Abstract
In trypanosomes, the production of mRNA relies on the synthesis of the spliced leader (SL) RNA. Expression of the SL RNA is initiated at the only known RNA polymerase II promoter in these parasites. In the pathogenic trypanosome, Trypanosoma brucei, transcription factor IIB (tTFIIB) is essential for SL RNA gene transcription and cell viability, but has a highly divergent primary sequence in comparison to TFIIB in well-studied eukaryotes. Here we describe the 2.3 Å resolution structure of the C-terminal domain of tTFIIB (tTFIIBC). The tTFIIBC structure consists of 2 closely packed helical modules followed by a C-terminal extension of 32 aa. Using the structure as a guide, alanine substitutions of basic residues in regions analogous to functionally important regions of the well-studied eukaryotic TFIIB support conservation of a general mechanism of TFIIB function in eukaryotes. Strikingly, tTFIIBC contains additional loops and helices, and, in contrast to the highly basic DNA binding surface of human TFIIB, contains a neutral surface in the corresponding region. These attributes probably mediate trypanosome-specific interactions and have implications for the apparent bidirectional transcription by RNA polymerase II in protein-encoding gene expression in these organisms.
Keywords: eukaryotic transcription, parasite, RNA polymerase II, trypanosome
Trypanosomes are flagellated protozoa, belonging to the order Kinetoplastida, that are exclusively parasitic (1). These organisms reside in insect vectors that transmit the parasites to mammals, birds, fish, and plants. Trypanosoma brucei and Trypanosoma cruzi are of particular medical concern because they cause debilitating and fatal diseases in millions of people annually in tropical regions of the world (2). Trypanosomes diverged from other eukaryotes early in evolution, and thus many biological processes that are well understood in metazoans are highly distinct in these parasites (3, 4). For example, little is known about how trypanosome RNA polymerase II (tRNAP-II) transcription is initiated. It is known that tRNAP-II transcribes most protein-encoding genes, which are arranged in tandem arrays, into polycistronic mRNAs (5). Thus far, the only tRNAP-II promoter that has been identified is the spliced leader (SL) RNA gene promoter (6). The SL RNA gene codes for the SL, which is capped and added in a trans-splicing reaction to the 5′ end of each ORF contained in a polycistronic mRNA. The addition of an SL to every mRNA is a universal requirement in trypanosomes; therefore, understanding transcription initiation at the SL RNA gene promoter is a crucial step toward understanding the control of tRNAP-II transcription in these parasites (4, 7).
To date, components of the SL RNA transcriptional machinery have been studied by using genomics or biochemistry to identify candidate proteins followed by a combination of phenotypic analysis of protein function using either RNAi-silencing of endogenous protein in parasites or depletion/add back studies of in vitro transcription systems. These methods have identified several players in tRNAP-II-dependent SL RNA transcription, including trypanosome TATA-box binding protein (tTBP), transcription factors IIB, -IIA, -IIH (tTFIIB, tTFIIA, tTFIIH), and tSNAPc (8–17). However, each trypanosome factor is only distantly related to its metazoan homolog and contains regions or subunits unique to trypanosomes. Indeed, a recent study of tTFIIH using single particle electron microscopy suggests that tTFIIH has a similar structure to human TFIIH but contains trypanosome-specific subunits (18). Thus, the mechanism of transcription initiation in trypanosomes cannot be deduced from metazoan transcription systems. Our work presented here is the primary venture to understand the mechanics of the SL RNA transcriptional machinery using high-resolution structural analysis.
Trypanosome TFIIB is essential for SL RNA transcription in T. brucei (13, 14). tTFIIB associates with tTBP, tRNAP-II, and an SL RNA gene promoter fragment in nuclear extracts. In yeast and mammals, TFIIB binds specifically to TBP and DNA and recruits RNAP-II into a minimal preinitiation complex (reviewed in ref. 19). TFIIB consists of N- and C-terminal domains (20, 21). NMR analysis of the N-terminal domain from Pyrococcus furiosus TFIIB reveals a Zn ribbon motif and an extended B finger loop (22, 23). In the cocrystal structure of Saccharomyces cerevisiae TFIIB and RNAP-II, the TFIIB N-terminal domain binds RNAP-II and uses the B finger loop to finely position RNAP-II at the transcription start site (24). NMR analysis of the human TFIIB C-terminal domain reveals 2 helical repeats (25). In the human and Pyrococcus woesei TFIIB/TBP/DNA ternary complexes, the TFIIB C-terminal domain recognizes TBP and the promoter (26–29). The C-terminal domain of the trypanosome TFIIB (tTFIIBC) contains 1 loosely conserved repeat module followed by a trypanosome-specific region (13, 14). Here, we report the 2.3 Å resolution structure of tTFIIBC. The tTFIIBC structure reveals 2, closely packed helical modules followed by a C-terminal extension of 32 aa. The trypanosome-specific region comprises the second helical module and the C-terminal extension. The overall tTFIIBC structure is similar to other TFIIBC structures (25–29), but contains additional loops and helices. In contrast to the highly basic DNA binding surface of human TFIIB, the structure reveals a neutral surface in the corresponding region. These attributes are probably functionally important, mediating trypanosome-specific interactions in a preinitiation complex.
