Abstract
The intracellular signals that convert fast and slow motor neuron activity into muscle fiber type specific transcriptional programs have only been partially defined. The calcium/calmodulin-dependent phosphatase calcineurin (Cn) has been shown to mediate the transcriptional effects of motor neuron activity, but precisely how 4 distinct muscle fiber types are composed and maintained in response to activity is largely unknown. Here, we show that 4 nuclear factor of activated T cell (NFAT) family members act coordinately downstream of Cn in the specification of muscle fiber types. We analyzed the role of NFAT family members in vivo by transient transfection in skeletal muscle using a loss-of-function approach by RNAi. Our results show that, depending on the applied activity pattern, different combinations of NFAT family members translocate to the nucleus contributing to the transcription of fiber type specific genes. We provide evidence that the transcription of slow and fast myosin heavy chain (MyHC) genes uses different combinations of NFAT family members, ranging from MyHC-slow, which uses all 4 NFAT isoforms, to MyHC-2B, which only uses NFATc4. Our data contribute to the elucidation of the mechanisms whereby activity can modulate the phenotype and performance of skeletal muscle.
Keywords: skeletal muscle, calcineurin, myosin, gene regulation
Slow and fast motor neuron activity triggers both muscle contraction and the transcription of activity-dependent genes, ensuring muscle growth and an adequate composition in fast and slow fibers. This characteristic, in turn, determines the specific performances of each muscle. Fiber type composition depends on developmental cues during embryogenesis, but activity is the major controller of fiber plasticity in the adult (1, 2). Four muscle fiber types have been distinguished based on the kinetic properties of their myosin heavy chain (MyHC) ATPase: 1 type1/slow and 3 type2/fast. Slow fibers express MyHC-slow, are oxidative and fatigue-resistant, whereas fast fibers display a graded range of functional properties according to the scheme 2A↔2X↔2B, with MyHC-2A having the slowest and 2B the fastest shortening velocity, and 2A being oxidative and fatigue-resistant and 2B glycolytic and easily fatigable. The slow and the various fast fiber phenotypes result from the concerted activation of complex gene programs comprising various contractile proteins and metabolic enzymes. Fast and slow motor unit firing patterns are transduced into different gene programs within the myofibers, but the specific effector signaling pathways have been only partially elucidated. Myogenic regulatory factors such as MyoD and myogenin are differentially expressed in fast and slow fibers (3), and a MyoD-binding E-box is necessary for the expression of MyHC-2B (4). Also, dephosphorylation of MyoD occurs in response to fast nerve activity and drives the expression of fast MyHCs (5). Myocyte enhancer factor (MEF)2 has also been linked to a slow fiber phenotype, because inhibition of MEF2 suppresses the formation of slow fibers (6). The phosphatase calcineurin (Cn) and its target, the nuclear factor of activated T cells (NFAT) (7, 8), have been linked to slow fibers, because Cn inhibition with the immunosuppressive drugs Cyclosporin A and FK506 results in a shift from a slow to a fast phenotype (9, 10). Also, inhibition of Cn/NFAT interaction by using the peptide VIVIT causes a slow to fast fiber transition (11), and transgenic mice with muscle-specific overexpression of Cn exhibit increased slow fiber content (12). During development, the slow fiber phenotype depends on the transcription factor Sox6 (13) and independent of Cn, yet becomes strictly dependent on Cn in the postnatal period, when motor neuron activity becomes prevalent (14). NFAT is a family of 4 transcription factors, NFATc1, c2, c3, and c4, regulated by Cn through dephosphorylation of multiple serine/threonine residues, leading to nuclear translocation and eventually DNA binding. Targeted disruption of NFATc1, c2, and c3 has demonstrated that these factors are fundamental in lymphocytes, but less information is available on their specific roles in muscle. NFATc1 knockout is embryonic lethal due to defects in cardiogenesis (15), whereas the knockouts of NFATc2 and c3 result in muscle atrophy and only minor, if any, alterations in fiber composition (16, 17). Mice with targeted disruption of NFATc4 exhibit no known phenotype (18).
The aim of the present study was to determine whether different NFAT family members have specific roles in fiber type specification in adult skeletal muscle. We have carried out an in vivo analysis of NFAT isoform-specific silencing by RNAi, by transfecting adult skeletal muscle in situ in living rats. We propose that graded activation of different NFAT family members is an important determining factor for activity-dependent muscle fiber specification.
Results
All NFAT Isoforms Are Expressed in Rat Skeletal Muscle.
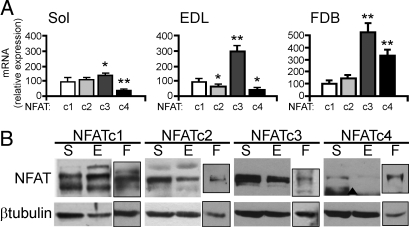
We analyzed the expression of NFAT family members in rat skeletal muscle by using qPCR. We compared a slow muscle, soleus, which in rat is composed of ≈90% type I/slow and 10% fast fibers, with a fast muscle, Extensor digitorum longus (EDL), which contains the opposite proportion of fast and slow fibers (Fig. S1). All four NFAT transcripts were detected, with differences in relative expression between soleus and EDL, particularly for NFATc3, which is more abundant in EDL (Fig. 1A). To confirm that the 4 NFATs are expressed in muscle fibers, rather than in nonmuscle cells present in crude extracts, we analyzed adult isolated muscle fibers from Flexor digitorum brevis (FDB), which are virtually devoid of contamination from nonmuscle cells (19). Fig. 1A shows that all 4 NFAT isoforms are expressed in adult muscle in vivo and ex vivo. The presence of all 4 NFATs was confirmed at the protein level by Western blotting (WB). All NFAT proteins were expressed in soleus, EDL, and isolated muscle fibers (Fig. 1B). Multiple bands likely indicate the presence of different isoforms and phosphorylation states as previously observed (20).
