Abstract
Raman spectroscopy is a newly developed, noninvasive preclinical imaging technique that offers picomolar sensitivity and multiplexing capabilities to the field of molecular imaging. In this study, we demonstrate the ability of Raman spectroscopy to separate the spectral fingerprints of up to 10 different types of surface enhanced Raman scattering (SERS) nanoparticles in a living mouse after s.c. injection. Based on these spectral results, we simultaneously injected the five most intense and spectrally unique SERS nanoparticles i.v. to image their natural accumulation in the liver. All five types of SERS nanoparticles were successfully identified and spectrally separated using our optimized noninvasive Raman imaging system. In addition, we were able to linearly correlate Raman signal with SERS concentration after injecting four spectrally unique SERS nanoparticles either s.c. (R2 = 0.998) or i.v. (R2 = 0.992). These results show great potential for multiplexed imaging in living subjects in cases in which several targeted SERS probes could offer better detection of multiple biomarkers associated with a specific disease.
Keywords: imaging in vivo, multiplex, SERS, nanoparticles
In recent years, the biomedical research community has come to realize that no single targeting agent is likely to provide sufficient information needed to characterize or detect a specific disease process. As a result, several efforts have been made toward the discovery of multiple biomarkers and targeting ligands in the hope of improving earlier detection and management of specific diseases. The ability to simultaneously detect multiple targets, sensitively and in vivo, is an attractive feat; but it is a task often difficult to accomplish. Thus far, nanoparticles have played an important role in this endeavor; however most nanostructure-based platforms for multiplex detection methods have been tailored for in vitro applications (1–6), leading to little progress in the field of in vivo multiplex imaging.
Recently, there has been an overwhelming interest in sensitive imaging of nanoparticles for both diagnostic and therapeutic applications (7–11). As a result, new preclinical imaging modalities optimized for nanoparticle imaging have been developed, further expanding the field of molecular imaging. Thus far, fluorescence and Raman spectroscopy, in conjunction with quantum dots and surface enhanced Raman scattering (SERS) nanoparticles, respectively, have been the predominant imaging modalities to evaluate in vivo multiplex imaging (12–14). Raman imaging, in particular, has generated quite a bit of interest recently; we have demonstrated its ability to detect picomolar concentrations in vivo along with its unique ability to multiplex using SERS nanoparticles and others have developed novel Raman nanoparticles with the potential to be used in vivo as well (14–17).
Both quantum dots and SERS nanoparticles have shown great potential as multiplexed imaging probes ex vivo, whether for cellular imaging or for biosensor applications; however, several limitations exist with quantum dots when attempting to translate their use in vivo. For instance, there are limited flavors of quantum dots available in the near-infrared window, where autofluorescence emanating from superficial tissue layers is minimized; thus, sensitivity is lowered and the depth to which fluorescence imaging can be used is restricted. Moreover, quantum dots have been shown to be cytotoxic under certain conditions, greatly limiting their application in living subjects (18). SERS nanoparticles, on the other hand, are not limited in terms of available spectral signatures (“flavors”) in the near-infrared window, because their size and Au core configuration are ideal for Raman scattering at 785 nm and because the spectral signature derives from vibrational modes of Au-bound molecules, the structures of which can be widely varied. In terms of cytotoxicity, the silica and Au composition of SERS nanoparticles make them fairly inert. In addition, SERS nanoparticles have narrower spectral peaks, which leads to little spectral overlap—unlike quantum dots, the emission spectra of which are broad in comparison. Furthermore, Raman spectroscopy, in conjunction with SERS nanoparticles, offers picomolar sensitivity in vivo (at limited depths), as opposed to the nanomolar sensitivity achievable using conventional fluorescence imaging in conjunction with quantum dots in vivo (14). Finally, quantum dots, although more resistant than conventional fluorophores, display a natural tendency to photobleach—unlike SERS nanoparticles, which consist of Raman-active molecules adsorbed to the surface of a 60-nm diameter Au core that are inherently insensitive to photodestruction.
This article will discuss the ability of our optimized preclinical Raman microscope to produce multiplexed in vivo images in conjunction with 10 of our unique SERS nanoparticle batches. Each SERS flavor emits its own unique spectral fingerprint when excited by a 785-nm laser, thereby allowing spectral identification and unmixing of various combinations of simultaneously injected SERS nanoparticle batches. In vivo multiplex Raman imaging using SERS nanoparticles will be evaluated in both superficial (skin) and deep (liver) tissue after s.c. and i.v. injections, respectively. Finally, we will evaluate the ability of our Raman imaging system to quantitatively multiplex increasing concentrations of various SERS batches.
Results
SERS Raman Nanoparticles.