Results
tTFIIB and tTFIIBC Are Stably Folded Monomers.
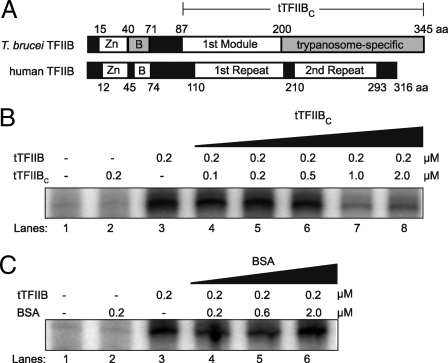
We performed limited proteolysis on recombinant tTFIIB to identify a stable fragment amenable to crystallization, because the full-length protein was refractory to crystallization. The human TFIIB C-terminal domain forms a protease-resistant core (20, 21). In contrast, tTFIIB was highly protease sensitive and did not yield any stable fragments (Fig. S1). Therefore, we defined the tTFIIB C-terminal domain (tTFIIBC) by comparing the amino acid sequence of tTFIIB with those of crystal structures of human and archaeal TFIIB C-terminal domains as shown schematically (Fig. 1A). tTFIIBC, which comprises the first module and the trypanosome-specific region (residues 87–345), was expressed and purified.
Fig. 1.
Primary structure and function of tTFIIB and tTFIIBC. (A) Schematic of T. brucei and human TFIIB amino acid sequence alignment. The Zn ribbon and the first module/first repeat are well conserved between trypanosomes and humans. The B finger and the trypanosome-specific/second repeat are not well conserved (gray background). tTFIIBC is 259 residues long, comprising the first module and the trypanosome-specific region (bracketed). (B) Activity of tTFIIB and tTFIIBC in in vitro SL RNA transcription assays. Transcription activity in tTFIIB-depleted extracts (lane 1) could not be restored by 0.2 μM tTFIIBC (lane 2), whereas tTFIIB restored maximally at that concentration (lane 3). tTFIIBC maximally inhibited restoration by tTFIIB when added in 5-fold molar excess (lanes 4–8). (C) In control assays, BSA did not restore transcription (lane 2) and did not inhibit restoration by tTFIIB (lanes 4–6).
Because the limited proteolysis results suggested that tTFIIB and tTFIIBC might not be stably folded, we characterized both proteins biophysically. In gel filtration experiments, each protein eluted as a single peak at a position between its calculated dimer and monomer mass (Fig. S2A). This is consistent with the extended monomer conformation of TFIIB and TFIIBC from other organisms (23–25). We found that the fluorescence spectrum of native tTFIIB was blue-shifted when compared to the spectrum under denaturing conditions, suggesting hydrophobic burial of Trp-260 in the native protein. Because Trp-260 is the only tryptophan in tTFIIB and resides in the C-terminal domain, unfolding of both tTFIIB and tTFIIBC could be monitored by fluorescence. Denaturant-induced unfolding of tTFIIB and tTFIIBC occurred in a cooperative and reversible fashion with a half-maximal Cm of unfolding between 2 and 3 M guanidinium chloride (Fig. S2B). tTFIIBC appears to be slightly more stable than full-length tTFIIB (Cm = 2.9 M for tTFIIBC versus 2.7 M for tTFIIB). Taken together, these results indicate that full-length tTFIIB and tTFIIBC are stably folded monomers.
Activity of Purified tTFIIB and tTFIIBC by in Vitro Transcription.
We showed previously that tTFIIB expressed and purified from Escherichia coli restores transcription in vitro when added to tTFIIB-immunodepleted nuclear extracts of T. brucei (14). Using this system, tTFIIB restored transcription maximally at 0.2 μM (Fig. 1B, lane 3, and Fig. S3). By contrast, tTFIIBC was unable to restore transcription (Fig. 1B, lane 2). Notably, tTFIIBC had a trans-dominant negative effect on transcription. tTFIIBC inhibited transcription in the presence of full-length tTFIIB when tTFIIBC was added in 5-fold molar excess (Fig. 1B, lane 7). This effect is specific, as transcription by tTFIIB was unaffected by molar excess of BSA (Fig. 1C). We hypothesize that tTFIIBC is able to bind to proteins and/or DNA in the preinitiation complex but requires the N-terminal domain to stimulate transcription, consistent with previous studies of human TFIIBC (21).
Structure of tTFIIBC and Comparison to Human tTFIIBC.