Fig. 1.
Expression and localization of NFAT isoforms in skeletal muscle. (A) Quantitative PCR of Soleus (Sol), EDL, and isolated fibers from FDB. Results are expressed as percentage of NFATc1 expression. (B) WB with antibodies specific for NFAT isoforms of total muscle extracts from Soleus (S), EDL (E), and FDB fibers (F). ▴, aspecific band.
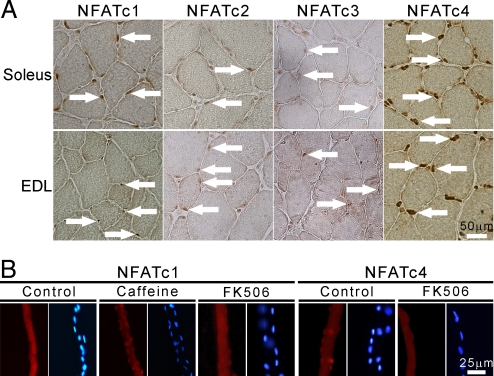
To localize all NFATs in adult muscle, we analyzed soleus and EDL by immunohistochemistry (IHC) with NFAT isoform-specific antibodies. The staining specificity was assessed by overexpressing in vivo each NFAT isoform fused to GFP and staining serial sections with all NFAT antibodies (Fig. S2a, Table S1). NFATc1 was localized in the nucleus in most fibers in soleus, whereas a very low nuclear signal was detected in EDL; thus, confirming our previous observations with NFATc1-GFP (21). NFATc2 and NFATc3 showed a low, but detectable nuclear signal both in soleus and EDL. Surprisingly, NFATc4 showed a very strong nuclear accumulation in both muscles (Fig. 2A), clearly detected also in longitudinal sections (Fig. S2b). Some interstitial nuclei were also stained with all NFAT antibodies.
Fig. 2.
Localization of NFAT isoforms in adult muscle. (A) Cross-sections of soleus and EDL stained with antibodies specific for all 4 NFAT isoforms as indicated. White arrows indicate some positive nuclei. (B) Cultured muscle fibers from FDB stained with anti-NFATc1 and anti-NFATc4 Abs, and corresponding counterstaining of nuclei with Hoechst; treatments, FK506 (1 μM), caffeine (10 mM), 2 h.
We also analyzed the nucleocytoplasmic distribution of each NFAT by cell fractionation (Fig. S2c). NFATc1, c2, and c3 could be detected both in nuclear and cytosolic fractions in both muscles, with NFATc1 being more nuclear in soleus. However, NFATc4 was highly enriched in the nuclei in both muscles, and showed only a minor cytosolic signal, confirming the IHC data. The difference in nuclear NFATc1 between soleus and EDL was stronger when analyzed by IHC than by WB. However, it should be kept in mind that also nonmuscle nuclei are abundant in nuclear fractions of skeletal muscle, and they likely give a background signal reducing the difference. To confirm the nuclear localization of NFATc4, we stained adult muscle fibers in culture with anti-NFATc4 Ab. Specific staining of NFATc4 in the nuclei of resting FDB fibers was confirmed by the decreased staining on treatment with the Cn inhibitor FK506. NFATc1 was cytosolic in resting FDB fibers in culture as previously reported (19), but translocated to the nucleus in response to caffeine (Fig. 2B).
Fiber Type Specific Transcriptional Effects of NFAT Isoforms.
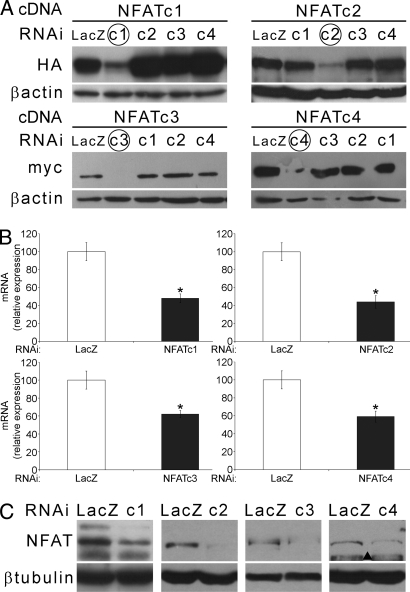
We selectively knocked down the expression of each NFAT isoform by RNAi to define their role in adult muscle. The selected siRNA sequences were cloned in the pSuper vector and proven to be specific only for the target NFAT while leaving all other isoforms unaffected (Fig. 3A; Fig. S3a). Transfection of the siRNAs in adult muscle caused down-regulation of endogenous NFATs both at the RNA and protein level (Fig. 3 B and C), which was also detected by a specific NFAT sensor (Fig. S3b).
Fig. 3.
RNAi of individual NFATs. (A) WB showing specific down-regulation of the target NFAT (HA or myc-tagged as indicated), but not of other isoforms obtained by cotransfection of NFAT cDNA and isoform-specific siRNAs in HEK293 cells. Down-regulation of endogenous NFATs in muscles transfected with NFAT isoform-specific and control siRNAs is shown by qPCR at the RNA (B) and by WB at the protein level (C). ▴, aspecific band.
We asked whether the 4 NFATs may be part of the machinery that translates different patterns of neuronal activity into fiber type specific gene programs. To verify this hypothesis, we used reporter constructs consisting of portions (>0.8 kb) of the promoters of the 4 adult MyHC isoforms, namely slow, fast-oxidative 2A, −2X, and fast-glycolytic 2B, respectively, linked to luciferase. These constructs are fiber type specific and activity-dependent (20, 22, 23). MyHCs are the main determinants of the contractile properties, and their fiber-specific expression ultimately defines fiber type. These constructs allow detection of rapid variations in MyHC gene transcriptional regulation. We monitored the activity of MyHC reporters cotransfected in adult muscles with the NFAT isoform-specific siRNAs.
