Abstract
BglG, which regulates expression of the β-glucoside utilization (bgl) operon in Escherichia coli, represents a family of RNA-binding transcriptional antiterminators that positively regulate transcription of sugar utilization genes in Gram-negative and Gram-positive organisms. BglG is negatively regulated by the β-glucoside phosphotransferase, BglF, by means of phosphorylation and physical association, and it is positively regulated by the general phosphoenolpyruvate phosphotransferase system (PTS) proteins, enzyme I (EI) and HPr. We studied the positive regulation of BglG both in vitro and in vivo. Here, we show that although EI and HPr are essential for BglG activity, this mode of activation does not require phosphorylation of BglG by HPr, as opposed to the phosphorylation-mediated activation of many BglG-like antiterminators in Gram-positive organisms. The effect of EI and HPr on BglG is not mediated by BglF. Nevertheless, the release of BglG from BglF, which is stimulated by the extracellular sugar in a sugar uptake-independent manner, is a prerequisite for BglG activation. Taken together, the results indicate that activation of BglG is a 2-stage process: a sugar-stimulated release from the membrane-bound sugar sensor followed by a phosphorylation-independent stimulatory effect exerted by the general PTS proteins.
Keywords: bgl system, E. coli
The BglG protein positively regulates expression of the β-glucoside utilization (bgl) operon in Escherichia coli by preventing premature termination of transcription within the bgl transcript (1, 2). It does so by binding to the emerging bgl mRNA at sites that partially overlap intrinsic terminators, thereby stabilizing an alternative conformation of the RNA chain (3). BglG represents a group of transcriptional antiterminators that positively regulate expression of genes that encode sugar-specific components of the phosphoenolpyruvate phosphotransferase system (PTS). BglG and its homologues, found in Gram-negative and Gram-positive organisms, consist of an RNA-binding domain followed by 2 homologous domains, PTS regulation domains (PRDs) 1 and 2. Each PRD contains 2 conserved histidines, which are important for regulation of the antitermination activity, although BglG lacks the fourth histidine (4). To bind to their RNA targets and accomplish antitermination, BglG and its homologues need to dimerize.
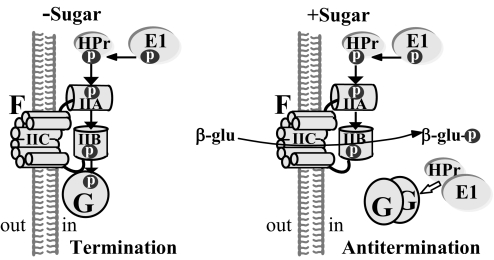
The dimeric state of BglG, and hence its activity, is regulated by BglF, the PTS β-glucoside phosphotransferase (EIIbgl), by reversible phosphorylation, depending on the availability of β-glucosides (5), as schematically described in Fig. 1. In the absence of β-glucosides, a phosphoryl group is delivered to BglF via the general PTS proteins, enzyme I (EI) and HPr; this phosphate is then transferred to a conserved histidine in BglG, thus inactivating it. Upon addition of β-glucosides, BglF dephosphorylates BglG, thus relieving its inhibition; nonphosphorylated BglG dimerizes and antiterminates bgl operon transcription while phosphorylated BglF catalyzes sugar phosphotransfer (6). Other PTS sugar phosphotransferases were either shown (7) or speculated to possess similar capabilities to regulate activity of antiterminator proteins by (de)phosphorylation, depending on the presence of their cognate sugars (4, 8–13). In addition, BglF was shown to recruit BglG to the membrane, where the 2 proteins form a precomplex that dissociates upon addition of β-glucosides to the growth medium (14). Hence, BglF regulates BglG both by reversible phosphorylation and by physical interaction (Fig. 1).
Fig. 1.
Model for BglG regulation. See text for details. Filled arrows indicate phosphorylation; open arrow, activation.
The activity of BglG homologues was reported to depend also on the general PTS proteins, EI and HPr. Some antiterminators from Gram-positive bacteria were shown to be activated by HPr-catalyzed phosphorylation on one or more of the conserved histidines (13, 15, 16). The HPr-catalyzed phosphorylation was proposed to be part of a mechanism of carbon catabolite repression (CCR) that operates in Gram-positive bacteria (reviewed in ref. 4). On the basis of the requirement of the general PTS proteins for BglG activity and the detection of some phosphorylated BglG (BglG-P) in the absence of BglF in vivo, it was suggested that HPr from E. coli acts similarly and phosphorylates BglG (17, 18). However, direct phosphorylation of BglG by HPr has never been observed in sensitive in vitro systems (e.g., refs. 19 and 20), and it is possible that cross-talk with other PTS sugar phosphotransferases could account for the residual phosphorylation of BglG in ΔbglF strains (see Discussion).
In this work, we set out to investigate the requirements for BglG activation. By observing the bgl operon transcripts generated in vivo and in vitro, we show that BglG-mediated antitermination requires the general PTS proteins. However, unlike the regulation of BglG-like antiterminators in Gram-positive organisms, HPr-mediated phosphorylation is not involved in the positive effect of the general PTS proteins on BglG activity. We also show that BglF is not essential for the positive effect of the general PTS proteins on BglG, because BglG can be activated in ΔbglF cells. Nevertheless, the release of BglG from BglF, which is stimulated by the extracellular sugar, is a prerequisite for BglG activation by the general PTS proteins. Interestingly, although the sugar exerts its effect through its phosphotransferase (i.e., BglF), sugar phosphotransfer is not required for inducing BglG activation. These results shed light on the mechanism by which sugar phosphotransferases of the PTS, such as BglF, are stimulated and uncover a link between these proteins and the general PTS proteins that does not involve phosphorylation.