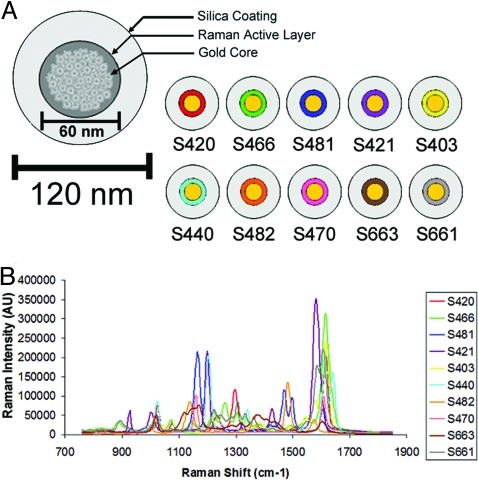
All experiments described herein were conducted using SERS nanotags (Oxonica Materials, Mountain View, CA) (19, 20). Ten spectrally unique SERS batches were used to evaluate multiplexed imaging in vivo. Each SERS batch consists of a unique Raman active molecular layer adsorbed onto a 60-nm diameter Au core coated with silica, making the entire diameter of the nanoparticle on the order of 120 nm (shown schematically in Fig. 1A). The Au nanoparticle core acts as a substrate for SERS and can increase the effective Raman scattering efficiency by several orders of magnitude (21), allowing more sensitive detection in deep tissue and making it ideal for in vivo imaging. Upon excitation with a 785-nm laser, each type of SERS nanoparticle emits a unique Raman spectrum based on the structure of the adsorbed molecule, called the “reporter.” All SERS nanotags in this work have the same 60-nm diameter Au core. To simplify nomenclature, each type of SERS nanotag is given a three-digit suffix (e.g., S-420) instead of the formal name of the adsorbed reporter molecule. The graph in Fig. 1B depicts all 10 color-coded spectra along with their corresponding SERS-xyz abbreviation. Chemical structures for the 10 reporter molecules are shown in Fig. S1. Because each flavor of SERS nanotag generates a unique Raman spectrum, our postprocessing software was able to spectrally unmix a combination of various SERS nanoparticles injected simultaneously using a least-squares curve-fitting algorithm.
Fig. 1.
Schematic representation of a SERS Raman nanoparticle and graph depicting unique Raman spectra associated with each of the 10 SERS nanoparticles used for in vivo multiplexed imaging. (A) Schematic of a SERS Raman nanoparticle consisting of a 60-nm gold core with a unique Raman active layer adsorbed onto the gold surface and coated with glass totaling 120 nm in diameter. The trade name of each SERS nanoparticle is depicted to the right, where a color has been assigned to the Raman active layer of each SERS nanoparticle. (B) Graph depicting Raman spectra of all 10 SERS nanoparticles; each spectra has been assigned a color corresponding to its unique Raman active layer as shown in (A).
Demonstration of in Vivo Multiplexed Imaging Using 10 SERS Nanoparticles.
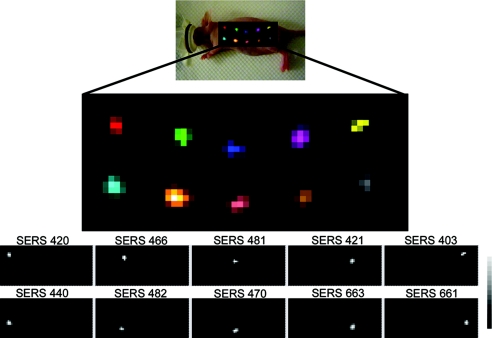
To initially evaluate our systems ability to correctly identify and spectrally unmix spectra from various SERS nanoparticles, we first administered 10 separate s.c. injections of each SERS flavor to a nude mouse and then mapped the entire area of interest with our Raman microscope using 750-μm steps at 1 s/frame. The map was then analyzed with our postprocessing software, where preassigned reference spectra of each SERS nanotag flavor were used to determine the best spectral fit with their corresponding SERS nanotag flavor. The software then separated out intensity maps into various channels showing where in the map each SERS nanotag flavor was detected. The resulting image showed all 10 s.c. injections correctly separated out into their corresponding spectral channel with minimal crosstalk among the channels (Fig. 2). This sets the stage for evaluating colocalization of multiple SERS tags in deep tissues that have been administered simultaneously.
Fig. 2.
Evaluation of multiplexing 10 different SERS nanoparticles in vivo. Raman map of 10 different SERS particles injected s.c. in a nude mouse. Arbitrary colors have been assigned to each unique SERS nanoparticle batch injected. Panels below depict separate channels associated with each of the injected SERS nanoparticles (S420, S466, S481, S421, S403, S440, S482, S470, S663, and S661, respectively). Grayscale bar to the right depicts the Raman intensity, where white represents the maximum intensity and black represents no intensity. The postprocessing software was able to successfully separate all 10 SERS nanoparticles into their respective channels with minimal crosstalk.
In Vivo Multiplexed Imaging in Liver.