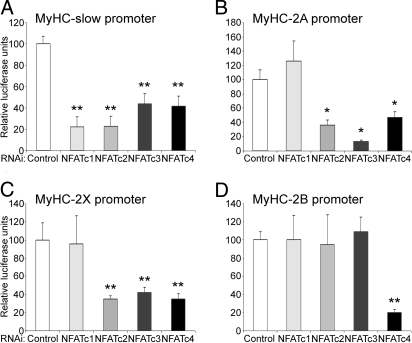
Fig. 4A shows the effect of NFAT gene silencing on the MyHC-slow promoter, which depends on slow nerve activity (23). Interestingly, the expression of MyHC-slow was controlled by NFAT in a more complex way than previously thought, because all 4 NFAT isoform-specific siRNA showed a strong inhibitory effect. This result suggests that the 4 NFAT isoforms are used to modulate the expression of MyHC-slow in adult muscle, and that their role is nonredundant under these conditions.
Fig. 4.
Role of individual NFATs in fiber type specific MyHC expression. The promoters of the MyHC isoforms linked to luciferase were cotransfected with siRNAs specific for either GFP or LacZ (control) or for the indicated NFAT isoforms. All experiments are means of at least 4 individual muscles per data point, repeated with 2 siRNA sequences.
We then analyzed the expression of the 3 fast isoforms of MyHC in EDL. MyHC-2A, which characterizes fast-oxidative fibers, does not need NFATc1 for its expression, as indicated by the fact that the siRNA against NFATc1 did not influence the expression of the MyHC-2A promoter-reporter construct. However, NFATc2, c3 and, c4 seem to be fundamental determinants of the expression of MyHC-2A, because the corresponding siRNAs were very powerful transcriptional inhibitors (Fig. 4B). Expression of MyHC-2X appears to be controlled by the same combination of NFAT family members needed for MyHC-2A (Fig. 4C). A difference was seen in the relative influence of NFATc3, which was stronger on MyHC-2A than 2X (compare Fig. 4 B and C). The observation that MyHC-2A and -2X are controlled by the same combination of NFATs is not unexpected, because rat EDL contains ≈20% of hybrid 2A/2X fibers expressing both MyHCs (24).
We finally analyzed the expression of MyHC-2B, which characterizes the fastest and fully glycolytic fibers. Our results indicate that this MyHC is under a unique regulatory control by NFAT, because only the NFATc4-specific siRNA had an inhibitory effect, whereas the siRNAs for the other isoforms yielded results indistinguishable from control (Fig. 4D). Thus, NFATc4 is the only necessary isoform for the expression of MyHC-2B in skeletal muscle in vivo.
Together, these results suggest that: (i) NFATc1 is a specific determinant of slow muscle gene expression; (ii) different combinations of NFAT family members are implicated in the activity-dependent transcription of the various MyHC isoforms; and (iii) there is a graded nuclear recruitment of NFAT transcription factors modulating the promoters of activity-dependent genes, ranging from MyHC-slow, which requires all 4 NFATs, to MyHC-2B, which uses only NFATc4. The regulation of MyHC promoters by NFAT may also be indirect, because mutation of a NFAT site in a short form of the MyHC-slow promoter did not abolish the sensitivity to the inhibitor peptide VIVIT (Fig. S4).
Activity-Dependence of NFAT Isoforms.
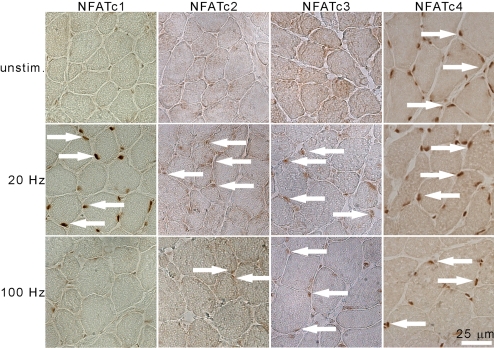
The nucleocytoplasmic distribution of NFAT is an indicator of its activation state, because nuclear accumulation is the result of dephosphorylation and activation by Cn. If NFAT isoforms are regulators of activity-dependent muscle gene programs, then their localization must be different in fast and slow muscles (Fig. 2), and differentially sensitive to the electrical activity of the plasma membrane that characterizes fast and slow muscle fibers. To address this issue, we analyzed the nucleocytoplasmic distribution of NFAT isoforms in response to electrical stimulation of the peroneal nerve in EDL. We used 2 stimulation patterns (25): (i) slow pattern (200 pulses, 20 Hz every 30 s); and (ii) fast high amount pattern, possibly corresponding to the firing patterns of motor neurons innervating 2A/2X fibers (25 pulses, 100 Hz every 60 s). The slow pattern characterizes slow motor units, which, in postural muscles such as soleus, have been estimated to be active ≈30% of the day. The fast pattern resembles the behavior of fast motor units and generates more force per twitch, but is comparatively a “low amount” type of activity (26). Slow/20 Hz stimulation readily induced the translocation of NFATc1, c2, and c3 to the nucleus (see arrows in Fig. 5A), whereas NFATc4 is already nuclear. Therefore, under this condition, all 4 NFAT isoforms are accumulated in the nucleus, which neatly complements the observation that all 4 NFATs are required for the expression of MyHC-slow. However, an EDL stimulated at 100 Hz showed a nuclear accumulation of NFATc3 and c4, and, in a fraction of nuclei, of NFATc2. This combination of NFAT isoforms found in the nucleus under 100-Hz stimulation is similarly complemented by results from the RNAi experiments for MyHC-2A/-2X (Fig. 4). It is intriguing that NFATc3 shows a clearly distinct activity-dependence from NFATc1, in that it translocates to the nucleus in response both to a slow and to a fast-like pattern of activity (Fig. 5; Fig. S5).