Results
BglG-Mediated Antitermination in Vitro Depends on EI and HPr.
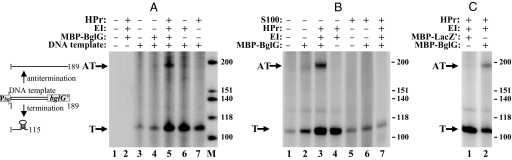
The function of BglG as a transcriptional antiterminator has been studied mainly in vivo (2, 21). Transcription termination at the bgl terminator was successfully reproduced in vitro by using E. coli RNA polymerase, with bgl DNA as a template (1). This reaction was not dependent on additional factors. We attempted to reproduce transcription antitermination in this minimal system by adding purified and soluble BglG fused to maltose-binding protein (MBP). MBP-BglG was shown to be active in bgl antitermination in vivo and to be regulated by BglF in vitro and in vivo (20, 22). As a template, we used a 532-bp PCR fragment containing the bgl promoter activated by a point mutation in the CAP-binding site (23), the leader region, which contains a transcription terminator, and the very beginning of the bglG gene (encoding the first 19 aa of BglG). The expected length of the transcript that terminates within the leader region is ≈115 bases. The expected length of a putative runoff transcript, generated by the RNA polymerase initiating at the bgl promoter and reading through the terminator, is 189 bases. The transcripts were radioactively labeled with [32P]-UTP and fractionated on denaturing polyacrylamide-urea gels. The products obtained in this in vitro transcription assay in the absence of BglG and in its presence were similar and consisted mainly of terminated transcripts (Fig. 2A, lanes 3 and 4, and B, lanes 1 and 2). The same results were obtained when longer PCR or restriction fragments were used as templates. These results suggest that additional cellular factors are required for antitermination.
Fig. 2.
BglG-mediated transcription antitermination in vitro is activated by EI and HPr. [α-32P]UTP-labeled RNA transcripts were produced in vitro and analyzed as described in Materials and Methods. Except for lanes 1 and 2 in A, all other reaction mixtures contained bgl DNA as a template. When indicated, MBP-BglG, MBP-LacZ′, His-tagged EI, His-tagged HPr, and S100 E. coli protein extract were added. The S100 extract was prepared from strain AD7333, which is defective in several nucleases and was shown to lack ribonuclease activity, as described in Materials and Methods. The scheme on the left describes the DNA template and the transcription products. T, terminated transcripts; AT, antiterminated transcripts; M, [γ-32P]ATP-labeled, denatured, HinfI-cleaved ΦX174 DNA fragments. The efficiency of BglG-mediated antitermination, inferred from the ratio between the terminated and antiterminated transcripts, was 4 ± 0.3 (4.0 in A, lane 5; 4.3 in B, lane 3; 3.7 in C, lane 2). To calculate this ratio, the intensity of the RNA bands on gel was determined by the TINA 2.0 software, and the values were corrected according to the number of the UTP nucleotides in the respective transcript. The intensity of the antiterminated transcripts produced because of BglG activity was normalized to the intensity in this area of the gel in lanes lacking BglG activity.
The general PTS proteins, EI and HPr, were shown to be required for the expression of a plasmid-based bgl′-lacZ fusion from a heterologous promoter (18). We therefore asked whether these proteins can assist BglG in antiterminating bgl operon transcription in vitro. The results presented in Fig. 2 show that antiterminated transcripts were detected when EI and HPr were added to the in vitro system that contained BglG (Fig. 2 A, lane 5, and B, lane 3). The general PTS proteins, added individually (Fig. 2A, lanes 6 and 7) or together (Fig. 2B, lane 4) to the reaction mixture, did not lead to antitermination of bgl transcription in the absence of BglG. A very low level of antiterminated transcripts was occasionally detected when BglG was added without the general PTS proteins, apparently because of the presence of EI and HPr in BglG preparations (see Discussion). Similar results were obtained with 3-fold lower and 3-fold higher levels of MBP-BglG. When longer PCR fragments, which extended further into the bglG gene, were used as templates, the size of the runoff transcripts corresponded to the increased length of the templates. The results in Fig. 2C show that BglG, and not the MBP moiety of MBP-BglG, is responsible for antitermination, because no antiterminated transcripts were detected when an MBP-LacZ′ protein was added to the in vitro system instead of MBP-BglG. The same was demonstrated by using commercial MBP.
Notably, the relative amount of the terminated transcripts detected in the presence of BglG, EI, and HPr was still high compared with the antiterminated transcripts (by 4 ± 0.3-fold, see legend to Fig. 2), indicating that antitermination in vitro is not very efficient. We tested the possibility that additional proteins are required to enable efficient antitermination by adding cellular extracts to the minimal in vitro transcription system. However, no BglG activity was detected in the presence of an S100 extract of E. coli proteins (Fig. 2B, lanes 5–7) or a ribosome-depleted S30 extract.
BglG-Mediated Antitermination in Vivo Is a Low-Efficiency Process That Requires EI and HPr.
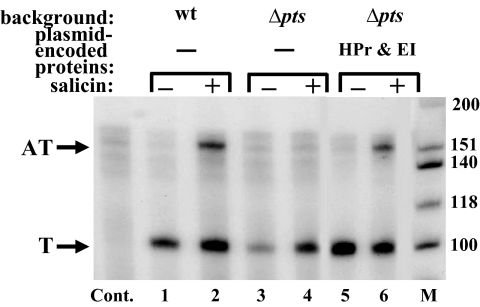
The results obtained in the in vitro transcription system encouraged us to examine the efficiency of the BglG-mediated antitermination process in vivo. To achieve this goal, we analyzed the cellular bgl transcripts by the S1 nuclease protection assay. To this end, total RNA was prepared from MA10, a bgl+ E. coli strain (21), grown with or without β-glucosides; hybridized to a radioactively labeled, single-stranded bgl probe; treated with S1 nuclease; and separated on a denaturing polyacrylamide-urea gel. As expected, antiterminated bgl transcripts were detected only when the MA10 cells were grown in the presence of a β-glucoside sugar (Fig. 3, compare lanes 1 and 2, growth without or with salicin, respectively). However, the ratio between the terminated and antiterminated bgl transcripts in these cells (2.2 ± 0.8, see legend to Fig. 3) indicates that antitermination of bgl operon transcription is quite inefficient in vivo. It is possible that the terminated transcripts are more stable than the antiterminated transcripts, but it is obvious that terminated transcripts are still produced in the presence of the sugar.