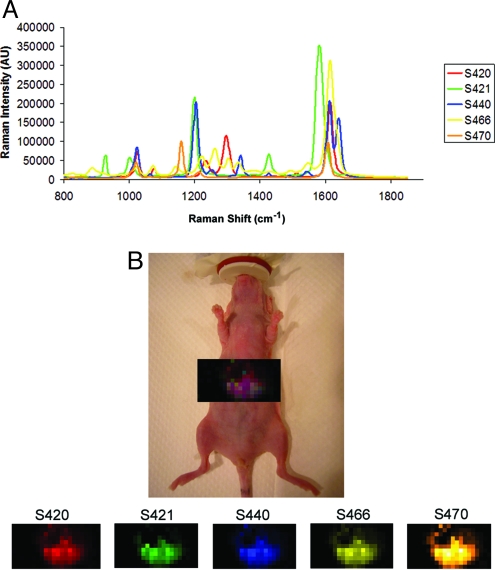
After successfully demonstrating multiplexed imaging in vivo using separate s.c. injections, we wanted to evaluate the capability of our Raman microscope used for this work to detect and deconvolve multiple SERS tags injected simultaneously into the tail vein. Because of the size of these SERS nanoparticles (≈120 nm in diameter), the particles tend to get taken up by the Kupffer cells of the reticuloendothelial system and, as a result, naturally accumulate in the liver. This allowed us to acquire images of the liver to evaluate multiplexing in deep tissue. Initially, several combinations of SERS nanoparticle batches were injected i.v. to evaluate in vivo multiplexing; however certain mismatches existed among particular SERS combinations because of spectral overlap. The number of SERS flavors that we were able to inject simultaneously was also limited because of the volume that we were allowed to inject in a single mouse. Therefore, we chose the five most spectrally unique SERS nanotags (S420, S421, S440, S466, and S470) with the least spectral overlap, ideal for deep-tissue imaging (Fig. 3A). We injected a mixture of equal amounts of all 5 SERS nanoparticle batches via the tail vein. Imaging commenced at 1, 24, and 48 h post injection to reveal relatively equal amounts of all five SERS tags accumulating in the liver at all time points. The postprocessing software correctly identified all five SERS batches accumulating in the liver after i.v. injection (Fig. 3B). Note that although all SERS batches were administered in equal concentrations, data analysis revealed non-uniform accumulation in the liver, probably because of variability in Raman intensity among each of the five SERS tags. As one can see from Fig. 3A, some of the SERS tags are just inherently less Raman intense than others, making it more difficult to detect at low signal-to-noise levels. Dimmer SERS particles would thus drop in and out of the detectable range during in vivo mapping, causing problems for quantification in deep tissue.
Fig. 3.
Demonstration of deep-tissue multiplexed imaging 24 h after i.v. injection of five unique SERS nanoparticle batches simultaneously. (A) Graph depicting five unique Raman spectra, each associated with its own SERS batch: S420 (red), S421 (green), S440 (blue), S466 (yellow), and S470 (orange). It is noteworthy that their peaks have very little spectral overlap, allowing easier spectral unmixing and resulting in better deep-tissue detection. (B) Raman image of liver overlaid on digital photo of mouse, showing accumulation of all five SERS batches accumulating in the liver after 24 h post i.v. injection. Panels below depict separate channels associated with each of the injected SERS nanoparticle batches. Individual colors have been assigned to each channel, and the resulting mixture shows a purple color that represents a mixture of the five SERS nanoparticle batches accumulating simultaneously. It should be noted that all channels show accumulation in the liver; however the channels are not all homogenous in their distribution throughout the liver.
Multiplexing Various Concentrations of SERS Nanoparticles.
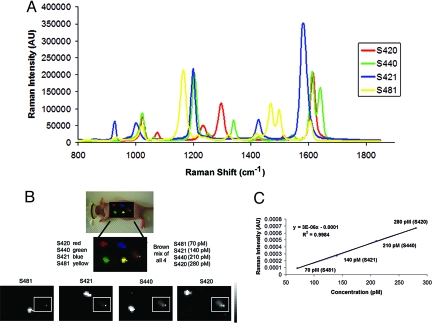
Quantification of signals from multiple spectra injected simultaneously is often difficult because of spectral overlap as well as the inherent variability in Raman intensity among varying SERS batches. To make a more quantitative assessment of multiplexing in vivo we chose four SERS batches (S420, S440, S421, and S481) that demonstrated little to no spectral overlap along with relatively elevated and uniform Raman intensities (Fig. 4A). Initially, we s.c. injected equal amounts of each of the four SERS nanotags separately and then performed a fifth injection containing a combination of all four SERS nanotags of varying concentrations. The mixture consisted of the following: 70 pM of S420, 140 pM of S440, 210 pM of S421, and 280 pM of S481. The entire area of interest was mapped with our Raman microscope using 750 μm steps at 1 s/frame. Each of the four SERS tags were correctly identified in their corresponding channels, and visually the Raman intensity of the mixture correlated well with increasing SERS concentration (Fig. 4B). Because numerical values are assigned to each pixel based on Raman intensity, we were able to draw a region of interest (ROI) around the mixed injection site to get a better quantitative estimate of Raman intensity versus SERS concentration. We took an average of the Raman intensity over the mixed injection site in each of the SERS channels to correlate concentration with Raman intensity. The data revealed a linear correlation (R2 = 0.998) in which Raman intensity increased with increasing SERS concentration (Fig. 4C).