Fig. 5.
Nucleocytoplasmic shuttling of NFAT isoforms in response to electrical activity. IHC staining of endogenous NFAT isoforms in cross-sections of EDL electrically stimulated with patterns of impulses at 20 and 100 Hz as indicated, for 2 h. Contralateral EDL from the anesthetized animal was used as unstimulated control (unstim). White arrows indicate NFAT-positive nuclei.
Effects of Silencing of NFAT Isoforms on Endogenous Genes.
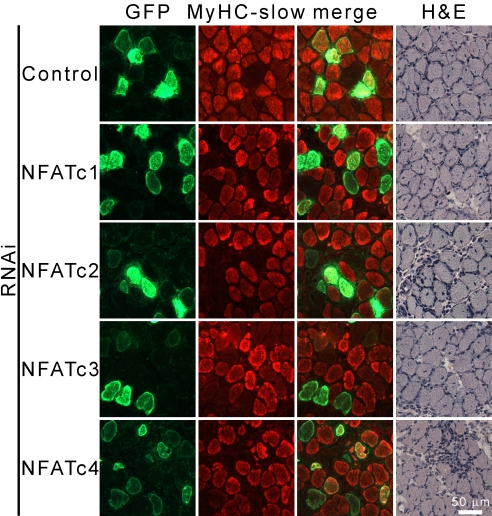
To prove that NFAT isoforms exert the same effects on endogenous genes, we analyzed the expression of MyHC-slow in regenerating muscle. An intramuscular injection of bupivacain was used to induce muscle necrosis, followed by a conservative regeneration recapitulating myogenesis within a few days. MyHCs are, thus, produced ex novo, which circumvents the problem of preexisting MyHC masking the effect of siRNAs. When regenerating soleus was transfected with the siRNA of the 4 NFAT isoforms, a clear inhibition in the expression of MyHC slow was seen in all cases (Fig. 6), whereas a control (LacZ-specific) siRNA had no effect. Not all fibers transfected with the NFAT-specific siRNAs were completely negative for MyHC-slow (see quantification in Fig. S6). This result may be due to different expression levels of the siRNA in the fibers. We obtained similar results with the general NFAT inhibitor peptide VIVIT (11). Interestingly, fibers in which MyHC-slow expression was strongly reduced showed a relative up-regulation of MyHC-2A, indicating fiber type transition, rather than just the inhibition of MyHC-slow (Fig. S6). Brightfield images show that morphology was comparable in muscles transfected with control and NFAT-specific siRNAs, indicating that down-regulation of MyHC was not due to inhibition of regeneration. These observations confirm the data obtained with the MyHC promoter constructs and further substantiate our hypothesis that MyHC gene expression and muscle fiber type depend on the specific nuclear import of NFAT family members in response to motor neuron activity.
Fig. 6.
All NFAT isoforms are necessary for the expression of MyHC-slow in regenerating muscle. Regenerating soleus was cotransfected with NFAT isoform-specific siRNAs and SNAP-GFP at day 3 after myotoxic damage induced with bupivacaine. Analyses were performed 1 week later. Serial cross-sections were stained with antibodies specific for MyHC-slow and GFP or with hematoxylin/eosin (H&E).
Discussion
Cn is an important regulator of muscle differentiation in cultured muscles cells and of fiber type diversification in adult muscle. However, the role of the different NFAT isoforms in these processes has been comparatively less investigated, and diverging results were reported (see also SI Materials Methods). Abbott et al. (27) found that thapsigargin causes NFATc3 nuclear translocation in myoblasts, but not myotubes, whereas NFATc1 and c2 translocate to the nucleus in myotubes, but not in myoblasts. In contrast, Delling et al. (28) reported that both thapsigargin and activated Cn cause nuclear translocation of NFATc3, but not c1 and c2 in myoblasts, and that NFATc3, but not c1 or c4, enhanced myogenic differentiation induced by MyoD. In adult muscle fibers, translocation of NFATc1 is induced by slow, but not fast electrical stimulation (19, 21), and the NFAT role in activity-dependent fast-to-slow fiber type switching was demonstrated by using transfection in vivo with constitutively active NFATc1 and with the NFAT inhibitor VIVIT (11). However, the role of the NFATc2, c3, and c4 has not been defined. Here, we examine the role of all 4 NFAT isoforms in adult skeletal muscle, and show that (i) nucleocytoplasmic localization of NFAT family members is differentially sensitive to the electrical activity of the plasma membrane, and that (ii) different combinations of NFAT family members are necessary to specify transcription not only of slow, but also of fast MyHCs.
NFATs are expressed in many embryonic and adult tissues, and examples of cooperation between NFAT family members both in a synergistic (29, 30) and antagonistic way (8, 31) have been reported. Some controversy exists as to whether all NFAT isoforms are expressed in skeletal muscle. The presence of all 4 NFATs has been shown at the RNA (32) and protein level (33). However, some studies could not detect the expression of NFATc3 and c4 (20, 27). Here, we document the expression of all 4 NFATs in adult skeletal muscle, and explore their individual function in activity-dependent gene regulation.