Fig. 3.
Antitermination of bgl operon transcription in vivo requires the general PTS proteins. Total RNA was prepared from 3 bgl+ E. coli strains, MA10 (wt), a Δpts derivative of MA10 (Δpts), and a version of the latter strain transformed with a pBR322-based plasmid encoding EI and HPr, which were grown with or without the β-glucoside salicin. The different RNA preparations were hybridized to a radioactively labeled, single-stranded bgl probe and analyzed by the S1 nuclease protection assay, as described in Materials and Methods. The samples were separated on a denaturing polyacrylamide-urea gel. T, terminated transcripts; AT, antiterminated transcripts; M, [γ-32P]ATP-labeled, denatured, HinfI-cleaved ΦX174 DNA fragments; Cont, total RNA prepared from MA110, which is isogenic to MA10 but lacks the bgl operon, containing pBR322, and analyzed by the S1 nuclease protection assay. The ratio between the terminated and antiterminated transcripts in MA10 (wt), calculated as described in Fig. 2, in 7 independent experiments was 2.2 ± 0.8. The ratio between the 2 transcripts in the Δpts derivative of MA10 (Δpts) transformed with a plasmid encoding EI and HPr, calculated from 3 independent experiments, was 2.4 ± 0.3.
To study the effect of the general PTS proteins on BglG-mediated antitermination in vivo, we constructed a Δpts derivative of the MA10 strain. Total RNA prepared from this Δpts mutant strain, grown with or without β-glucosides, was analyzed by the S1 nuclease protection assay and compared to the RNA prepared from the isogenic pts+ strain. Contrary to the pts+ strain, no antiterminated bgl transcripts were detected in the Δpts strain independent of sugar addition (Fig. 3, lanes 3 and 4). Expression of EI and HPr from a plasmid restored bgl antitermination in the Δpts strain grown with β-glucosides (Fig. 3, compare lanes 5 and 6) at an efficiency comparable to antitermination in MA10 (2.4 ± 0.3-fold terminated vs. antiterminated transcripts, see legend to Fig. 3). Hence, the results obtained in vivo, which are in complete agreement with the in vitro results, indicate that BglG-mediated antitermination is an inefficient process that depends on the general PTS proteins.
To further investigate BglG-mediated antitermination in vivo, we made use of the bgl+ strain MA200, which carries a chromosomal bgl-lacZ transcriptional fusion (21), and its Δpts derivative, which was constructed as described in Materials and Methods. In MA200, transcription of lacZ from the chromosomal fusion depends on antitermination by BglG and is induced by β-glucosides, as indicated by the growth of red colonies on MacConkey lactose plates and the production of increased levels of β-galactosidase in the presence of the inducing sugar (Table 1 and Fig. S1, no. 1). In the Δpts strain, lacZ is not expressed, as indicated by the pale color of colonies on MacConkey lactose plates and the low β-galactosidase produced, independently of β-glucoside addition (Table 1 and Fig. S1, no. 2). Expression of EI and HPr from a plasmid that complements the Δpts mutation restored lacZ expression in the presence of β-glucosides, as indicated by the growth of red colonies on MacConkey lactose plates and the increase in β-galactosidase units (Table 1 and Fig. S1, no. 7). Complementation of the Δpts mutation by the plasmid-encoded EI and HPr was demonstrated by the ability of the cells to use β-glucosides; i.e., growth of red colonies on MacConkey salicin plates (Table 1, no. 7). Expression of either HPr or EI alone did not restore lacZ expression (Table 1 and Fig. S1, nos. 3 and 4, respectively), indicating that both general PTS proteins are required for the BglG-dependent expression of lacZ.
Table 1.
BglG-mediated transcription through the bgl terminator in vivo depends on the general PTS proteins but not on HPr-catalyzed phosphorylation
| No. | Strain‡ | Plasmid-encoded PTS protein§ | β-Gal activity, units* |
Color on Mac-lactose† |
Color on Mac-salicin¶ | ||
|---|---|---|---|---|---|---|---|
| −Inducer | +Inducer | −Inducer | +Inducer | ||||
| 1 | MA200 | − | 20 ± 2.5 | 215 ± 11 | W | R | R |
| 2 | MA200Δpts | − | 11 ± 3.0 | 15 ± 2.0 | W | W | W |
| 3 | MA200Δpts | HPr | 10 ± 2.0 | 11 ± 1.0 | W | W | W |
| 4 | MA200Δpts | EI | 6 ± 0.4 | 14 ± 0.4 | W | W | W |
| 5 | MA200Δpts | HPr(H15A) | 7 ± 0.3 | 10 ± 0.9 | W | W | W |
| 6 | MA200Δpts | EI(H189A) | 8 ± 0.3 | 7 ± 0.3 | W | W | W |
| 7 | MA200Δpts | HPr&EI | 11 ± 2.0 | 137 ± 11 | W | R | R |
| 8 | MA200Δpts | HPr(H15A)&EI | 7 ± 1.0 | 169 ± 20 | W | R | W |
| 9 | MA200Δpts | HPr&EI(H189A) | 7 ± 0.2 | 8 ± 3.0 | W | W | W |
| 10 | MA200Δpts | HPr(H15A)&EI(H189A) | 6 ± 0.3 | 9 ± 1.4 | W | W | W |
| 11 | MA200Δpts | HPr&EI-N′ | 15 ± 5.0 | 10 ± 1.7 | W | W | W |
| 12 | MA200Δpts | HPr(H15A)&EI-N′ | 7 ± 2.0 | 8 ± 1.5 | W | W | W |
| 13 | MA200Δpts | HPr&EI-C′ | 9 ± 3.2 | 9 ± 2.0 | W | W | W |
| 14 | MA200Δpts | HPr(H15A)&EI-C′ | 14 ± 3.7 | 10 ± 3.5 | W | W | W |
*Expression of lacZ was tested by measuring β-galactosidase activity in the absence and presence of the β-glucoside salicin as inducer. IPTG was added to a final concentration of 0.5 mM. The values represent the average of 4 independent experiments.