Fig. 4.
Evaluation of multiplexing various concentrations of SERS nanoparticles in s.c. injection model. (A) Graph depicting four unique Raman spectra, each associated with its own SERS batch (S420, S440, S421, and S481). It may be noted that their peaks have very little spectral overlap, and their maximum Raman intensities are all fairly similar which makes them ideal for evaluating various concentrations of different SERS flavors. (B) Raman image depicting multiplexing various concentrations of SERS nanoparticles after s.c. injection. Upper shows a Raman map of four different SERS particles injected s.c., each assigned a separate color: red for S420, green for S440, blue for S421, and yellow for S481. The fifth s.c. injection, represented by a brown color at the far right, is a mixture of the four unique SERS batches of varying concentrations. Lower shows separate channels in which each of the individual SERS bathes were detected. Grayscale bar to the right depicts the Raman intensity, where white represents the maximum intensity and black represents no intensity. All s.c. injections were correctly identified. It may be noted that the fifth s.c. mixture (in white box) becomes visually more intense as the concentration of SERS nanoparticles increases, allowing one to qualitatively determine which SERS nanoparticle batch is more prevalent in the mixture, from least to most. (C) This graph represents a more quantitative assessment of how the Raman intensity taken directly from the Raman images is linearly related to the SERS concentration injected in the mixture.
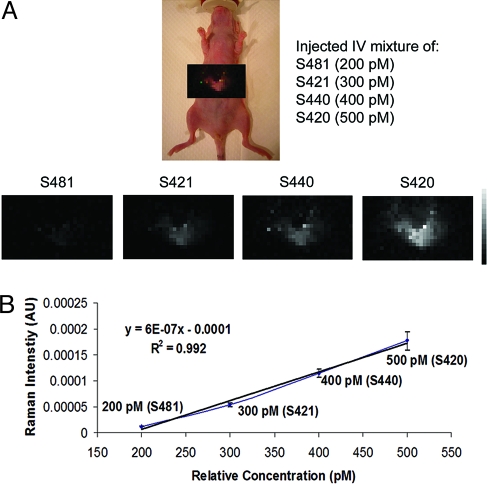
Next we multiplexed varying concentrations of the same four SERS nanoparticle batches in the liver after simultaneously injecting them i.v. (n = 3). The varying concentrations were as follows: 200 pM of S481, 300 pM of S421, 400 pM of S440, and 500 pM of S420. After 2 h post injection, the entire liver area was mapped using 1-mm steps at 3 s/frame. The resulting liver image correctly revealed increasing Raman intensity in the liver with increased concentrations of the corresponding SERS batch (Fig. 5A). Average ROIs were taken over the liver area within each SERS channel and revealed a linear correlation (R2 = 0.992) whereby Raman intensity increased with increasing SERS concentration (Fig. 5B). Both the s.c. and deep-tissue results support the use of SERS nanoparticles for in vivo multiplexed imaging applications.
Fig. 5.
Evaluation of multiplexing various concentrations of SERS nanoparticles in a deep-tissue model. (A) Raman map of the entire liver taken 2 h post i.v. injection containing a mixture of four unique SERS batches of varying concentrations (200 pM of S481, 300 pM of S421, 400 pM of S440, and 500 pM of S420). Bottom shows separate channels in which each of the individual SERS bathes were detected. Grayscale bar to the right depicts the Raman intensity, where white represents the maximum intensity and black represents no intensity. Liver images reveal a consistent pattern with the concentration of SERS nanoparticles injected; as the concentration of SERS nanoparticles injected increased, the Raman intensity on the Raman maps increased. It may be noted that the liver images become more intense or visible with increased SERS concentration injected, allowing a correct qualitative assessment of which SERS nanoparticle batch is more prevalent in the mixture, from least to most. (B) This graph represents a more quantitative assessment of how the Raman intensity taken directly from the Raman images is linearly related to the SERS concentration injected in the mixture (n = 3; error bars represent standard deviation).