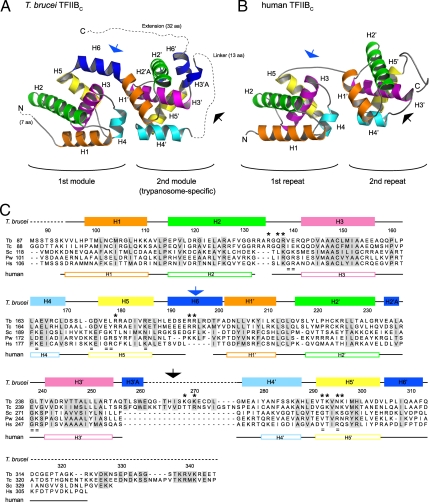
We crystallized and solved the structure of tTFIIBC at 2.3 Å resolution (Rwork = 20.3%; Rfree = 24.9%) (Table S1 and Table S2). The tTFIIBC structure reveals 2, closely packed helical modules followed by a C-terminal extension of 32 aa (Fig. 2A). The trypanosome-specific region comprises the second helical module and the C-terminal extension. The first helical module contains 6 helices (H1–H6), while the second module has 8 helices (H1′–H6′ and 2 310 helices, H2′A and H3′A). Both helical modules contain the canonical 5-helix cyclin fold characteristic of TFIIB proteins (helices H1–H5 in the first module and H1′–H5′ in the second module). The cyclin fold in each module aligns well to the cyclin folds of human TFIIBC (rmsd = 1.8 Å on 90 of 96 matched Cα atoms for the first module; rmsd = 2.3 Å on 86 of 100 matched Cα atoms for the second module). The cyclin fold of the second module has a slightly greater deviation from the human cyclin folds owing to divergent regions discussed below. The 32-residue C-terminal extension of tTFIIBC (residues 314–345) is not visible in the crystal, suggesting that it is unstructured. The prevalence of Pro, Glu, Ser, and Lys residues in the C-terminal extension of the tTFIIBC is consistent with this conclusion (30).
Fig. 2.
Structure of the C-terminal domain of T. brucei TFIIB (tTFIIBC) and comparison to human TFIIBC. (A) Structure of tTFIIBC (this work). Each helix of the 5-helix cyclin fold is colored identically in each of the 2 modules. Additional motifs distinct in tTFIIB (H6, H2′A, H3′A, and H6′) are blue. H2′A is behind H2 in this view. Dashed lines denote amino acids not visible in the crystal, and the number of residues is in parentheses. Arrows denote regions for comparison between tTFIIBC and human TFIIBC in panel B and their position in the sequence in panel C as discussed in the text: Helix H6 (blue arrow) and the linker between H3′A and H4′ (black arrow). (B) Structure of human TFIIBC (PDB ID 1c9b) (29). (C) The amino acid sequence of tTFIIBC (Tb; accession no. EAN76636) is aligned with that of T. cruzi (Tc; XP 806216), S. cerevisiae (Sc; P29055), and the sequences of known TFIIBC structures from P. woesei (Pw; 1d3u), and Homo sapiens (Hs; 1c9b). Sequence gaps are denoted by dashes in panel C. tTFIIBC amino acid numbers and secondary structure elements are indicated above the alignment, and human TFIIBC secondary structure elements are indicated below the alignment. Helices are depicted as boxes, intervening segments as lines, gaps in the structural alignment are blank, and residues not visible in the electron density are denoted by dashes. Amino acids that were changed to alanine in tTFIIB variants are indicated by asterisks (see Figs. 3 and 4). Structurally equivalent residues in human TFIIB that affected function when mutated are denoted by equal signs (19). Residues Asp-229 to Thr-237 are omitted from the S. cerevisiae sequence for clarity.
The overall tTFIIBC structure is similar to other TFIIBC structures (25–29); however, the additional loops and helices between the modules and within the second module of tTFIIBC explain why primary sequence analysis did not reveal the second cyclin fold (13, 14) (Fig. 2 A and B). For instance, the first module has an additional helix H6 that links the 2 modules together by connecting H5 and H1′. Helix H6 essentially forms a bent, 24-residue helix with H1′ of the second module (Fig. 2A, blue arrow). In human TFIIBC, the helical repeats are connected by a short, random-coil segment that interacts with TBP (26) (Fig. 2B, blue arrow). In tTFIIBC, the presence of a helix at this position instead of a random coil creates a larger surface that would allow for additional contacts with tTBP.
The second module in tTFIIBC contains 2 significant insertions within its cyclin fold that are not observed in other TFIIB structures. The first insertion is the helix H2′A, which is a short 310 helix inserted into the loop connecting helices H2′ and H3′. The second insertion is a much longer segment within the loop between helices H3′ and H4′. This insertion consists of another 310 helix, H3′A, that is followed by a 13-residue linker (Fig. 2A, black arrow). In the tTFIIBC structure, H3′A veers ≈90 ° from H3′ and away from H4′. The linker (residues 162–174), which connects H3′A to H4′, is not visible in the electron density, suggesting that it is unstructured. T. cruzi TFIIBC also contains a linker in this region of different length but with similar amino acid composition to T. brucei TFIIB (Fig. 2C, black arrow). There appears to be no precedent for a long unstructured segment between helices H3′ and H4′ in either TFIIB or cyclin structures in the Protein Data Bank, as assessed by a search using the DALI structural alignment server (31). In these well-studied proteins, the region connecting H3′ and H4′ is typically a structured loop of 5–8 residues (Fig. 2B, black arrow). Unstructured protein segments often contain short recognition motifs that mediate protein-protein or protein-nucleic acid interactions (32). We predict that the tTFIIBC unstructured linker is functionally important, mediating trypanosome-specific interactions in the preinitiation complex.