†Expression of lacZ was tested by comparing colony color on MacConkey lactose plates that either contained or did not contain 7 mM β-glucoside salicin as inducer. R, red colonies, W, white colonies. All plates contained 0.5 mM IPTG. The plates are shown in Fig. S1.
‡Strains used in this experiment were MA200, a bgl+ strain that carries a chromosomal bgl-lacZ transcriptional fusion (21), and MA200Δpts.
§Each strain contained a plasmid that encodes either no PTS proteins or the specified PTS proteins.
¶Utilization of β-glucosides was indicated by the growth of red colonies on MacConkey salicin plates.
Taken together, the results obtained with the bgl-lacZ fusion and the data obtained from the S1 analysis of the bgl operon transcripts indicate that BglG requires EI and HPr to allow the polymerase to read through the bgl terminator, independent of the genes downstream of the terminator.
Activation of BglG by the General PTS Proteins Does Not Require Phosphorylation by HPr.
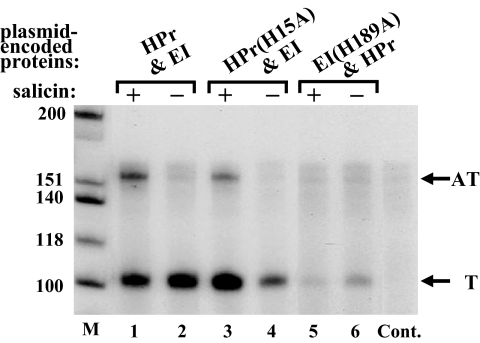
Phosphorylation of BglG by HPr was suggested to underlie the requirement for the PTS proteins for bgl antitermination (17, 18). These results were in conflict with the inability of the general PTS proteins to phosphorylate BglG in sensitive in vitro systems (e.g., refs. 19 and 20). To resolve this conflict, we asked whether HPr(H15A), which is an HPr derivative mutated in its phosphorylation site, can restore BglG-mediated antitermination in a Δpts strain. First, by using the S1 nuclease protection assay, we compared the bgl transcripts produced in strain MA10Δpts, which was transformed with a plasmid encoding EI together with either wild-type HPr or HPr(H15A). As shown in Fig. 4, expression of EI together with HPr(H15A) restored BglG-mediated antitermination in the Δpts strain.
Fig. 4.
Positive regulation of BglG-mediated antitermination does not require phosphorylation by HPr. Total RNA was prepared from the Δpts derivative of MA10 that has been transformed with a plasmid encoding either EI and HPr (lanes 1 and 2) or EI and HPr(H15A) (lanes 3 and 4) or EI(H189A) and HPr (lanes 5 and 6), grown with or without the β-glucoside salicin. The different RNA preparations were analyzed by the S1 nuclease protection assay, as in Fig. 3. The samples were separated on a denaturing polyacrylamide-urea gel. Abbreviations Cont., M, T, and AT are as in Fig. 3. The ratio between the terminated and antiterminated transcripts in Δpts MA10 transformed with a plasmid encoding EI and HPr(H15A), calculated as described in Fig. 2, in 2 independent experiments was 4.2 ± 1 (3.5 in this experiment, lane 3). The ratio between the 2 transcripts in the same strain transformed with a plasmid encoding wild-type EI and HPr, calculated from 3 independent experiments, was 2.4 ± 0.3 (2.6 in this experiment, lane 1).
Next, we tested expression of the chromosomal bgl-lacZ fusion in MA200Δpts cells producing EI and HPr(H15A). The fusion was expressed in these cells similarly to its expression in cells producing EI and wild-type HPr; i.e., growth of red colonies on MacConkey lactose plates and high β-galactosidase units in the presence of β-glucosides (Table 1 and Fig. S1, compare no. 8 to no. 7). Notably, the cells expressing the mutant HPr protein were defective in β-glucoside utilization, as indicated by the growth of white colonies on MacConkey salicin plates (Table 1, no. 8). Based on the results obtained with the 2 in vivo systems, phosphorylation by HPr is not essential and cannot explain the requirement for the general PTS proteins for BglG-mediated antitermination. An interesting observation that emerged from the 2 experimental systems was that activation of BglG by the HPr(H15A) protein was still dependent on the presence of β-glucosides (Fig. 4, compare lanes 3 and 4, with and without salicin, respectively; Table 1; and Fig. S1, compare minus and plus inducer for no. 8). Because BglF is not phosphorylated and is not active in sugar uptake in cells expressing the HPr(H15A) protein, these results indicate that BglF inhibits BglG activity independently of its phosphorylation state and that β-glucosides relieve this inhibition without a need to enter the cell (see Discussion).