Discussion
Current literature suggests that a multimarker approach could potentially improve the sensitivity and specificity of detecting certain disease processes. Several researchers have successfully conjugated various nanoparticles to several kinds of peptides, proteins, and oligonucleotides (22–30). Thus far, quantum dots have been the nanoparticle predominantly used to evaluate in vivo multiplexed imaging (12, 31). However, these nanoprobes share similar limitations with other commonly used fluorescent probes, including autofluorescence, susceptibility to photobleaching, and relatively broad emission spectra. Now, with the development of a noninvasive Raman imaging strategy, we are able to assess the potential of SERS nanoparticles as multiplexing imaging agents.
The unique spectral properties associated with SERS nanoparticles make them ideal for ultrasensitive and noninvasive multiplexed imaging applications in living subjects. First, their narrow peaks are easily resolved, allowing simultaneous identification and colocalization of several SERS nanoparticles. In this study, we have shown the ability of our optimized Raman microscope to spectrally separate and correctly identify 10 different SERS nanoparticle batches in an s.c. murine model. This could serve as a usefully way to mimic what would happen if one interrogated an s.c. tumor xenograft model in a live mouse with multiple targeted SERS. In addition, we have been able to watch multiple SERS batches naturally accumulate in the liver simultaneously, and our software was able to correctly identify which five SERS batches were injected i.v.. Finally, our postprocessing software was able to linearly correlate SERS concentration with Raman intensity using the Raman images taken after both s.c. and i.v. injections of a mixture of various concentrations of four unique SERS nanoparticles.
To date, only a few research groups have demonstrated noninvasive multiplexed imaging in live animal models. Our group showed multiplexed imaging with embryonic stem cells labeled with six different quantum dots (12). The six quantum dot/embryonic stem cell batches were individually injected s.c. and imaged, revealing correct localization and identification of each set of quantum dots/embryonic stem cells. However, there was inhomogeneity in the energy absorbed for each quantum dot because of their ability to produce different light levels at the same excitation wavelengths. This phenomenon could make it difficult to relate light signal to the amount of probe that actually localized. One significant problem that we experienced with multiplexed imaging of multiple quantum dots was their inability to be spectrally separated when they colocalized in vivo (for more details see SI Text). Another group showed simultaneous visualization of five separate quantum dots accumulating in various lymph nodes after interstitial injection (31). This method was able to correctly identify the draining patterns and mixing of five adjacent lymphatic basins.
Although SERS nanoparticles would not likely be optimal for lymphatic drainage evaluation because of their relatively large size (120 nm) and the limited depth of penetration associated with our Raman microscope (5 mm), they have the potential to play an important role in more localized diagnostic applications, such as tumor/disease detection during laparoscopic/intraoperative surgeries or endoscopic procedures such as colonoscopies. Endoscopic Raman probes are currently being developed to overcome these limitations while maintaining its ultrasensitive and multiplexing properties (32, 33).
Another potential multiplexing application involves trafficking of various cells, such as T cells, dendritic cells, or even stem cells, as demonstrated in a preliminary s.c. model by Lin et al. (12). The ability to uniquely label multiple cell types and watch them simultaneously localize is a powerful tool that could add a wealth of knowledge to the biomedical community and that could potentially offer a better way of guiding the practice and progress of medicine.
Liu et al. have recently communicated another intriguing prospect: use of isotopically modified single-walled nanotubes (SWNTs) as multiplexing nanoparticles in conjunction with Raman spectroscopy (27). The group successfully multiplexed three forms of carbon-based nanotubes (C12 SWNTs, C13 SWNTs, and C12 and C13 SWNTs mixed) in cell culture. Each set of SWNTs was conjugated to a different targeting ligand, each of which would ideally bind to its respective biomarker, found on one of three different cell lines. Each targeted SWNT batch was successfully multiplexed using Raman imaging, and specific labeling of cells by targeted SWNTs was shown in each cell line, with minimal nonspecific binding. A mixture of all three cell lines was also evaluated for colocalization of each SWNT batch to its respective cell line. Raman imaging showed the ability of these isotopically different SWNTs to simultaneously localize to their corresponding cell line. It was also suggested that more multiplexers could be developed by combining different ratios of C12 and C13, creating Raman tags that could easily be applied to a tumor-bearing animal model and evaluated in vivo with our optimized Raman imaging microscope.
The ability to colocalize increasing numbers of individual SERS particles is limited because of both spectral overlap and the variability in the inherent Raman intensity arising from each of the unique Raman active layers associated with the SERS nanoparticles. Larger sets of tags must be optimized for spectral uniqueness, uniformity, and increased signal strength, which can be controlled during the synthesis process described herein. Thus far, we have been able to successfully multiplex four different SERS tags at varying concentrations. We observed a linear correlation between increased Raman signal and increased concentration of SERS nanoparticles that allowed us to semiquantitatively predict the amount of SERS nanoparticles accumulating in the liver. An important factor that limits our capacity to fully quantitate our images is the inability to acquire a complete map in deeper tissues, as our system is limited to a 5-mm depth of penetration. Because the total depth of the liver exceeds 5 mm, we were unable to acquire maps of the entire liver, and we therefore had an inaccurate representation of how many total SERS nanoparticles had accumulated in the organ. However, one can see that the Raman signal was fairly reproducible and consistent across all three animals that we evaluated for multiplexing various concentrations (Fig. 5B). We are currently working to optimize our Raman microscope to increase the depth of penetration and possibly to look at tomographic capabilities for better quantification.