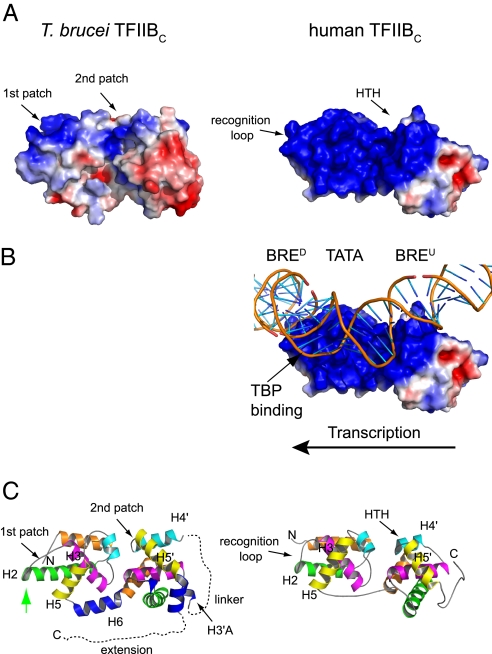
The overall conformation of tTFIIBC is closer to human TFIIBC bound in the TFIIBC/TBP/DNA complex rather than to the unbound protein (25, 29). This allows us to predict how tTFIIBC might interact with DNA and tTBP. Strikingly, the electrostatic potential of the putative DNA binding surface of tTFIIBC is markedly different from the DNA binding surface of human TFIIBC (Fig. 3A). In the human TFIIBC/TBP/DNA complex, TFIIBC contacts DNA through a large positive surface (Fig. 3B). This surface stabilizes the TBP-induced deformation of the DNA by interacting both upstream and downstream of the TATA box. In the TFIIBC/TBP/DNA complex, the DNA tracks along the large positive surface of TFIIBC (Fig. 3B). In stark contrast, the analogous surface of tTFIIBC is relatively neutral and contains only 2 small patches of positive charge (Fig. 3A, Left). Thus the trypanosome TFIIBC/TBP/DNA complex probably has a different conformation than the canonical ternary complex.
Fig. 3.
Putative DNA binding surface and electrostatic potential surface properties of tTFIIBC compared to human TFIIBC. (A) The putative DNA binding face of tTFIIBC (Left) reveals a more neutral surface than the highly basic DNA binding surface of human tTFIIBC (Right). Areas colored in blue, white, and red denote positive, neutral, and negative potential contoured at +3, 0, −3 kT/e, respectively. Black arrows in panels A and C indicate the basic patches in tTFIIBC and the corresponding regions (recognition loop and HTH motif) in human TFIIBC. (B) In the human TFIIBC/TBP/DNA complex, TFIIBC contacts the DNA at BRED and BREU through its basic surface (29). TBP, which binds the minor groove, is omitted for clarity. (C) Ribbon diagram of putative DNA binding surface. The green arrow indicates the additional turn on H2 unique to the trypanosome protein. View in this figure is rotated 180° around the horizontal axis relative to Fig. 2.
tTFIIBC likely interacts with DNA through the 2 small positive patches found on the electrostatic surface of the structure (Fig. 3A, Left). The first patch corresponds to the loop between helices H2 and H3 (Fig. 3C, Left). In human TFIIBC, the loop between helices H2 and H3, known as the recognition loop, binds to the DNA downstream of the TATA box at the downstream TFIIB recognition element (BRED) (Fig. 3 A–C, Right) (26, 29). tTFIIBC contains a similar loop that is preceded by an additional helical turn at the C-terminal end of H2 (Fig. 3C, Left, green arrow). This helical turn extends out from the first module into solvent, placing basic residues in the loop, namely Arg-135 and Arg-138, in position to interact with the DNA. Thus, tTFIIBC may use the extra helical turn of H2 as well as its loop for DNA binding.
The second positive patch in tTFIIBC corresponds to H4′ and H5′ (Fig. 3 A and C, Left). In human TFIIBC, H4′ and H5′ form a helix-turn-helix (HTH) motif that binds the DNA upstream of the TATA box at the upstream TFIIB recognition element (BREU) (Fig. 3 B and C, Right) (29, 33). In the HTH motif, helix H4′ uses a conserved glutamine to stabilize a conserved arginine in H5′ that contacts DNA (29). Interestingly, tTFIIBC lacks these conserved residues. Moreover, H4′ in tTFIIBC is 1 turn longer than the first helix of the HTH motif of human TFIIBC. This would not allow H4′ to fit in the DNA major groove. On the other hand, human TFIIBC uses residues in H5′, the recognition helix of the HTH motif, to make base-specific contacts with the major groove (29, 33). tTFIIBC contains similar residues in H5′ (Thr-292, Lys-293, Asn-295, and Lys-296) that may function analogously in DNA recognition.