We also tested the effect of a mutation in the EI phosphorylation site (H189A) on BglG-mediated antitermination. Interestingly, the EI mutant impaired BglG-mediated antitermination in both experimental systems (Fig. 4, lane 5; Table 1; and Fig. S1, no. 9). In the experiment shown in Fig. 4, the mutated EI protein seemed to reduce bgl transcription overall, although in other experiments the effect of the mutated EI protein on bgl transcription was less dramatic. Also, the basal level of the LacZ protein produced from the bgl-lacZ fusion in the presence of the mutated EI protein was similar to the basal level observed with wild-type EI (Table 1). The effect of the mutant EI protein was the same when expressed with wild-type HPr or with the mutant HPr protein that cannot be phosphorylated (Table 1 and Fig. S1, compare nos. 9 and 10). These results raised the possibility that EI needs to be phosphorylated to efficiently interact with HPr. To test this possibility, we asked whether the N′-terminal domain of EI (EI-N′), which interacts with HPr (24), is sufficient for BglG-mediated antitermination. Although EI-N′ cannot be phosphorylated, it can still interact with HPr (24). Our results show that neither EI-N′ nor EI-C′ could activate BglG in MA200Δpts, independent of the coexpressed HPr protein (Table 1 and Fig. S1, compare nos.11–14). Hence, although phosphorylation of BglG by EI was ruled out in previous studies (19, 20), phosphorylated EI still seems to be required for BglG activation (see Discussion).
The Effect of EI and HPr on BglG Activity Is Not Mediated via BglF.
To examine whether the general PTS proteins exert their effect on BglG activity via its regulator BglF, we asked whether BglF is required for this regulation. To answer this question, we constructed a ΔptsΔbglF MA10 double mutant (for strain construction, see Materials and Methods). We then compared the expression of a plasmid-based bgl-lacZ fusion in MA10 and in its 2 derivatives, the Δpts mutant and a ΔptsΔbglF double mutant, by comparing colony color on MacConkey lactose plates and by measuring β-gal units in the absence and presence of the β-glucoside salicin. The results are presented in Table 2 and in Fig. S2. The background level of β-galactosidase produced by the plasmid-based bgl-lacZ fusion in the absence of the inducing sugar was higher than that produced by the chromosomal fusion (compare Table 1, no. 2, to Table 2, no. 2). Still, expression of lacZ increased more than 3-fold in the presence of β-glucosides only when EI and HPr were expressed together in MA10 Δpts (Table 2, no. 5), indicating that expression of the plasmid-based fusion (Table 2), like that of the chromosomal fusion (Table 1), requires the general PTS proteins. Activation of BglG by EI and HPr also was observed in the double mutant, which also was deleted for the bglF gene (ΔptsΔbglF MA10), although in this case the activation did not depend on the presence of β-glucosides in the growth medium (Table 2 and Fig. S2, compare minus and plus inducer in no. 9). Notably, unlike the Δpts mutant, utilization of β-glucosides could not be restored in the ΔptsΔbglF mutant by expressing EI and HPr from a plasmid (Table 2, compare color on MacConkey salicin plates in nos. 5 and 9). Taken together, these results indicate that BglF is not needed for the activation of BglG by the general PTS proteins.
Table 2.
Activation of BglG by the general PTS proteins is BglF-independent
| No. | Strain‡ | Plasmid-encoded PTS protein§ | β-Gal activity, units* |
Color on Mac-lactose† |
Color on Mac-salicin¶ | ||
|---|---|---|---|---|---|---|---|
| −Inducer | +Inducer | −Inducer | +Inducer | ||||
| 1 | MA10 | − | 32 ± 2 | 94 ± 2 | W | R | R |
| 2 | MA10Δpts | − | 30 ± 3 | 33 ± 1 | W | W | W |
| 3 | MA10Δpts | HPr | 35 ± 3 | 40 ± 1 | W | W | W |
| 4 | MA10Δpts | EI | 28 ± 5 | 43 ± 10 | W | W | W |
| 5 | MA10Δpts | HPr&EI | 27 ± 3 | 108 ± 5 | W | R | R |
| 6 | MA10ΔptsΔbglF | − | 34 ± 2 | 37 ± 1 | W | W | W |
| 7 | MA10ΔptsΔbglF | HPr | 37 ± 2 | 40 ± 3 | W | W | W |
| 8 | MA10ΔptsΔbglF | EI | 24 ± 1 | 24 ± 1 | W | W | W |
| 9 | MA10ΔptsΔbglF | HPr&EI | 99 ± 4 | 119 ± 6 | R | R | W |
*Expression of lacZ was tested by measuring β-galactosidase activity, as in Table 1. The values represent the average of 4 independent experiments.
†Expression of lacZ was tested by comparing colony color on MacConkey lactose plates, as in Table 1. The plates are shown in Fig. S2.
‡Strains used in this experiment were MA10, a bgl+ E. coli strain (21), and 2 derivatives of MA10, a Δpts mutant and a ΔptsΔbglF double mutant. All strains contained pANS200, which encodes a bgl-lacZ fusion.
§Each strain contained a second plasmid that encodes either no PTS proteins or the specified PTS proteins.
¶Utilization of β-glucosides was indicated by growth of red colonies on MacConkey salicin plates.
