Abstract
To get mechanistic insight into the DNA strand-exchange reaction of homologous recombination, we solved a filament structure of a human Rad51 protein, combining molecular modeling with experimental data. We build our structure on reported structures for central and N-terminal parts of pure (uncomplexed) Rad51 protein by aid of linear dichroism spectroscopy, providing angular orientations of substituted tyrosine residues of Rad51-dsDNA filaments in solution. The structure, validated by comparison with an electron microscopy density map and results from mutation analysis, is proposed to represent an active solution structure of the nucleo-protein complex. An inhomogeneously stretched double-stranded DNA fitted into the filament emphasizes the strategic positioning of 2 putative DNA-binding loops in a way that allows us speculate about their possibly distinct roles in nucleo-protein filament assembly and DNA strand-exchange reaction. The model suggests that the extension of a single-stranded DNA molecule upon binding of Rad51 is ensured by intercalation of Tyr-232 of the L1 loop, which might act as a docking tool, aligning protein monomers along the DNA strand upon filament assembly. Arg-235, also sitting on L1, is in the right position to make electrostatic contact with the phosphate backbone of the other DNA strand. The L2 loop position and its more ordered compact conformation makes us propose that this loop has another role, as a binding site for an incoming double-stranded DNA. Our filament structure and spectroscopic approach open the possibility of analyzing details along the multistep path of the strand-exchange reaction.
Keywords: homologous recombination, HsRad51
Homologous recombination, the process of exchanging DNA strands of homologous sequences, is important for both the repair of damaged DNA and the maintenance of genomic diversity. In eukaryotes, from yeast to human, Rad51 is involved in homology search and DNA strand-exchange, central processes for recombination (1, 2). The mechanism of DNA strand-exchange appears evolutionarily conserved (3), and has been extensively studied with the bacterial RecA as a model protein (4–8). The nucleo-protein filament structures appear similar among the RecA and RecA-like proteins, including Rad51 (3, 9, 10). The first structure of a RecA filament was solved >15 years ago (11) and more recently structures of Rad51 proteins have been reported (12–14). Despite these efforts, though, no detailed filamentous structure of the human Rad51 protein has been reported and only the principal steps of the recombination process are known.
RecA and Rad51 protein filaments formed in presence of DNA and ATP, or a nonhydrolysable ATP analogue, all show an extended conformation with a helical pitch of ≈90–100 Å. The RecA and Rad51 filament structures vary significantly depending on the presence of DNA and type of nucleotide cofactor (10, 15–19) suggesting that the filament structure may change during the strand-exchange reaction, possibly related to its function. Thus, a structure of the human Rad51 protein filament in its complex with DNA and ATP may yield essential mechanistic information for the recombination reaction, in analogy with insight gained from crystal structures of RecA in its active filament with DNA and ATPγS (15).
Here, we present a filamentous structure of the human Rad51 protein obtained by combining molecular modeling with spectroscopic studies of the protein-dsDNA-ATP complex in solution, believed to represent the final product of strand-exchange reaction. The modeling was partly based on already known domain structures providing a model of the full-length protein monomer. The protein filament was assembled using angular data from polarized-light spectroscopy and protein engineering, so-called site-specific linear dichroism (SSLD) (16), applied to HsRad51 in its complex with dsDNA and ATP in solution. The SSLD approach is based on molecular replacement of amino acid chromophores inside a retained protein structure to determine their angular coordinates and is well suited for studying filamentous structures (16, 20, 21). We believe that our model is a good representation of a solution structure of HsRad51-dsDNA complex.
HsRad51 Monomer and Filament Structures Modeling.
Two structures of HsRad51 domains are available: a crystal structure of the central domain (14), as a fusion complex with a Rad51-binding motif of the BRCA2 protein, and an NMR structure of the N-terminal domain (12). These structures were used as a basis to model the monomer structure of the HsRad51 protein including residues 16 to 324. However, when merged together they do not result in a complete monomeric structure. Two putative DNA binding loops and, most importantly, the polymerization motif—a flexible interdomain linker—are missing. To merge the domain structures and fill in the gaps not covered by the domains, homology modeling was performed using ScRad51, the Rad51 protein homologue from yeast Saccharomyces cerevisiae, as a template structure (13).