Our data show that transcription of a slow muscle-specific gene, MyHC-slow, is controlled in a cooperative way by all 4 NFAT family members in vivo. This result was obtained both by using a MyHC-slow reporter construct (Fig. 4A) and by analyzing endogenous MyHC (Fig. 6). We have previously shown that a constitutively active NFATc1 is sufficient for the induction of MyHC-slow, and that it also represses MyHC-2B (11). We hypothesize that nuclear import of NFATc1, which is driven by slow nerve activity, shifts the specificity of the transcriptional machinery toward slow muscle genes by adding a necessary combinatorial partner to the preexisting assembly of NFATc2, c3, and c4, which specifies fast gene transcription (Fig. 4 B–D).
It remains to be established whether the cooperative interactions between NFAT family members occur by protein–protein interaction between NFATs or with other transcriptional partners, such as MEF2, AP1 (34), or MyoD (35). We show here that NFATs may act indirectly, because mutation of 1 NFAT consensus sequence in a short stretch of the MyHC promoter does not change its sensitivity to the inhibitor peptide VIVIT (Fig. S4). NFAT could act by protein–protein interaction and/or through the expression of other transcription factors that bind to the MyHC promoters. Indeed, NFAT transcription factors can form heterodimers, due to the structural homology of their DNA binding domain to the Rel domain, but also homodimers at quasi-palindromic sites that resemble NF-κB binding motifs (36, 37). Also, it is possible that different NFAT elements in muscle gene promoters preferentially bind one or another NFAT isoform (7).
Intriguingly, our data show that a common trait of all fast MyHC genes is their independence from NFATc1. In the presence of siRNAs specific for NFATc1, the expression of MyHC-2A, -2X, and -2B is indistinguishable from control (Fig. 4 B–D). It is tempting to speculate that the lack of nuclear NFATc1, and of its repressive action on fast genes, is also a generally permissive state for the expression of fast MyHCs (Fig. 4). Indeed, NFATc1 represses troponin I-fast (TnIf) in slow muscle fibers by binding to a single NFAT consensus sequence. However, in that study, silencing of NFATc1 results in ectopic expression of TnIf in a slow muscle (38). We do not observe similar ectopic effects for MyHCs, which may be because, in contrast to the TnIf enhancer, the promoters that we analyze contain numerous NFAT consensus sequences.
We also observe differences among fast fibers in the requirement for NFAT isoforms. In particular, the expression of MyHC-2A and -2X is inhibited by siRNAs for NFATc2, c3, and c4 (Fig. 4 B and C), whereas fast-glycolytic MyHC-2B is only inhibited by siRNAs for NFATc4 (Fig. 4D). We hypothesize that fiber type transitions, occurring in the sequence IIB↔IIX↔IIA↔type I, are paralleled by a graded recruitment of NFAT isoforms ranging from NFATc4 alone to a combination of 4 members (Fig. S7). Such a combinatorial mechanism requires that NFATs have graded sensitivity to nerve activity. Indeed, the data in Fig. 5 provide evidence that NFAT family members respond differently to activity. When EDL is stimulated with a 20-Hz pattern, similar to that of slow motor neurons, nuclear accumulation of all 4 NFATs can be observed. Conversely, on fast-like stimulation at 100 Hz, NFATc1 is cytosolic, whereas the remaining isoforms are nuclear and, presumably, contributing to the transcription of fast MyHCs. In terms of calcium increases, fast-like stimulation causes higher peak calcium concentrations than slow-like stimulation (39, 40). However, slow stimulation is essentially continuous (26), and the calcium reuptake phase is presumably not completed before the beginning of the next transient. As a result, summation occurs in slow (but not fast) fibers leading to a low, but sustained increase in average calcium concentration, which is known to strongly activate Cn (41). Indeed, NFATs have been shown to act as integrators of calcium signals and to decode variations in the frequency of calcium spikes (42, 43). Our results suggests that NFATc4, c3, and c2 are transcriptionally active in fast muscles, i.e., in response to less frequent calcium transients and, presumably, lower Cn activity.
Our observation that NFATc4 is nuclear in skeletal muscle under all activity conditions analyzed was unexpected. One could speculate that NFATc4 is implicated in the default 2X/2B-fiber-based transcriptional program. The existence of a default-fast program independent of nerve activity has been demonstrated (44) by showing that denervation of newborn rats blocks the expression of MyHC-slow, but not of -2X and -2B, and the same is true in regenerating muscle (45). However, the signals that control this activity-dependent switch are still unknown. We propose that NFATc4 could be a candidate, because it is mostly nuclear in skeletal muscle and controls the expression of fast genes. We did observe a predominantly cytosolic localization of NFATc4 in other tissues with our staining protocols (Fig. S2e), as reported. Nuclear accumulation of NFATc4 in skeletal muscle might reflect the combination of rapid import and/or slow export. Differences in import/export mechanisms between NFAT isoforms have been documented (46). NFATc1 is cytosolic in cerebellar granules in the presence of growth factors and KCl, whereas c4 accumulates in the nucleus under these conditions (47). It was also reported that NFATc4 remains in the nucleus of hippocampal neurons after a brief calcium pulse, whereas in lymphocytes, it needs a sustained calcium signal (48). These observations point to a tissue-specificity in the localization and activation of NFAT isoforms. Also, nuclear residence of NFATc4 is controlled not only by the phosphatase Cn, but also by the kinase RSK2, which directly causes NFATc4 nuclear import and stabilizes its interaction with DNA (49). Importantly, a specific nuclear localization of NFATc4, but not other isoforms under resting conditions, has also been shown in cultured human myotubes (50).
In conclusion, we have found one of the mechanisms whereby different patterns of activity are converted into muscle fiber type specific gene programs. We propose that specificity is achieved by sequentially recruiting NFAT isoforms to the nucleus in predictable combinations (Fig. S7). A fundamental open question remains as to how different patterns of activity, and presumably different levels of Cn activity, can specifically activate individual NFATs (51). It has been shown that the kinetics of NFAT dephosphorylation and nuclear residence vary between individual NFATs (41). Different NFAT isoforms undergo calcium-induced nuclear translocation at specific stages of myoblast differentiation, suggesting not only specificity among NFAT isoforms in gene regulation, but, because these isoforms are all expressed during all stages, also in their import/export mechanism (27).