Discussion
The results presented in this manuscript demonstrate that the activity of BglG depends on the general PTS proteins, EI and HPr, both in vivo and in vitro. In this sense, BglG resembles many of its homologues that cannot lead to expression of the genes they control in pts− strains (25). However, unlike these antiterminators, the activity of BglG does not depend on phosphorylation by HPr. Indeed, numerous trials to phosphorylate BglG in vitro by HPr and EI from various sources (E. coli, Salmonella typhimurim, and Bacillus subtilis) failed (e.g., refs. 19 and 20). The possibility that BglF is required for the phosphorylation of BglG by HPr was ruled out by the demonstration that BglG was not phosphorylated by the general PTS proteins in the presence of 2 different BglF variants (20). The results presented here, showing that BglF is dispensable for the general PTS-dependent activation, support this conclusion. The possibility that HPr-like proteins, such as the diphosphoryl transfer protein, substitute for HPr in activating BglG by phosphorylation in the absence of a phosphorylatable HPr protein is ruled out by our current observation that BglG is inactive in cells lacking an HPr-encoding allele (but containing an EI-encoding allele), whereas it is active in cells expressing the HPr(H15A) protein. Previous speculation that HPr phosphorylates BglG was based on the requirement of the general PTS proteins for BglG activity and the detection of some BglG-P in the absence of BglF in vivo (17, 18). Our results show that this is not the case. We suggest that cross-talk accounts for the residual phosphorylation of BglG observed in ΔbglF strains. In support of this idea, we have detected a low level of in vitro phosphorylation of BglG by the trehalose phosphotransferase from E. coli, which resembles BglF in structure and function.
The HPr-catalyzed phosphorylation of BglG homologues was proposed to be part of a CCR mechanism that operates in Gram-positive bacteria (reviewed in ref. 4). Importantly, the major CCR mechanisms that operate in Gram-positive bacteria are quite different from those in Gram-negative bacteria, although components of the PTS play a major role in both types of organisms (26). One major difference is that these global responses rely on the ability of the PTS to sense the availability of carbohydrates and the metabolic capacity of the cell to metabolize these carbohydrates via the phosphorylation state of a different PTS component; namely, IIAglc in enteric bacteria and HPr in low-GC Gram-positive bacteria and lactic acid bacteria. Another significant difference is that in most Gram-positive bacteria, HPr is also phosphorylated on a seryl residue by an ATP-dependent protein kinase. This reaction plays a regulatory function; depending on its phosphorylation state, HPr regulates carbon metabolism by steric interactions, analogous to the role of IIAglc in Gram-negative bacteria (reviewed in ref. 27). It might be that these differences account for the differences in the mechanism by which the HPr proteins activate BglG-like antiterminators. In support of this hypothesis, GlcT, a BglG homologue from Staphylococcus carnosus, was shown to be efficiently phosphorylated by the B. subtilis HPr, whereas HPr from E. coli failed to phosphorylate it (28). Our study points at another difference between the mechanisms of transcription antitermination in Gram-positive and Gram-negative bacteria. Whereas BglG-mediated antitermination is shown in the experiments presented here, which were supported by Northern blot analysis of the bgl transcripts, to be an inefficient process in vivo (i.e., a high level of terminated transcripts is produced even in the presence of β-glucosides), sugar-induced antitermination of the bglPH operon in B. subtilis seems quite efficient (29).
What are the nature and the role of the PTS-mediated activation of BglG? In Gram-positive bacteria, HPr-mediated phosphorylation of BglG homologues drives dimer formation, thus modulating their nucleic acid-binding activity. However, even among these organisms, the details of this regulation vary greatly from one regulator to another. It has been argued that the cellular population of these molecules is heterogeneous with respect to their conformation, and that various factors can modify the relative amounts of the different conformers, thereby affecting their activity (4). It is reasonable to assume that a similar mechanism operates in E. coli; i.e., the general PTS proteins provoke a conformational change that stabilizes the BglG active dimer, albeit not via phosphorylation but perhaps by physical interaction. Indeed, the general PTS proteins copurify with BglG from pts+ strains in pull-down assays. This explains the low level of antiterminated transcripts infrequently detected in the in vitro system when only BglG, and not EI and HPr, is added.
The failure of the individual EI domains and of the nonphosphorylatable EI mutant to activate BglG might reflect a requirement for phosphorylated EI for this mode of regulation. This, in turn, might be due to the need for phosphorylated EI to elicit HPr-mediated activation of BglG. Alternatively, it might be that by interacting with phosphorylated EI or with a complex of phosphorylated EI and HPr, BglG senses the phosphorylation state of the general PTS proteins, which is low when glucose or other efficiently metabolized carbon sources are present. If this interpretation is correct, it suggests a new CCR mechanism. Then again, the requirement for both EI domains for BglG activation might reflect the need of EI to interact with both HPr and BglG. The results with the nonphosphorylatable EI mutant do not disagree with this possibility. Structural studies have shown that the activity of EI relies on swiveling between a compact and a relaxed conformation (24), whereas the EI(H189A) mutant is locked in the compact conformation (30). Therefore, the failure of the mutant to activate BglG might stem from its inability to interact with its partners. Studies to distinguish between these possibilities are underway.
The prevailing belief has been that phosphorylation on PRD1 by the cognate sugar phosphotransferases inactivates BglG-like antiterminators, whereas phosphorylation by EI and HPr in PRD2 stimulates their activity (25). We have identified previously a conserved histidine in PRD2 of BglG as the target for BglF phosphorylation (20). It has been suggested that HPr, present in the phosphorylation mixture, was actually responsible for PRD2 phosphorylation, rather than BglF (25). The results presented here rule out this hypothesis. We propose that the 2 consecutive homologous PRDs, which apparently arose from an ancestral gene duplication (31), have the potential to play similar roles in regulation; i.e., in different antiterminators, a different PRD is negatively phosphorylated. The requirement for the conserved histidines in PRD1 for the EII-catalyzed phosphorylation on PRD2, observed in the case of BglG and other antiterminators, might reflect the importance of these residues for BglG folding into a phosphorylatable conformation. Indeed, PRD1 and PRD2 of BglG could be cross-linked in vitro and in vivo, indicating that a fraction of BglG folds into a compact conformation in which these domains are in a very close proximity (22).