A major portion of the neutral electrostatic surface of tTFIIBC derives from surface residues on helix H5. In the human TFIIBC/TBP/DNA complex, H5 stabilizes the TBP-induced deformation of the DNA through interactions between Lys-189 and Arg-193 and the DNA phosphate backbone (29). In tTFIIBC, the corresponding region of H5 does not contain basic residues. However, the basic residues adjacent to this region, including Arg-179 in H5 and Arg-194 and Arg-195 in H6, may assist in stabilizing the trypanosome TFIIBC/TBP/DNA complex.
Activity of tTFIIB Mutants by in Vitro Transcription.
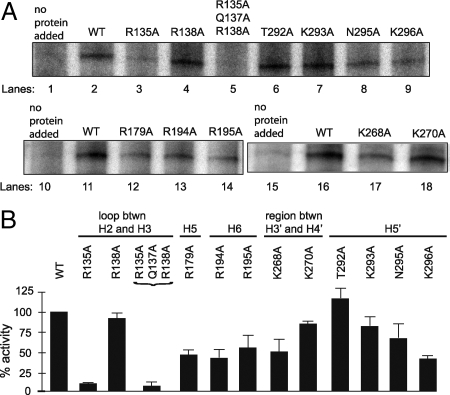
To determine if specific residues in tTFIIBC are important for function, we assayed variants of full-length tTFIIB for their ability to support transcription. We made alanine substitutions in Arg-135 and Arg-138 and the triple substitution Arg-135/Gln-137/Arg-138 in the loop linking H2 and H3. We also made single alanine substitutions in Arg-179 in H5, Arg-194 and Arg-195 in H6, Lys-268 and Lys-270 in the linker between H3′ and H4′, and Thr-292, Lys-293, Asn-295, and Lys-296 in H5′ (Fig. S4).
Single alanine substitutions in the loop linking H2 and H3 reduced activity minimally in the case of Arg-138 but decreased activity to ≈10% in the case of Arg-135 (Fig. 4). The triple variant, containing alanine substitutions of Arg-138, Arg-135 along with Gln-137, reduced activity to ≈7%. The single mutation in helix H5 reduced activity to ≈50%. Mutations of basic residues near the center of helix H5′ had a more severe effect on transcription than those near the N-terminal edge of the helix. Specifically, Lys-296 reduced activity to ≈40%, whereas Lys-293 reduced activity to ≈80%. The Asn-295 to Ala mutation reduced activity to ≈65% and the Thr-292 to Ala mutation appeared to stimulate tTFIIB activity. Overall, mutations in trypanosome-specific regions reduced activity of tTFIIB. The mutations of basic residues in H6 and Lys-268 in the linker between H3′ and H4′ reduced activity to 40–50%. Mutation of Lys-270 had only a small effect (≈85% activity). Taken together, these data suggest that basic residues within the loop linking H2 and H3, the helices H5 and H5′, and in the trypanosome-specific regions (H6 and the linker between H3′ and H4′) are important for in vitro transcription, probably by stabilizing tTFIIB interactions at the SL RNA gene promoter.
Fig. 4.
Activity of tTFIIB mutants with alanine substitutions in in vitro SL RNA transcription assays. (A) Transcription activity in tTFIIB-depleted extracts (lanes 1 and 10) was restored upon addback of wild-type (WT) tTFIIB (lanes 2 and 11). In the loop linking H2 and H3, the R138A mutant had near WT activity (lane 4), but both R135A and the triple mutant nearly abolished activity. Mutations in H5 and H6 (R179A, R194A, R195A; lanes 12–14) reduced activity ≈50%. In the linker between H3′ and H4′, K168A also reduced activity ≈50% (lane 17), while K270 had near WT activity (lane 18). In helix H5′, T292A had increased activity (lane 6), while K293A, N295A, and K296A toward the center of the helix had reduced activity (lane 7–9). (B) Histogram represents averages of 3 replicates for each mutant. Error bars indicate 1 standard deviation.
Discussion
This work describes the high-resolution structure of a trypanosome transcription factor. The structure of trypanosome TFIIBC shares important features with its human counterpart. tTFIIBC contains 2 cyclin folds that align well to human TFIIBC even though the second module was not apparent in tTFIIBC from primary sequence information. In T. brucei, the loop between helices H2 and H3 is functionally important, as is the corresponding recognition loop in the human protein. Specifically, mutation of basic amino acids in this region completely compromised TFIIB function in the human and trypanosome proteins. Thus, our work supports a fundamental role of the loop between helices H2 and H3 in transcription.