Following the discovery of any new disease/tumor targeting agent comes the task of being able to sensitively determine the binding and reporter potential of that agent. We have demonstrated a high degree of noninvasive multiplexing, using 10 unique SERS nanoparticles in live animals, and have shown colocalization of five different SERS nanoparticles within deep tissue after i.v. injection. In conclusion, we believe that combining the ultrasensitive properties of Raman spectroscopy with the multiplexing capabilities of SERS nanoparticles will allow more molecular targets to be interrogated simultaneously with a single noninvasive image, thereby improving disease detection.
Methods
Raman Imaging Apparatus.
We used a Renishaw InVia Raman microscope optimized for noninvasive in vivo imaging as recently described by our group (14). The microscope included a 785-nm near-infrared laser operating at 60 mW. Light was guided through a collimator onto a series of mirrors that focused the light through an open-field 12× microscope lens. The area of interest on the mouse was illuminated with the laser beam. Scattered light from the illuminated spot was collected with a lens and then was sent through a holographic edge filter to filter out the Rayleigh scattering close to the laser line. The remaining inelastic (Raman) scattered light was then focused through a slit (100-μm width) and dispersed by a diffraction grating (600 lines/mm) onto a cooled CCD detector, where the resulting Raman spectrum was sent to a workstation for further processing.
SERS Nanoparticles.
SERS nanotags were provided by Oxonica Materials; each nanotag comprised a 60-nm diameter Au core coated with a monolayer of Raman-active organic molecule and encapsulated with a 30-nm diameter silica shell, making the entire particle on the order of ≈120 nm (Fig. 1A). The Raman active material varied for each of the 10 particle types that were used in this study. (For more details on the chemical structures of the Raman active material on these SERS nanoparticles, please see Fig. S1.) The encapsulated SERS particle process developed by Oxonica Materials overcomes the inherent variability of SERS by optimizing and controlling the gold colloid used, as well as the addition of the reporter molecule and the formation of the silica encapsulant. With this optimization, any changes in SERS spectral intensity or the relative heights of the major spectral peaks for a given particle across lots is minimized. Any artifacts introduced by these changes are eliminated, as a new reference spectrum is created for each new lot of particles. In addition, reproducibility of these SERS nanoparticles was evaluated in our laboratory, which revealed a 1.9% coefficient of variance among multiple sample measurements (14).
Animal Experiments.
Female 8-week-old nude mice (Charles River Labortories) were used for all Raman spectroscopy studies. All procedures performed on the animals were approved by the university's Institutional Animal Care and Use Committee, and were conducted within the guidelines for humane care of laboratory animals.
Animal Injections.
All s.c. injections consisted of a 1:1 ratio of SERS to Matrigel, a gelatinous protein that resembles the complex extracellular environment found in many tissues. The Matrigel was used to keep the SERS nanoparticles from diffusing quickly out of the skin and showed no inherent Raman spectra. Initial evaluation of all 10 SERS nanoparticles consisted of 10 separate s.c. injections (6.6 fmol of SERS in 5 μl and 5 μl of Matrigel, equivalent to 700 pM). The s.c. injections to evaluate multiplexing various concentrations consisted of four SERS nanoparticle batches injected individually (6.6 fmol of SERS in 5 μl and 5 μl of Matrigel) and then a mixture of all four simultaneously, varying by 70-pM increments and totaling a volume of 10 μl of SERS and 10 μl of Matrigel. Deep-tissue multiplexing analysis consisted of i.v. injections of a mixture of varying batches of SERS nanoparticles. To evaluate multiplexing in vivo with multiple SERS batches, mice were injected via tail vein with 390 fmol of five equally mixed SERS nanoparticle flavors in a 300-μl volume using a 26-gauge needle. To evaluate multiplexing various concentrations of SERS batches in vivo, three mice were injected via tail vein with varying concentrations (100-pM increments) of SERS nanoparticles in a total volume of 500 μl using a 26-gauge needle.
Raman Spectroscopic Imaging in Living Mice.
Raman measurements were performed with a Renishaw microscope system. A semiconductor diode near-infrared laser operating at λ = 785 nm was used as the excitation source with a laser power of 60 mW measured at the surface of the mouse's skin. Raman images were obtained by using a Raman point mapping method. A computer-controlled x–y translation stage was used to raster scan the mouse, creating a spectral image by measuring the Raman spectrum of each individual pixel in the area of interest with a 750-μm or 1-mm step size. Integration times of 1 s per step were used to acquire Raman maps in s.c. models and three s per step for deep-tissue Raman maps. The objective lens used was a ×12 open field in a dimly lit room.