A key observation in this work is that β-glucosides are required for the induction of BglG activation by the general PTS proteins—even in cells expressing a nonphosphorylatable HPr protein—where BglF, the negative regulator of BglG, is not phosphorylated, and sugar uptake is precluded (Fig. 4 and Table 1). Notably, in a bglF null strain, activation of BglG was sugar-independent (Table 2). These observations can be explained by a conformational change that PTS sugars were suggested to induce in EIIs (25), including in a nonphosphorylatable BglF protein (32). This change is propagated to BglG, which is found in a precomplex with BglF near the membrane. Interaction of the sugar with BglF was shown to release BglG from this complex (14). Apparently, formation of the complex plays a central role in keeping BglG fully inactive, and the sugar-induced change is absolutely necessary for BglG activation. An important and far-reaching outcome of our observations is that environmental sugars do not have to enter the cell to have an impact on metabolism. The implication of such a sensing mechanism is that the cell is ready to use less-preferable sugars once better sugars are exhausted.
Materials and Methods
Strains, plasmids, and proteins are described in SI Materials and Methods.
In Vitro Transcription.
In vitro transcription was carried by incubating a DNA template (extending 343 bp upstream of the transcription start site of the bgl operon and ending 189 bp past +1 of the bgl operon) with E. coli holoenzyme RNA polymerase, CAP, cAMP, RNasin, and ribonucleotides, including [α-32P]UTP, under the conditions detailed in SI Materials and Methods. RNase-free DNase was added to end the transcription reaction and to digest the DNA template. MBP-BglG, His-tagged EI, His-tagged HPr MBP-LacZ′, or S100 protein extract was added as indicated. The S100 extract was tested for lack of RNase activity by incubating it with the products of the in vitro transcription system. The samples were heated and separated on a denaturing acrylamide-urea gel. Labeled RNAs were detected by autoradiography. HinfI-cleaved φX174 DNA fragments (Fermentas), which were end-labeled by [γ-32P]ATP using T4 polynucleotide kinase (PNK; NEB), were denatured the same as the RNAs and fractionated alongside to serve as size markers.
Preparation of Radioactively Labeled DNA Probe.
A 166-bp DNA fragment, initiating 8 bp before the transcription start site of the bgl operon and terminating 158 bp past +1 of the bgl operon, was synthesized by PCR using pMN25 as a template. A single-stranded, radioactively labeled DNA probe, complementary to the bgl transcript, was produced in a second PCR that contained [α-32P]dCTP. Details on the PCRs and products purification are given in SI Materials and Methods.
S1 Nuclease Protection Assay.
Total RNA was incubated with a radioactively labeled, single-stranded DNA probe at 90 °C for 10 min (for denaturation of secondary structures) and then at 50 °C for 3–4 h to allow hybridization. After treating the mixture with S1 nuclease, DNA was extracted and analyzed on a denaturing polyacrylamide-urea gel. The details are given in SI Materials and Methods. When the same amount of radioactively labeled, single-stranded DNA was incubated with increased amounts of mRNA, stronger signals were observed, demonstrating that the probe is in excess relative to the bgl transcripts.
Supplementary Material
Acknowledgments.
This work was supported by the Israel Science Foundation founded by the Israel Academy of Sciences and Humanities.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902559106/DCSupplemental.
References
- 1.Mahadevan S, Wright A. A bacterial gene involved in transcription antitermination: Regulation at a rho-independent terminator in the bgl operon of E coli. Cell. 1987;50:485–494. doi: 10.1016/0092-8674(87)90502-2. [DOI] [PubMed] [Google Scholar]
- 2.Schnetz K, Rak B. Regulation of the bgl operon of E. coli by transcriptional termination. EMBO J. 1988;7:3271–3277. doi: 10.1002/j.1460-2075.1988.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houman F, Diaz-Torres MR, Wright A. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell. 1990;62:1153–1163. doi: 10.1016/0092-8674(90)90392-r. [DOI] [PubMed] [Google Scholar]
- 4.van Tilbeurgh H, Declerck N. Structural insights into the regulation of bacterial signalling proteins containing PDRs. Curr Opin Struct Biol. 2001;11:685–693. doi: 10.1016/s0959-440x(01)00267-6. [DOI] [PubMed] [Google Scholar]
- 5.Amster-Choder O, Wright A. Modulation of the dimerization of a transcriptional antiterminator protein by phosphorylation. Science. 1992;257:1395–1398. doi: 10.1126/science.1382312. [DOI] [PubMed] [Google Scholar]
- 6.Amster-Choder O. The bgl sensory system: A transmembrane signaling pathway controlling transcriptional antitermination. Curr Opin Microbiol. 2005;8:127–134. doi: 10.1016/j.mib.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 7.Schmalisch MH, Bachem S, Stülke J. Control of the B. subtilis antiterminator protein GlcT by phosphorylation. Elucidation of the phosphorylation chain leading to inactivation of GlcT. J Biol Chem. 2003;278:51108–51115. doi: 10.1074/jbc.M309972200. [DOI] [PubMed] [Google Scholar]