Trypanosome TFIIBC deviates in potentially significant ways from its human counterpart. The 2 cyclin folds are connected by a helix (H6) rather than a random coil, and in between helices H3′ and H4′, there is an additional helix (H3′A) followed by a disordered 13-residue linker. Importantly, mutations in these regions reduced transcriptional activity of tTFIIB, indicating that these regions play a role in tTFIIB function. Moreover, the presence of a potentially protease-sensitive linker between H3′ and H4′ may explain why tTFIIB lacks a protease-resistant core. In addition to the 13 disordered residues in the linker, the C-terminal 32 residues of the tTFIIBC structure are also disordered. These regions are common to all trypanosome TFIIB proteins sequenced thus far, and while the sequences vary in length between each protein, they have similar amino acid composition. It is likely that these unstructured segments contain short, peptide motifs involved in recognizing DNA or other trypanosome-specific protein partners, possible adopting secondary structure upon binding. These segments will be the object of future studies.
Trypanosome TFIIB lacks the conserved Gln and Arg residues in helices H4′ and H5′ that stabilize the interaction of HTH motifs with the DNA major groove. In addition, tTFIIB H4′ has an additional helical turn that would hinder major groove binding. Mutations in H5′ (Thr-292 and Asn-295) did not knock out function as they did in the corresponding residues (Val-283 and Arg-286) in human TFIIB (33). These traits in the trypanosome protein may be related to the lack of a BREU in the SL RNA gene promoter. In human TATA box-containing promoters, TFIIB binding to the BREU directs assembly of the preinitiation complex in the correct orientation. The majority of gene promoters do not contain a TATA box or an associated BREU, thus they rely on multiple interactions from transcription-associated factors for preinitiation complex orientation. In trypanosome TFIIB, the linker between H3′ and H4′, as well as the C-terminal extension, may recruit factors that help determine preinitiation orientation at the SL RNA gene promoter.
tTFIIBC contains a large neutral surface in place of the highly basic surface on the human protein (Fig. 3). The large neutral surface might be required to stabilize TBP binding to DNA. Whereas human TBP has 4 phenylalanines that deform the TATA box upon binding, only 2 phenylalanines are present in trypanosome TBP (10, 34). Thus, tTFIIB may help stabilize weak tTBP-promoter interactions. In support of this argument, our previous studies showed that tTBP and tTFIIB tightly associate (14).
In well-studied eukaryotic transcription, the basic surface of TFIIB specifically binds at gene promoters to orient RNAP-II so that transcription proceeds in the correct direction. The lack of an extensive basic surface on trypanosome TFIIB may reflect a relaxed specificity of tRNAP-II orientation at promoters in trypanosomes. Most trypanosome protein-encoding genes are transcribed in large arrays that appear to initiate bidirectionally from strand switch regions (35–37). Weak specificity of preinitiation complex orientation could cause bidirectional transcription from strand switch regions. Thus, trypanosome TFIIB would be relieved of the function to orient tRNAP-II for unidirectional transcription.
Methods
Protein Expression and Purification.
Full-length T. brucei TFIIB (tTFIIB), the C-terminal domain (tTFIIBC), and variants were recombinantly expressed and purified from E. coli as His6-MBP fusion proteins following the protocol of Tropea et al. (38). TFIIB and variants were purified by sequential affinity and gel filtration chromatography. For tTFIIBC, the latter step was replaced by ion exchange chromatography. The selenomethionine labeled tTFIIBC (SeMet-tTFIIBC) was expressed as described (39) and purified as described above for native tTFIIBC. A detailed protocol is provided in the SI Text.
Limited Proteolysis.
tTFIIB (20 μM) was digested with trypsin or subtilisin in 50 mM Tris-HCl, pH 8.0, 50 mM NaCl at 25 °C for 1 h in 50 μL with an enzyme: substrate molar ratio of 1:400. Aliquots (10 μL) were removed at 5-, 10-, 15-, 30-, 60-min intervals, and the reaction in the aliquot was stopped by addition of 4% SDS, 100 mM DTT, 20 mM EDTA, 2 mM PMSF, and flash-freezing. Samples were heated to 95 °C for 10 min and analyzed by SDS/PAGE.
Gel Filtration and Chemical Denaturation.
Gel filtration was performed on a Sephacryl S-200 column equilibrated in 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM DTT, at 4 °C with 1 mg of tTFIIB and tTFIIBC. Chemical denaturation experiments monitoring tryptophan fluorescence were performed at 25 °C (QuantaMaster spectrofluorometer; Photon Technology International). Samples of 3 μM tTFIIB or tTFIIBC were prepared at each guanidinium chloride (GdmCl) concentration in 25 mM potassium phosphate, pH 7.5, 200 mM KCl, and 1 mM DTT and incubated overnight at room temperature. Fluorescence emission spectra were measured by exciting the samples at 265 nm and recording spectra from 300 to 400 nm (0.1-s integration time) and the center-of-mass of the spectral peak was calculated. The GdmCl concentration was determined by refractive index using a Bausch & Lomb refractometer. GdmCl denaturation of tTFIIB or tTFIIBC was reversible as judged by the recovery of center-of-mass from dilution of the 4 M GdmCl sample to 2 M GdmCl. Curve fitting was performed in Kaleidagraph (Synergy Software).