The proteins in the RecA-Rad51 family are conserved among species regarding both sequence and structure. A ClustalW (22) comparison shows 89% sequence homology, with 68% identical residues between HsRad51 and ScRad51, and eukaryotic Rad51 proteins share a homologous core with bacterial RecA (23). Crystal structures of ScRad51 and RecA, both in filamentous forms, have been reported (11, 13, 15). A higher extent of sequence homology, and yeast and human being both eukaryotic species with similar structural domain organization, may justify our use of ScRad51 as a scaffold for the molecular modeling of a HsRad51 monomer unit. The assembled monomer model was subjected to a structure optimization to avoid sterical clashes before filament construction.
To correctly assemble the HsRad51 filament, information about orientations of protein monomers is crucial. Although crystal structures of RecA-ssDNA and RecA-dsDNA complexes (called “active”) are overall similar to that of a pure RecA filament (“inactive”), we notice significant variations of the orientation of monomers relative to filament axis. Based on the homology with RecA we shall assume that also the Rad51 protein in DNA-bound (active) and pure (inactive) forms has overall similar conformations but differ mainly regarding monomers orientation. To assist the assignment of orientation of Rad51 monomers in the nucleo-protein filament, we exploit polarized-light spectroscopic information about the angular orientations of substituted Tyr residues. The La transition moments (Fig. 1) are used as indicators of both overall orientation of monomer units and local conformational changes upon filament formation and DNA binding. Using SSLD (see below) we have collected orientation data for tyrosine-substituted Rad51 mutants for the modeling of the filamentous Rad51-dsDNA complex, believed to be the final product of the recombination reaction.
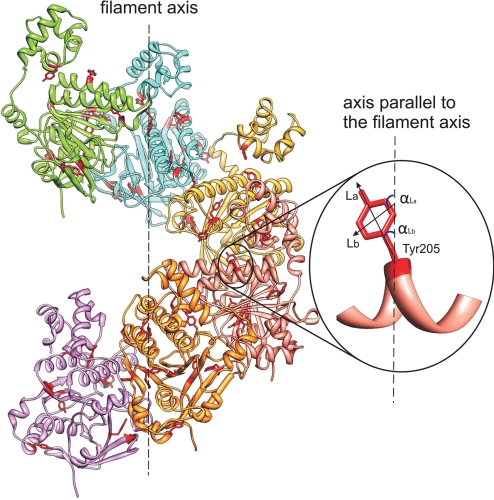
Fig. 1.
Model structures of the HsRad51 helical filament constructed by replication of the monomer structure with respect to 6.39 monomers per 1 helical turn with a pitch of 99 Å (27). The angular orientations of 2 orthogonal tyrosine transition moments La and Lb, relative to the filament axis are illustrated for Tyr-205.
To find the axis around which the filament could be built, the angular orientations of 8 tyrosine residues included in the SSLD study were compared with the angles of the transition moments of these residues, in the monomer model structure, relative to a trial filament axis. The trial axis was rotated to minimize a functional defined as a square root of the squared differences between angles calculated from the model and angles measured by SSLD (see Methods, Eq. 1 and 2). The functional has 2 local minima: They correspond to the same orientation of trial axes in 3D space but with opposite polarity (see Methods). The trial axis giving the best fit with the experimental angles was set as the filament axis. To define the inner orientations of the monomers in the filament relative to each other and the filament axis, additional information from tryptophan-scanning mutagenesis of strategically positioned amino acid residues was used (18, 24–26). The filament was constructed to have a 99 Å pitch and 6.39 monomers per turn (10, 27).