Also, it has been demonstrated that individual NFATs can bind specific transcriptional interactors (9, 52–55), which could, in turn, affect their kinetics of nuclear residence. The complex relationship between calcium transients after fast and slow nerve activity, Cn activation, and nuclear import of individual NFATs awaits further mechanistic definition.
Materials and Methods
In Vivo Transfection and Electrostimulation.
In vivo transfection was carried out essentially as described (21); also, see SI Materials Methods. All experimental protocols were reviewed and supervised by the Animal Care Committee of the University of Padova.
Cell Culture and Transfection.
HEK293 cells were maintained in culture in DMEM with 10% fetal bovine serum in a humidified incubator and transfected by using Lipofectamine2000 (Invitrogen) as recommended by the manufacturer.
RNAi-Mediated Gene Silencing.
We have selected at least 2 sequences with high silencing efficiency for each gene and performed every experiment with both of them. As control, to avoid reported off-target effects (56), we used 2 siRNAs, designed to silence GFP and lacZ, respectively. The procedures and sequences used can be found in SI Materials Methods.
Dual Luciferase Assay.
A commercially available dual luciferase assay system was used (E1960; Promega).
FDB Fiber Preparation.
FDB muscles were excised from 4- to 6-weeks-old Wistar rats and dissociated by using collagenase as described (19). Fibers were processed immediately for RNA or protein isolation. For immunolabeling, vital fibers were counted by trypan blue exclusion and plated for 24 h on laminin-coated culture dishes.
IHC and Microscopy.
NFAT antibodies are listed in Table S1. These mouse mAbs produced in our laboratory were used: BA-D5 (MyHC-slow), SC-71 (MyHC-2A). Anti-HA 16B12 (Covance), and anti-myc 9E10 (Roche). Images were collected with an epifluorescence Leica DMR microscope equipped with a Leica DFC300 FX digital charge-coupled device.
Quantitative Real-Time PCR.
RNA was purified (SV Total RNA Isolation; Promega) and characterized by electrophoresis (Agilent); 400 ng of RNA was converted to cDNA by using random hexamers and SuperScript II (Invitrogen). Amplification was carried out in triplicates with an IQ5 real-time PCR system (Bio-Rad) by using SYBR green and a standard 2-step protocol. Experiments were performed at least twice, on 2 individually prepared cDNAs. Primers are listed in SI Materials Methods.
Data Analysis.
Data are expressed as mean ± SEM (error bars). Comparisons were made by using a t test, with P < 0.05 being considered statistically significant. *, P < 0.05; **, P < 0.01.
Supplementary Material
Acknowledgments.
We thank A. Rao (Harvard Medical School, Boston), N.A. Clipstone (Loyola University Chicago, Maywood, IL), K.G. Farrance (University of Maryland School of Medicine, Baltimore), K. Esser (University of Kentucky, Lexington, KY), S. Swoap (Williams College, Williamstown, MA), S. Williams (Duke University Medical School, Durham, NC), J. Molkentin (Children's Hospital Medical Center, University of Cincinnati, Cincinnati), R.N. Kitsis (Albert Einstein College of Medicine, New York), K. Hasegawa (Kyoto Medical Center, Kyoto) for plasmids; G.R. Crabtree (Stanford University, Stanford, CA), V. Mouly (Université Pierre et Marie Curie-Paris 06, Paris), and T. Pozzan (University of Padova, Padova, Italy) for antibodies; and L. Agatea and S. Furlan for their precious help. This work was supported by the European Commission Grants NoE MYORES, LSHG-CT-2004-511978, and IP EXGENESIS, LSHM-CT-2004-005272; the Ministero dell'Universitá e della Ricerca Scientifica e Technologica of Italy Grant PRIN 2006; and the Telethon Grant GGP04227, Agenzia Spaziale Italiana Osteoporosis and Muscular Atrophy Project.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812911106/DCSupplemental.