Quantitative Spectral Analysis.
The direct classical least-squares (DCLS) method, also called the linear un-mixing method and K-matrix method, was used in this work to perform a quantitative analysis of Raman spectroscopy (34, 35). DCLS finds the linear combination of spectra from the pure components contained in the sample that most closely matches the Raman spectrum of the sample. Pure component spectra of various SERS nanoparticles were acquired from a pure 3-μl sample aliquoted onto a piece of Parafilm under the microscope. The multiplicative constants derived by the DCLS analysis are proportional to the concentration of the pure components. The DCLS method was chosen because all of the Raman spectra of the pure components, background autofluorescence, and SERS were available and because those components have considerable spectral overlap. This spectral overlap makes it impossible to quantify the contribution of one component independently of the others. For our quantitative analysis, the Nanoplex software (Oxonica Materials) was used (for more details, see SI Text). Before every scan, pure spectra components were taken from the SERS nanoparticles, along with the murine autofluorescence that was used as a background component. The DCLS method gave very accurate results because pure spectral components did not change when mixed together or when injected into a living organism, nor did they change as a function of tissue depth.
Supplementary Material
Acknowledgments.
We thank Dr. Zhenhuan Chi (Rensihaw) for his support with this project. This work was funded in part by National Cancer Institute CCNE U54 CA119367 (S.S.G.), National Institutes of Biomedical Imaging and Bioengineering BRP 5-RO1-EBB000312 (S.S.G.), and In Vivo Cancer Molecular Imaging Centers CMIC P50 CA114747 (S.S.G.). C.Z. received support from National Institutes of Health Training Grant T32 CA09695-15 Advanced Techniques for Cancer Imaging.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.M.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813327106/DCSupplemental.
References
- 1.Xing Y, et al. Bioconjugated quantum dots for multiplexed and quantitative immunohistochemistry. Nat Protoc. 2007;2:1152–1165. doi: 10.1038/nprot.2007.107. [DOI] [PubMed] [Google Scholar]
- 2.Young SH, Rozengurt E. Qdot nanocrystal conjugates conjugated to bombesin or ANG II label the cognate G protein-coupled receptor in living cells. Am J Physiol. 2006;290:C728–C732. doi: 10.1152/ajpcell.00310.2005. [DOI] [PubMed] [Google Scholar]
- 3.Kim JH, et al. Nanoparticle probes with surface enhanced Raman spectroscopic tags for cellular cancer targeting. Anal Chem. 2006;78:6967–6973. doi: 10.1021/ac0607663. [DOI] [PubMed] [Google Scholar]
- 4.Chan WC, Maxwell DJ, Gao X, Bailey RE, Han M, Nie S. Luminescent quantum dots for multiplexed biological detection and imaging. Curr Opin Biotechnol. 2002;13:40–46. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 5.Sun L, Yu C, Irudayaraj J. Surface-enhanced Raman scattering based nonfluorescent probe for multiplex DNA detection. Anal Chem. 2007;79:3981–3988. doi: 10.1021/ac070078z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu KN, et al. Multiplex targeting, tracking, and imaging of apoptosis by fluorescent surface enhanced Raman spectroscopic dots. Bioconjug Chem. 2007;18:1155–1162. doi: 10.1021/bc070011i. [DOI] [PubMed] [Google Scholar]
- 7.Fortina P, Kricka LJ, Surrey S, Grodzinski P. Nanobiotechnology: The promise and reality of new approaches to molecular recognition. Trends Biotechnol. 2005;23:168–173. doi: 10.1016/j.tibtech.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Michalet X, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain KK. Nanomedicine: Application of nanobiotechnology in medical practice. Med Princ Pract. 2008;17:89–101. doi: 10.1159/000112961. [DOI] [PubMed] [Google Scholar]
- 10.Jain KK. Recent advances in nanooncology. Technol Cancer Res Treat. 2008;7:1–13. doi: 10.1177/153303460800700101. [DOI] [PubMed] [Google Scholar]
- 11.Hartman KB, Wilson LJ, Rosenblum MG. Detecting and treating cancer with nanotechnology. Mol Diagn Ther. 2008;12:1–14. doi: 10.1007/BF03256264. [DOI] [PubMed] [Google Scholar]