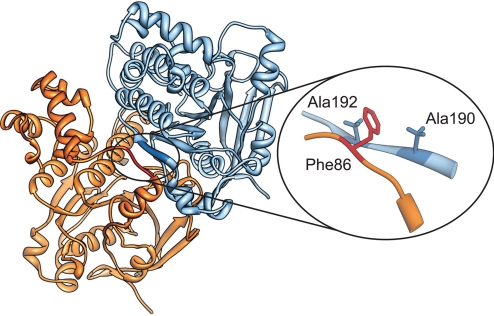
A molecular dynamics simulation of 3 monomers from the designed filament was performed to include effects from subunit-subunit interactions. The refined structure of the central monomer was then kept as the final monomeric structure. We found that in presence of a consecutive monomeric unit, the polymerization motif (the interdomain linker) of the preceeding monomer changed its conformation to arrange a short β-strand region parallel to the neighboring promoter's β-sheet (see below). These 2 β-strands appear to interact in a “key-lock” manner with each other. The Phe-86 residue, in the polymerization joint, is intercalated between Ala-190 and Ala-192 of the following monomer. Interestingly, the same key-lock pattern with an aromatic residue intercalating between 2 small nonpolar residues is also found in several other recombinase structures: the HsRad51 central domain as a fusion complex with a Rad51-binding motif of the BRCA2 protein (14), HsDms1 (28), ScRad51 (13), and in PfRad51 (29).
Minor adjustments of the orientations of the monomers in the filament were made to obtain a refined angular fit of the final structure relative to the experimental SSLD data (Table 1). The resulting structure of HsRad51 filament is shown in Fig. 1.
Table 1.
Angles of tyrosine transition moments in HsRad51 structures
| Transition moment | Wavelength/nm | Angle α/°, from SSLD* | Angle α/°, from model structure | Angle α/°, from crystal structure (1NOW) |
|---|---|---|---|---|
| Tyr 54-La | 227 | 54(54–54) | 60 | r. m. |
| Tyr 54-Lb | 278 | 40(38–44) | 47 | r. m. |
| Tyr 159-La | 227 | 35(33–41) | 36 | 40 |
| Tyr 159-Lb | 278 | 53(53–53) | 59 | 59 |
| Tyr 178-La | 227 | 71(73–65) | 30 | 27 |
| Tyr 178-Lb | 278 | 79(85–70) | 64 | 63 |
| Tyr 205-La | 227 | 43(42–47) | 53 | 54 |
| Tyr 205-Lb | 278 | 41(39–45) | 45 | 38 |
| Tyr 216-La | 227 | 56(56–55) | 76 | 75 |
| Tyr 216-Lb | 278 | 30(27–37) | 38 | 47 |
| Tyr 228-La | 227 | 28(25–36) | 29 | 41 |
| Tyr 228-Lb | 278 | 30(27–37) | 72 | 57 |
| Tyr 232-La | 227 | 40(38–44) | 48 | r. m. |
| Tyr 232-Lb | 278 | 45(44–48) | 44 | r. m. |
| Tyr 315-La | 227 | 60(61–59) | 58 | 60 |
| Tyr 315-Lb | 278 | 29(26–37) | 41 | 53 |
r. m., the residues are missing in the crystal structure.
*Angles of transition moments determined according to Eq. 3 in SI Text assuming the DNA base orientation is 80° ± 10°
As model verification a 3-dimensional reconstruction of an electron micrograph of the HsRad51-DNA filament was used as a reference (27). Our filament model structure was aligned to optimally overlap with the density map of the filament reconstruction, to compare monomer orientation relative to helical axis. Most encouragingly, an excellent agreement is found between the monomer orientations in our filamentous model and in the electron micrograph (see below). The good fit can be seen as a verification of the high quality of our model.
Modeling of Loops and DNA Binding.
In bacterial RecA and in eukaryotic Rad51 proteins 2 flexible loops, L1 and L2, have been identified and proposed to be involved in DNA binding (11, 18, 24). These loops are not visible in the crystal structures of either the central domain of HsRad51 or the ScRad51 filament and therefore particular care was taken to model these parts of the structure.
