells were activated by anti-CD3/CD28-coated Dynabeads and transduced with control HIV-derived HDV expressing red fluorescent protein (RFP) lentiviral vectors (Clontech Laboratories) or encoding GARP gene as previously described (10) and expanded for 7 days in 200 U/mL IL-2 culture medium. The cells were then restimulated for 48 h with plate-bound anti-CD3/CD28 and analyzed for surface expression of GARP and LAP. The Jurkat cells were transduced with the same vectors and maintained in regular RPMI media with 10% FCS.
Supplementary Material
Acknowledgments.
We thank Carol Henry, Tom Moyer, and Calvin Eigsti in the National Institute of Allergy and Infectious Diseases Flow Cytometry Section for sorting our cells; Cynthia Matthews in the Department of Transfusion Medicine for the leukapheresis; and Lily Koo in the National Institute of Allergy and Infectious Diseases Biological Imaging Facility for performing the confocal microscopy. This work was supported by the National Institute of Allergy and Infectious Diseases Intramural Research Program and National Institutes of Health Grant R01 AI065303 (to D.U.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901944106/DCSupplemental.
References
- 1.Wan YY, Flavell RA. TGF-beta and regulatory T cell in immunity and autoimmunity. J Clin Immunol. 2008;28:647–659. doi: 10.1007/s10875-008-9251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gleizes PE, et al. TGF-beta latency: Biological significance and mechanisms of activation. Stem Cells. 1997;15:190–197. doi: 10.1002/stem.150190. [DOI] [PubMed] [Google Scholar]
- 3.Lawrence DA. Latent-TGF-beta: An overview. Mol Cell Biochem. 2001;219:163–170. doi: 10.1023/a:1010819716023. [DOI] [PubMed] [Google Scholar]
- 4.Grainger DJ, Wakefield L, Bethell HW, Farndale RW, Metcalfe JC. Release and activation of platelet latent TGF-beta in blood clots during dissolution with plasmin. Nat Med. 1995;1:932–937. doi: 10.1038/nm0995-932. [DOI] [PubMed] [Google Scholar]
- 5.Gandhi R, Anderson DE, Weiner HL. Cutting edge: Immature human dendritic cells express latency-associated peptide and inhibit T cell activation in a TGF-beta-dependent manner. J Immunol. 2007;178:4017–4021. doi: 10.4049/jimmunol.178.7.4017. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura K, et al. TGF-beta 1 plays an important role in the mechanism of CD4+CD25+ regulatory T cell activity in both humans and mice. J Immunol. 2004;172:834–842. doi: 10.4049/jimmunol.172.2.834. [DOI] [PubMed] [Google Scholar]
- 7.Andersson J, et al. CD4+ FoxP3+ regulatory T cells confer infectious tolerance in a TGF-beta-dependent manner. J Exp Med. 2008;205(9):1975–1981. doi: 10.1084/jem.20080308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran DQ, et al. Selective expression of latency-associated peptide (LAP) and IL-1 receptor type I/II (CD121a/CD121b) on activated human FOXP3+ regulatory T cells allows for their purification from expansion cultures. Blood. 2009;113:5125–5133. doi: 10.1182/blood-2009-01-199950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macaulay IC, et al. Comparative gene expression profiling of in vitro differentiated megakaryocytes and erythroblasts identifies novel activatory and inhibitory platelet membrane proteins. Blood. 2007;109:3260–3269. doi: 10.1182/blood-2006-07-036269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang R, Wan Q, Kozhaya L, Fujii H, Unutmaz D. Identification of a regulatory T cell specific cell surface molecule that mediates suppressive signals and induces Foxp3 expression. PLoS ONE. 2008;3:e2705. doi: 10.1371/journal.pone.0002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ollendorff V, Szepetowski P, Mattei MG, Gaudray P, Birnbaum D. New gene in the homologous human 11q13–q14 and mouse 7F chromosomal regions. Mamm Genome. 1992;2:195–200. doi: 10.1007/BF00302877. [DOI] [PubMed] [Google Scholar]
- 12.Ollendorff V, Noguchi T, deLapeyriere O, Birnbaum D. The GARP gene encodes a new member of the family of leucine-rich repeat-containing proteins. Cell Growth Differ. 1994;5:213–219. [PubMed] [Google Scholar]
- 13.Roubin R, et al. Structure and developmental expression of mouse Garp, a gene encoding a new leucine-rich repeat-containing protein. Int J Dev Biol. 1996;40:545–555. [PubMed] [Google Scholar]
- 14.Chen W, et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller DM, et al. Characterization of the binding of transforming growth factor-beta 1, -beta 2, and -beta 3 to recombinant beta 1-latency-associated peptide. Mol Endocrinol. 1992;6:694–702. doi: 10.1210/mend.6.5.1603080. [DOI] [PubMed] [Google Scholar]