- 12.Lin S, et al. Quantum dot imaging for embryonic stem cells. BMC Biotechnol. 2007;7:67. doi: 10.1186/1472-6750-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao J, Dragulescu-Andrasi A, Yao H. Fluorescence imaging in vivo: Recent advances. Curr Opin Biotechnol. 2007;18:17–25. doi: 10.1016/j.copbio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Keren S, et al. Noninvasive molecular imaging of small living subjects using Raman spectroscopy. Proc Natl Acad Sci USA. 2008;105:5844–5849. doi: 10.1073/pnas.0710575105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De la Zerda A, et al. Carbon nanotubes as photoacoustic molecular imaging agents in living mice. Nat Nanotechnol. 2008;3:557–562. doi: 10.1038/nnano.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian X, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 17.Zavaleta C, et al. Noninvasive Raman spectroscopy in living mice for evaluation of tumor targeting with carbon nanotubes. Nano Lett. 2008;8:2800–2805. doi: 10.1021/nl801362a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medintz IL, Mattoussi H, Clapp AR. Potential clinical applications of quantum dots. Int J Nanomed. 2008;3:151–167. doi: 10.2147/ijn.s614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doering WE, Piotti ME, Natan MJ, Freeman RG. SERS as a foundation for nanoscale, optically detected biological labels. Adv Mater. 2007;19:3100–3108. [Google Scholar]
- 20.Sha MY, Xu H, Natan MJ, Cromer R. Surface-enhanced Raman scattering tags for rapid and homogeneous detection of circulating tumor cells in the presence of human whole blood. J Am Chem Soc. 2008;130:17214–17215. doi: 10.1021/ja804494m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleischmann M, Hendra PJ, McQuillan AJ. Raman spectra of pyridine adsorbed at a silver electrode. Chem Phys Lett. 1974;26:163–166. [Google Scholar]
- 22.Dijkraaf I, Wester HJ. Peptides, multimers and polymers. Handb Exp Pharmacol. 2008:61–92. doi: 10.1007/978-3-540-77496-9_4. [DOI] [PubMed] [Google Scholar]
- 23.Hoshino A, et al. Use of fluorescent quantum dot bioconjugates for cellular imaging of immune cells, cell organelle labeling, and nanomedicine: Surface modification regulates biological function, including cytotoxicity. J Artif Organs. 2007;10:149–157. doi: 10.1007/s10047-007-0379-y. [DOI] [PubMed] [Google Scholar]
- 24.Kirpotin DB, et al. Antibody targeting of long-circulating lipidic nanoparticles does not increase tumor localization but does increase internalization in animal models. Cancer Res. 2006;66:6732–6740. doi: 10.1158/0008-5472.CAN-05-4199. [DOI] [PubMed] [Google Scholar]
- 25.Smith BR, et al. Real-time intravital imaging of RGD-quantum dot binding to luminal endothelium in mouse tumor neovasculature. Nano Lett. 2008;8:2599–2606. doi: 10.1021/nl080141f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, et al. In vivo biodistribution and highly efficient tumour targeting of carbon nanotubes in mice. Nature Nanotechnol. 2007;2:47–52. doi: 10.1038/nnano.2006.170. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z, et al. Multiplexed multicolor Raman imaging of live cells with isotopically modified single walled carbon nanotubes. J Am Chem Soc. 2008;130:13540–1351. doi: 10.1021/ja806242t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noble CO, et al. Development of ligand-targeted liposomes for cancer therapy. Expert Opin Ther Targets. 2004;8:335–353. doi: 10.1517/14728222.8.4.335. [DOI] [PubMed] [Google Scholar]
- 29.Park JW, Benz CC, Martin FJ. Future directions of liposome- and immunoliposome-based cancer therapeutics. Semin Oncol. 2004;31:196–205. doi: 10.1053/j.seminoncol.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Cai W, et al. Peptide-labeled near-infrared quantum dots for imaging tumor vasculature in living subjects. Nano Lett. 2006;6:669–676. doi: 10.1021/nl052405t. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi H, et al. Simultaneous multicolor imaging of five different lymphatic basins using quantum dots. Nano Lett. 2007;7:1711–1716. doi: 10.1021/nl0707003. [DOI] [PubMed] [Google Scholar]
- 32.Huang Z, Zeng H, Hamzavi I, McLean DI, Lui H. Rapid near-infrared Raman spectroscopy system for real-time in vivo skin measurements. Opt Lett. 2001;26:1782–1784. doi: 10.1364/ol.26.001782. [DOI] [PubMed] [Google Scholar]
- 33.Short MA, et al. Development and preliminary results of an endoscopic Raman probe for potential in vivo diagnosis of lung cancers. Opt Lett. 2008;33:711–713. doi: 10.1364/ol.33.000711. [DOI] [PubMed] [Google Scholar]
- 34.Haaland DM, Easterling RG. Improved sensitivity of infrared spectroscopy by the application of least squares methods. Appl Spectrosc. 1980;34:539–548. [Google Scholar]
- 35.Pelletier MJ. Quantitative analysis using Raman spectroscopy. Appl Spec. 2003;57:20A–42A. doi: 10.1366/000370203321165133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.