References
- 1.Buckingham M, et al. The formation of skeletal muscle: from somite to limb. J Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schiaffino S, Sandri M, Murgia M. Activity-dependent signaling pathways controlling muscle diversity and plasticity. Physiology. 2007;22:269–278. doi: 10.1152/physiol.00009.2007. [DOI] [PubMed] [Google Scholar]
- 3.Hughes SM, et al. Selective accumulation of MyoD and myogenin mRNAs in fast and slow adult skeletal muscle is controlled by innervation and hormones. Development. 1993;118:1137–1147. doi: 10.1242/dev.118.4.1137. [DOI] [PubMed] [Google Scholar]
- 4.Wheeler MT, Snyder EC, Patterson MN, Swoap SJ. An E-box within the MHC IIB gene is bound by MyoD and is required for gene expression in fast muscle. Am J Physiol. 1999;276:C1069–C1078. doi: 10.1152/ajpcell.1999.276.5.C1069. [DOI] [PubMed] [Google Scholar]
- 5.Ekmark M, Rana ZA, Stewart G, Hardie DG, Gundersen K. De-phosphorylation of MyoD is linking nerve-evoked activity to fast myosin heavy chain expression in rodent adult skeletal muscle. J Physiol. 2007;584:637–650. doi: 10.1113/jphysiol.2007.141457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potthoff MJ, et al. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J Clin Invest. 2007;117:2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 8.Wu H, Peisley A, Graef IA, Crabtree GR. NFAT signaling and the invention of vertebrates. Trends Cell Biol. 2007;17:251–260. doi: 10.1016/j.tcb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Chin ER, et al. A Cn-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serrano AL, et al. Cn controls nerve activity-dependent specification of slow skeletal muscle fibers but not muscle growth. Proc Natl Acad Sci USA. 2001;98:13108–13113. doi: 10.1073/pnas.231148598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCullagh KJ, et al. NFAT is a nerve activity sensor in skeletal muscle and controls activity-dependent myosin switching. Proc Natl Acad Sci USA. 2004;101:10590–10595. doi: 10.1073/pnas.0308035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naya FJ, et al. Stimulation of slow skeletal muscle fiber gene expression by Cn in vivo. J Biol Chem. 2000;275:4545–4548. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- 13.Hagiwara N, Yeh M, Liu A. Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Dev Dyn. 2007;236:2062–2076. doi: 10.1002/dvdy.21223. [DOI] [PubMed] [Google Scholar]
- 14.Oh M, et al. Cn is necessary for the maintenance but not embryonic development of slow muscle fibers. Mol Cell Biol. 2005;25:6629–6638. doi: 10.1128/MCB.25.15.6629-6638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Pompa JL, et al. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature. 1998;392:182–186. doi: 10.1038/32419. [DOI] [PubMed] [Google Scholar]
- 16.Horsley V, et al. Regulation of the growth of multinucleated muscle cells by an NFATC2-dependent pathway. J Cell Biol. 2001;153:329–338. doi: 10.1083/jcb.153.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kegley KM, Gephart J, Warren GL, Pavlath GK. Altered primary myogenesis in NFATC3(−/−) mice leads to decreased muscle size in the adult. Dev Biol. 2001;232:115–126. doi: 10.1006/dbio.2001.0179. [DOI] [PubMed] [Google Scholar]
- 18.Graef IA, Chen F, Chen L, Kuo A, Crabtree GR. Signals transduced by Ca(2+)/Cn and NFATc3/c4 pattern the developing vasculature. Cell. 2001;105:863–875. doi: 10.1016/s0092-8674(01)00396-8. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Cseresnyes Z, Randall WR, Schneider MF. Activity-dependent nuclear translocation and intranuclear distribution of NFATc in adult skeletal muscle fibers. J Cell Biol. 2001;155:27–39. doi: 10.1083/jcb.200103020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swoap SJ, et al. The Cn-NFAT pathway and muscle fiber-type gene expression. Am J Physiol Cell Physiol. 2000;279:C915–C924. doi: 10.1152/ajpcell.2000.279.4.C915. [DOI] [PubMed] [Google Scholar]
- 21.Tothova J, et al. NFATc1 nucleocytoplasmic shuttling is controlled by nerve activity in skeletal muscle. J Cell Sci. 2006;119:1604–1611. doi: 10.1242/jcs.02875. [DOI] [PubMed] [Google Scholar]
- 22.Allen DL, Weber JN, Sycuro LK, Leinwand LA. Myocyte enhancer factor-2 and serum response factor binding elements regulate fast Myosin heavy chain transcription in vivo. J Biol Chem. 2005;280:17126–17134. doi: 10.1074/jbc.M501207200. [DOI] [PubMed] [Google Scholar]
- 23.Murgia M, et al. Ras is involved in nerve-activity-dependent regulation of muscle genes. Nat Cell Biol. 2000;2:142–147. doi: 10.1038/35004013. [DOI] [PubMed] [Google Scholar]
- 24.Schiaffino S, Murgia M, Serrano AL, Calabria E, Pallafacchina G. How is muscle phenotype controlled by nerve activity? Ital J Neurol Sci. 1999;20:409–412. doi: 10.1007/s100720050060. [DOI] [PubMed] [Google Scholar]
- 25.Ausoni S, Gorza L, Schiaffino S, Gundersen K, Lomo T. Expression of myosin heavy chain isoforms in stimulated fast and slow rat muscles. J Neurosci. 1990;10:153–160. doi: 10.1523/JNEUROSCI.10-01-00153.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hennig R, Lomo T. Firing patterns of motor units in normal rats. Nature. 1985;314:164–166. doi: 10.1038/314164a0. [DOI] [PubMed] [Google Scholar]
- 27.Abbott KL, Friday BB, Thaloor D, Murphy TJ, Pavlath GK. Activation and cellular localization of the cyclosporine A-sensitive transcription factor NF-AT in skeletal muscle cells. Mol Biol Cell. 1998;9:2905–2916. doi: 10.1091/mbc.9.10.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delling U, et al. A Cn-NFATc3-dependent pathway regulates skeletal muscle differentiation and slow myosin heavy-chain expression. Mol Cell Biol. 2000;20:6600–6611. doi: 10.1128/mcb.20.17.6600-6611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cante-Barrett K, Winslow MM, Crabtree GR. Selective role of NFATc3 in positive selection of thymocytes. J Immunol. 2007;179:103–110. doi: 10.4049/jimmunol.179.1.103. [DOI] [PubMed] [Google Scholar]