Immunodepletion and in Vitro Transcription.
T. brucei nuclear extracts were prepared as described (11, 40). Immunodepletion and in vitro transcription assays were performed as described (14) with the following exceptions. Transcription was monitored by detecting radiolabeled run-off transcript (172 bases) from a linearized plasmid rather than by primer extension. The plasmid pJP10, which contains the −125- to +120 region of the SL RNA gene was linearized with EcoRV. In the reaction, 250 μM CTP and GTP, 1 mM ATP, and 10 μCi alpha-32P-labeled UTP were incubated at 28 °C with other reaction components for 10 min, after which cold UTP was added to a final concentration of 50 μM. The potassium glutamate concentration in the reaction was 130 mM.
Structure Determination of tTFIIBC.
Crystals of native tTFIIBC were obtained by vapor diffusion against a reservoir containing 0.1 M Bis-Tris, pH 5.5, and 24% polyethylene glycol 3350. SeMet- tTFIIBC protein crystallized in the same space group (P43212) but required slightly higher concentrations of polyethylene glycol (32%) to form. The structure was solved using 3-wavelength multiple anomalous dispersion from data collected at National Synchrotron Light Source beamline X29 and refined against a native data set collected at the PHRI X-Ray Crystallography Core Facility. The final model consists of residues 94–261 and 275–313, 56 water molecules, and an ethylene glycol molecule, with values of Rwork = 0.204 and Rfree = 0.249. Details are provided in SI Text, and data collection and refinement statistics are given in Table S1 and Table S2.
Supplementary Material
Acknowledgments.
We thank the staff of beamline X29 at the National Synchrotron Light Source; Mary Ann Gawinowicz at the Columbia University Protein Core Facility; Tara Kurumaddali for assistance with site-directed mutagenesis; and Karl Drlica, David Dubnau, Phil Jeffrey, Leonard Mindich, Matthew Neiditch, David Perlin, and Issar Smith for helpful discussions. This work was supported by National Institutes of Health Grant AI-29478 (to V.B.), American Heart Association Fellowship 0425791T (to J.B.P.), American Heart Association Scientist Development Grant 0735532T (to A.D.), and Ministerio de Educacion y Ciencia (Spain) Grant CTQ2007-68014-C02-02 (to D.V.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3H4C).
This article contains supporting information online at www.pnas.org/cgi/content/full/0904309106/DCSupplemental.
References
- 1.Simpson AG, Stevens JR, Lukes J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;22:168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Despommier DD, Gwadz RW, Hotez PJ, Knirsch CA. Parasitic Diseases. New York, NY: Apple Trees Productions; 2005. [Google Scholar]
- 3.Clayton CE. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palenchar JB, Bellofatto V. Gene transcription in trypanosomes. Mol Biochem Parasitol. 2006;146:135–141. doi: 10.1016/j.molbiopara.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Liang XH, Haritan A, Uliel S, Michaeli S. trans and cis splicing in trypanosomatids: Mechanism, factors, and regulation. Eukaryot Cell. 2003;2:830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilinger G, Bellofatto V. Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 2001;29:1556–1564. doi: 10.1093/nar/29.7.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell DA, Thomas S, Sturm NR. Transcription in kinetoplastid protozoa: Why be normal? Microbes Infect. 2003;5:1231–1240. doi: 10.1016/j.micinf.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Das A, Bellofatto V. RNA polymerase II-dependent transcription in trypanosomes is associated with a SNAP complex-like transcription factor. Proc Natl Acad Sci USA. 2003;100:80–85. doi: 10.1073/pnas.262609399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schimanski B, Laufer G, Gontcharova L, Gunzl A. The Trypanosoma brucei spliced leader RNA and rRNA gene promoters have interchangeable TbSNAP50-binding elements. Nucleic Acids Res. 2004;32:700–709. doi: 10.1093/nar/gkh231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruan JP, Arhin GK, Ullu E, Tschudi C. Functional characterization of a Trypanosoma brucei TATA-binding protein-related factor points to a universal regulator of transcription in trypanosomes. Mol Cell Biol. 2004;24:9610–9618. doi: 10.1128/MCB.24.21.9610-9618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das A, et al. Trypanosomal TBP functions with the multisubunit transcription factor tSNAP to direct spliced-leader RNA gene expression. Mol Cell Biol. 2005;25:7314–7322. doi: 10.1128/MCB.25.16.7314-7322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schimanski B, Nguyen TN, Gunzl A. Characterization of a multisubunit transcription factor complex essential for spliced-leader RNA gene transcription in Trypanosoma brucei. Mol Cell Biol. 2005;25:7303–7313. doi: 10.1128/MCB.25.16.7303-7313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schimanski B, Brandenburg J, Nguyen TN, Caimano MJ, Gunzl A. A TFIIB-like protein is indispensable for spliced leader RNA gene transcription in Trypanosoma brucei. Nucleic Acids Res. 2006;34:1676–1684. doi: 10.1093/nar/gkl090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palenchar JB, Liu W, Palenchar PM, Bellofatto V. A divergent transcription factor TFIIB in trypanosomes is required for RNA polymerase II-dependent spliced leader RNA transcription and cell viability. Eukaryot Cell. 2006;5:293–300. doi: 10.1128/EC.5.2.293-300.