- 8.Bachem S, Stülke J. Regulation of the Bacillus subtilis GlcT antiterminator protein by components of the phosphotransferase system. J Bacteriol. 1998;180:5319–5326. doi: 10.1128/jb.180.20.5319-5326.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crutz AM, Steinmetz M, Aymerich S, Richter R, Le Coq D. Induction of levansucrase in Bacillus subtilis: An antitermination mechanism negatively controlled by the phosphotransferase system. J Bacteriol. 1990;172:1043–1050. doi: 10.1128/jb.172.2.1043-1050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Idelson M, Amster-Choder O. SacY, a transcriptional antiterminator from B. subtilis, is regulated by phosphorylation in vivo. J Bacteriol. 1998;180:660–666. doi: 10.1128/jb.180.3.660-666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Coq D, Lindner C, Krüger S, Steinmetz M, Stülke J. New beta-glucoside (bgl) genes in B. subtilis: The bglP gene product has both transport and regulatory functions similar to those of BglF, its E. coli homolog. J Bacteriol. 1995;177:1527–1535. doi: 10.1128/jb.177.6.1527-1535.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin-Verstraete I, et al. Antagonistic effects of dual PTS-catalysed phosphorylation on the B. subtilis transcriptional activator LevR. Mol Microbiol. 1998;28:293–303. doi: 10.1046/j.1365-2958.1998.00781.x. [DOI] [PubMed] [Google Scholar]
- 13.Tortosa P, et al. Sites of positive and negative regulation in the B. subtilis antiterminators LicT and SacY. Mol Microbiol. 2001;41:1381–1393. doi: 10.1046/j.1365-2958.2001.02608.x. [DOI] [PubMed] [Google Scholar]
- 14.Lopian L, Nussbaum-Shochat A, O'Day-Kerstein K, Wright A, Amster-Choder O. The BglF sensor recruits the BglG transcription regulator to the membrane and releases it on stimulation. Proc Natl Acad Sci USA. 2003;100:7099–7104. doi: 10.1073/pnas.1037608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnaud M, et al. Regulation of the sacPA operon of Bacillus subtilis: Identification of phosphotransferase system components involved in SacT activity. J Bacteriol. 1992;174:3161–3170. doi: 10.1128/jb.174.10.3161-3170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindner C, Galinier A, Hecker M, Deutscher J. Regulation of the activity of the B. subtilis antiterminator LicT by multiple PEP-dependent, enzyme I- and HPr-catalysed phosphorylation. Mol Microbiol. 1999;31:995–1006. doi: 10.1046/j.1365-2958.1999.01262.x. [DOI] [PubMed] [Google Scholar]
- 17.Görke B. Regulation of the E. coli antiterminator protein BglG by phosphorylation at multiple sites and evidence for transfer of phosphoryl groups between monomers. J Biol Chem. 2003;278(47):46219–46229. doi: 10.1074/jbc.M308002200. [DOI] [PubMed] [Google Scholar]
- 18.Görke B, Rak B. Catabolite control of E. coli regulatory protein BglG activity by antagonistically acting phosphorylations. EMBO J. 1999;18:3370–3379. doi: 10.1093/emboj/18.12.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amster-Choder O, Houman F, Wright A. Protein phosphorylation regulates transcription of the beta-glucoside utilization operon in E. coli. Cell. 1989;58:847–855. doi: 10.1016/0092-8674(89)90937-9. [DOI] [PubMed] [Google Scholar]
- 20.Chen Q, Arents JC, Bader R, Postma PW, Amster-Choder O. BglF, the sensor of the E. coli bgl system, uses the same site to phosphorylate both a sugar and a regulatory protein. EMBO J. 1997;16:4617–4627. doi: 10.1093/emboj/16.15.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahadevan S, Reynolds AE, Wright A. Positive and negative regulation of the bgl operon in E. coli. J Bacteriol. 1987;169:2570–2578. doi: 10.1128/jb.169.6.2570-2578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fux L, Nussbaum-Shochat A, Amster-Choder O. A fraction of the BglG transcriptional antiterminator from Escherichia coli exists as a compact monomer. J Biol Chem. 2003;278:50978–50984. doi: 10.1074/jbc.M308085200. [DOI] [PubMed] [Google Scholar]
- 23.Reynolds AE, Mahadevan S, LeGrice SF, Wright A. Enhancement of bacterial gene expression by insertion elements of by mutation in a CAP-cAMP binding site. J Mol Biol. 1986;191:85–95. doi: 10.1016/0022-2836(86)90424-9. [DOI] [PubMed] [Google Scholar]
- 24.Teplyakov A, et al. Structure of phosphorylated enzyme I, the phosphoenolpyruvate:sugar phosphotransferase system sugar translocation signal protein. Proc Natl Acad Sci USA. 2006;103:16218–16223. doi: 10.1073/pnas.0607587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol. 2008;11:87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Görke B, Stülke J. Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 28.Knezevic I, et al. Regulation of the glucose-specific phosphotransferase system (PTS) of Staphylococcus carnosus by the antiterminator protein GlcT. Microbiology. 2000;146:2333–2342. doi: 10.1099/00221287-146-9-2333. [DOI] [PubMed] [Google Scholar]
- 29.Krüger S, Gertz S, Hecker M. Transcriptional analysis of bglPH expression in Bacillus subtilis: Evidence for two distinct pathways mediating carbon catabolite repression. J Bacteriol. 1996;178:2637–2644. doi: 10.1128/jb.178.9.2637-2644.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel HV, Vyas KA, Savtchenko R, Roseman S. The monomer/dimer transition of enzyme I of the E. coli phosphotransferase system. J Biol Chem. 2006;281:17570–17578. doi: 10.1074/jbc.M508965200. [DOI] [PubMed] [Google Scholar]
- 31.Reizer J, Saier MH., Jr Modular multidomain phosphoryl transfer proteins of bacteria. Curr Opin Struct Biol. 1997;7:407–415. doi: 10.1016/s0959-440x(97)80059-0. [DOI] [PubMed] [Google Scholar]
- 32.Yagur-Kroll S, Amster-Choder O. Dynamic membrane topology of the E. coli beta-glucoside transporter BglF. J Biol Chem. 2005;280:19306–19318. doi: 10.1074/jbc.M410896200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.