- 30.Kaminuma O, et al. Differential contribution of NFATc2 and NFATc1 to TNF-alpha gene expression in T cells. J Immunol. 2008;180:319–326. doi: 10.4049/jimmunol.180.1.319. [DOI] [PubMed] [Google Scholar]
- 31.Robbs BK, Cruz AL, Werneck MB, Mognol GP, Viola JP. Dual Roles for NFAT Transcription Factors as Oncogenes and Tumor Suppressors. Mol Cell Biol. 2008;28:7168–7181. doi: 10.1128/MCB.00256-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoey T, Sun YL, Williamson K, Xu X. Isolation of two new members of the NF-AT gene family and functional characterization of the NF-AT proteins. Immunity. 1995;2:461–472. doi: 10.1016/1074-7613(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 33.Parsons SA, Wilkins BJ, Bueno OF, Molkentin JD. Altered skeletal muscle phenotypes in Cn Aalpha and Abeta gene-targeted mice. Mol Cell Biol. 2003;23:4331–4343. doi: 10.1128/MCB.23.12.4331-4343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meissner JD, Umeda PK, Chang KC, Gros G, Scheibe RJ. Activation of the beta myosin heavy chain promoter by MEF-2D, MyoD, p300, and the Cn/NFATc1 pathway. J Cell Physiol. 2007;211:138–148. doi: 10.1002/jcp.20916. [DOI] [PubMed] [Google Scholar]
- 35.Armand AS, et al. Cooperative synergy between NFAT and MyoD regulates myogenin expression and myogenesis. J Biol Chem. 2008;283:29004–29010. doi: 10.1074/jbc.M801297200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giffin MJ, et al. Structure of NFAT1 bound as a dimer to the HIV-1 LTR kappa B element. Nat Struct Biol. 2003;10:800–806. doi: 10.1038/nsb981. [DOI] [PubMed] [Google Scholar]
- 37.Bates DL, et al. Crystal structure of NFAT bound to the HIV-1 LTR tandem kappaB enhancer element. Structure. 2008;16:684–694. doi: 10.1016/j.str.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rana ZA, Gundersen K, Buonanno A. Activity-dependent repression of muscle genes by NFAT. Proc Natl Acad Sci USA. 2008;105:5921–5926. doi: 10.1073/pnas.0801330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chin ER, Allen DG. The role of elevations in intracellular [Ca2+] in the development of low frequency fatigue in mouse single muscle fibers. J Physiol. 1996;491:813–824. doi: 10.1113/jphysiol.1996.sp021259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westerblad H, Allen DG. Changes of myoplasmic calcium concentration during fatigue in single mouse muscle fibers. J Gen Physiol. 1991;98:615–635. doi: 10.1085/jgp.98.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feske S, Draeger R, Peter HH, Eichmann K, Rao A. The duration of nuclear residence of NFAT determines the pattern of cytokine expression in human SCID T cells. J Immunol. 2000;165:297–305. doi: 10.4049/jimmunol.165.1.297. [DOI] [PubMed] [Google Scholar]
- 42.Tomida T, Hirose K, Takizawa A, Shibasaki F, Iino M. NFAT functions as a working memory of Ca2+ signals in decoding Ca2+ oscillation. EMBO J. 2003;22:3825–3832. doi: 10.1093/emboj/cdg381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colella M, et al. Ca2+ oscillation frequency decoding in cardiac cell hypertrophy: role of Cn/NFAT as Ca2+ signal integrators. Proc Natl Acad Sci USA. 2008;105:2859–2864. doi: 10.1073/pnas.0712316105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butler-Browne GS, Bugaisky LB, Cuenoud S, Schwartz K, Whalen RG. Denervation of newborn rat muscle does not block the appearance of adult fast myosin heavy chain. Nature. 1982;299:830–833. doi: 10.1038/299830a0. [DOI] [PubMed] [Google Scholar]
- 45.Jerkovic R, Argentini C, Serrano-Sanchez A, Cordonnier C, Schiaffino S. Early myosin switching induced by nerve activity in regenerating slow skeletal muscle. Cell Struct Funct. 1997;22:147–153. doi: 10.1247/csf.22.147. [DOI] [PubMed] [Google Scholar]
- 46.Shen T, et al. Activity- and Cn-independent nuclear shuttling of NFATc1, but not NFATc3, in adult skeletal muscle fibers. Mol Biol Cell. 2006;17:1570–1582. doi: 10.1091/mbc.E05-08-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benedito AB, et al. The transcription factor NFAT3 mediates neuronal survival. J Biol Chem. 2005;280:2818–2825. doi: 10.1074/jbc.M408741200. [DOI] [PubMed] [Google Scholar]
- 48.Graef IA, et al. L-type calcium channels and GSK-3 regulate the activity of NF-ATc4 in hippocampal neurons. Nature. 1999;401:703–708. doi: 10.1038/44378. [DOI] [PubMed] [Google Scholar]
- 49.Yang TT, Xiong Q, Graef IA, Crabtree GR, Chow CW. Recruitment of the extracellular signal-regulated kinase/ribosomal S6 kinase signaling pathway to the NFATc4 transcription activation complex. Mol Cell Biol. 2005;25:907–920. doi: 10.1128/MCB.25.3.907-920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacquemin V, Furling D, Bigot A, Butler-Browne GS, Mouly V. IGF-1 induces human myotube hypertrophy by increasing cell recruitment. Exp Cell Res. 2004;299:148–158. doi: 10.1016/j.yexcr.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 51.Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 52.Musaro A, McCullagh KJ, Naya FJ, Olson EN, Rosenthal N. IGF-1 induces skeletal myocyte hypertrophy through Cn in association with GATA-2 and NF-ATc1. Nature. 1999;400:581–585. doi: 10.1038/23060. [DOI] [PubMed] [Google Scholar]
- 53.Wu H, et al. Activation of MEF2 by muscle activity is mediated through a Cn-dependent pathway. EMBO J. 2001;20:6414–6423. doi: 10.1093/emboj/20.22.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dai YS, Xu J, Molkentin JD. The DnaJ-related factor Mrj interacts with nuclear factor of activated T cells c3 and mediates transcriptional repression through class II histone deacetylase recruitment. Mol Cell Biol. 2005;25:9936–9948. doi: 10.1128/MCB.25.22.9936-9948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Molkentin JD, et al. A Cn-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tschuch C, et al. Off-target effects of siRNA specific for GFP. BMC Mol Biol. 2008;9:60. doi: 10.1186/1471-2199-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.