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas S, Yu MC, Sturm NR, Campbell DA. A non-universal transcription factor? The Leishmania tarentolae TATA box-binding protein LtTBP associates with a subset of promoters. Int J Parasitol. 2006;36:1217–1226. doi: 10.1016/j.ijpara.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Lecordier L, et al. Characterization of a TFIIH homologue from Trypanosoma brucei. Mol Microbiol. 2007;64:1164–1181. doi: 10.1111/j.1365-2958.2007.05725.x. [DOI] [PubMed] [Google Scholar]
- 17.Lee JH, Nguyen TN, Schimanski B, Gunzl A. Spliced leader RNA gene transcription in Trypanosoma brucei requires transcription factor TFIIH. Eukaryot Cell. 2007;6:641–649. doi: 10.1128/EC.00411-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JH, Jung HS, Gunzl A. Transcriptionally active TFIIH of the early-diverged eukaryote Trypanosoma brucei harbors two novel core subunits but not a cyclin-activating kinase complex. Nucleic Acids Res. 2009;37:3811–3820. doi: 10.1093/nar/gkp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng W, Roberts SG. TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma. 2007;116:417–429. doi: 10.1007/s00412-007-0113-9. [DOI] [PubMed] [Google Scholar]
- 20.Barberis A, Muller CW, Harrison SC, Ptashne M. Delineation of two functional regions of transcription factor TFIIB. Proc Natl Acad Sci USA. 1993;90:5628–5632. doi: 10.1073/pnas.90.12.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik S, Lee DK, Roeder RG. Potential RNA polymerase II-induced interactions of transcription factor TFIIB. Mol Cell Biol. 1993;13:6253–6259. doi: 10.1128/mcb.13.10.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen HT, Legault P, Glushka J, Omichinski JG, Scott RA. Structure of a (Cys3His) zinc ribbon, a ubiquitous motif in archaeal and eucaryal transcription. Protein Sci. 2000;9:1743–1752. doi: 10.1110/ps.9.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu W, et al. The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon. Nat Struct Biol. 1996;3:122–124. doi: 10.1038/nsb0296-122. [DOI] [PubMed] [Google Scholar]
- 24.Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: An RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science. 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 25.Bagby S, et al. Solution structure of the C-terminal core domain of human TFIIB: Similarity to cyclin A and interaction with TATA-binding protein. Cell. 1995;82:857–867. doi: 10.1016/0092-8674(95)90483-2. [DOI] [PubMed] [Google Scholar]
- 26.Nikolov DB, et al. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 27.Kosa PF, Ghosh G, DeDecker BS, Sigler PB. The 2.1-A crystal structure of an archaeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box. Proc Natl Acad Sci USA. 1997;94:6042–6047. doi: 10.1073/pnas.94.12.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Littlefield O, Korkhin Y, Sigler PB. The structural basis for the oriented assembly of a TBP/TFB/promoter complex. Proc Natl Acad Sci USA. 1999;96:13668–13673. doi: 10.1073/pnas.96.24.13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai FT, Sigler PB. Structural basis of preinitiation complex assembly on human pol II promoters. EMBO J. 2000;19:25–36. doi: 10.1093/emboj/19.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romero P, et al. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 31.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v. 3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohan A, et al. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 33.Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: Sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juo ZS, et al. How proteins recognize the TATA box. J Mol Biol. 1996;261:239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Calvillo S, et al. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol Cell. 2003;11:1291–1299. doi: 10.1016/s1097-2765(03)00143-6. [DOI] [PubMed] [Google Scholar]
- 36.Respuela P, Ferella M, Rada-Iglesias A, Aslund L. Histone acetylation and methylation at sites initiating divergent polycistronic transcription in Trypanosoma cruzi. J Biol Chem. 2008;283:15884–15892. doi: 10.1074/jbc.M802081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siegel TN, et al. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tropea JE, Cherry S, Nallamsetty S, Bignon C, Waugh DS. A generic method for the production of recombinant proteins in Escherichia coli using a dual hexahistidine-maltose-binding protein affinity tag. Methods Mol Biol. 2007;363:1–19. doi: 10.1007/978-1-59745-209-0_1. [DOI] [PubMed] [Google Scholar]
- 39.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 40.Huie JL, He P, Bellofatto V. In vitro transcription of the Leptomonas seymouri SL RNA and U2 snRNA genes using homologous cell extracts. Mol Biochem Parasitol. 1997;90:183–192. doi: 10.1016/s0166-6851(97)00146-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.