- 16.Murphy-Ullrich JE, Poczatek M. Activation of latent TGF-beta by thrombospondin-1: Mechanisms and physiology. Cytokine Growth Factor Rev. 2000;11:59–69. doi: 10.1016/s1359-6101(99)00029-5. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z, et al. Absence of integrin-mediated TGFbeta1 activation in vivo recapitulates the phenotype of TGFbeta1-null mice. J Cell Biol. 2007;176:787–793. doi: 10.1083/jcb.200611044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran DQ, Ramsey H, Shevach EM. Induction of FOXP3 expression in naive human CD4+FOXP3- T cells by T-cell receptor stimulation is transforming growth factor-beta dependent but does not confer a regulatory phenotype. Blood. 2007;110:2983–2990. doi: 10.1182/blood-2007-06-094656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allan SE, Song-Zhao GX, Abraham T, McMurchy AN, Levings MK. Inducible reprogramming of human T cells into Treg cells by a conditionally active form of FOXP3. Eur J Immunol. 2008;38:3282–3289. doi: 10.1002/eji.200838373. [DOI] [PubMed] [Google Scholar]
- 20.Thornton AM, Piccirillo CA, Shevach EM. Activation requirements for the induction of CD4+CD25+ T cell suppressor function. Eur J Immunol. 2004;34:366–376. doi: 10.1002/eji.200324455. [DOI] [PubMed] [Google Scholar]
- 21.Piccirillo CA, et al. CD4(+)CD25(+) regulatory T cells can mediate suppressor function in the absence of transforming growth factor beta1 production and responsiveness. J Exp Med. 2002;196:237–246. doi: 10.1084/jem.20020590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu L, Kitani A, Fuss I, Strober W. Cutting edge: Regulatory T cells induce CD4+CD25-Foxp3- T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-beta. J Immunol. 2007;178:6725–6729. doi: 10.4049/jimmunol.178.11.6725. [DOI] [PubMed] [Google Scholar]
- 23.Bonne-Tamir B, et al. Usher syndrome in the Samaritans: Strengths and limitations of using inbred isolated populations to identify genes causing recessive disorders. Am J Phys Anthropol. 1997;104:193–200. doi: 10.1002/(SICI)1096-8644(199710)104:2<193::AID-AJPA5>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 24.Janssens K, et al. Mutations in the gene encoding the latency-associated peptide of TGF-beta 1 cause Camurati-Engelmann disease. Nat Genet. 2000;26:273–275. doi: 10.1038/81563. [DOI] [PubMed] [Google Scholar]
- 25.O'Connor MN, et al. Functional genomics in zebrafish permits rapid characterization of novel platelet membrane proteins. Blood. 2009;113:4754–4762. doi: 10.1182/blood-2008-06-162693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feroze-Zaidi F, et al. Role and regulation of the serum- and glucocorticoid-regulated kinase 1 in fertile and infertile human endometrium. Endocrinology. 2007;148:5020–5029. doi: 10.1210/en.2007-0659. [DOI] [PubMed] [Google Scholar]
- 27.Liu CJ, Lin SC, Chen YJ, Chang KM, Chang KW. Array-comparative genomic hybridization to detect genomewide changes in microdissected primary and metastatic oral squamous cell carcinomas. Mol Carcinog. 2006;45:721–731. doi: 10.1002/mc.20213. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Cardus A, et al. Pharmacogenomic approach for the identification of novel determinants of acquired resistance to oxaliplatin in colorectal cancer. Mol Cancer Ther. 2009;8:194–202. doi: 10.1158/1535-7163.MCT-08-0659. [DOI] [PubMed] [Google Scholar]
- 29.Lassmann S, et al. Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. J Mol Med. 2007;85:293–304. doi: 10.1007/s00109-006-0126-5. [DOI] [PubMed] [Google Scholar]
- 30.Bekri S, et al. Detailed map of a region commonly amplified at 11q13–>q14 in human breast carcinoma. Cytogenet Cell Genet. 1997;79:125–131. doi: 10.1159/000134699. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez C, et al. Amplification of the BRCA2 pathway gene EMSY in sporadic breast cancer is related to negative outcome. Clin Cancer Res. 2004;10:5785–5791. doi: 10.1158/1078-0432.CCR-03-0410. [DOI] [PubMed] [Google Scholar]
- 32.Maire G, et al. 11q13 alterations in two cases of hibernoma: Large heterozygous deletions and rearrangement breakpoints near GARP in 11q13.5. Genes Chromosomes Cancer. 2003;37:389–395. doi: 10.1002/gcc.10223. [DOI] [PubMed] [Google Scholar]
- 33.Schraml P, et al. Combined array comparative genomic hybridization and tissue microarray analysis suggest PAK1 at 11q13.5-q14 as a critical oncogene target in ovarian carcinoma. Am J Pathol. 2003;163:985–992. doi: 10.1016/S0002-9440(10)63458-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwards J, Krishna NS, Witton CJ, Bartlett JM. Gene amplifications associated with the development of hormone-resistant prostate cancer. Clin Cancer Res. 2003;9:5271–5281. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.