The longer L2 loop was modeled according to a protocol described by Simmerling et al. (30), whereas the conformation of the shorter L1 loop was selected from a set of low energy structures provided by the loop modeling program MODELLER (31) with an extra criterion that the orientation of Tyr-232 located in this loop should satisfy the SSLD results. For the L2 loop the modeling resulted in a fairly compact structure including some ordered α-helical region. Both loops are facing the interior of the protein filament helix (Fig. 2).
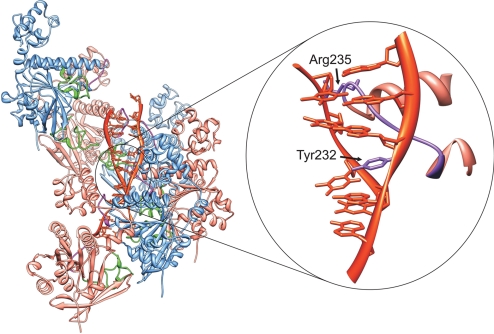
Fig. 2.
Loop orientation and DNA binding. The modeled filament structure allows the housing of a double-stranded DNA, as seen in side view. Both the L1 (magenta) and L2 (green) loops are facing the interior of the filament, available for interaction with DNA: Tyr-232 is intercalating between 2 DNA bases of one of the strands, and Arg-235 is located close to the sugar-phosphate backbone of the other DNA strand.
Our filament model structure allows housing of an inhomogenously stretched double-stranded DNA molecule, whose structure was adopted from that in the RecA-dsDNA crystal (15), since Benson et al. (32) demonstrated that HsRad51 protein unwinds and stretches a DNA molecule in a way similar to RecA. The DNA structure, fitted into our protein filament, was unwound to 19 b.p. per DNA-helical turn and elongated by 60% in an inhomogeneous manner as theoretically predicted by Prévost and Takahashi (5). The L1 loop contains 2 important residues according to mutation analysis (24, 25): Tyr-232, shown to be essential for the DNA binding, and Arg-235, facilitating the strand-exchange reaction. In our model, both residues appear in direct proximity to DNA: Tyr-232 is intercalating between 2 DNA bases of one of the strands, whereas Arg-235 is located close to the sugar-phosphate backbone of the other DNA strand. The structural details should be taken for what they are worth, recalling that the DNA-protein interactions were not subjected to energy minimization.
SSLD Spectra of HsRad51-DNA Complex.
To obtain structural information about the HsRad51-DNA complex in solution, we applied site-specific linear dichroism (SSLD) spectroscopy. SSLD is based on molecular replacement of individual aromatic residues in the protein with other aromatic residues providing angular orientations of the replaced residues relative to the filament axis (16, 21). These angles are indicative of the local structure around the residue and of the orientation of the whole protein subunit. A collection of angular coordinates for a set of aromatic residues, thus, greatly reinforces the structural information about the protein organization in the filament. The HsRad51 protein lacks tryptophan, which is otherwise the aromatic residue that provides the most distinct SSLD signal, but it contains 10 tyrosine residues (23) that were chosen as targets for this study. To apply the SSLD technique these residues were replaced, one at a time, by phenylalanine using site-directed mutagenesis (for the oligonucleotide sequences, see Table S1). The tyrosine to phenylalanine modification corresponds structurally to the elimination of an oxygen atom, and is expected to cause minimal or negligible conformational change of the protein. The expression level of soluble protein differed somewhat among the mutant proteins and was generally lower for mutants compared with that of wild-type protein. The lowest expression levels were observed for mutants Y191F and Y301F, resulting in very low total yield of purified protein. These 2 protein modifications were therefore excluded from the study.
To verify that the replacement of tyrosine residues by phenylalanine did not significantly affect the ability of the proteins to form active filaments, the strand-exchange activities of wild-type and modified HsRad51 proteins were compared. All modified proteins included in the study were shown to be active, although some decrease in activity was observed for Y159F, Y205F, and Y228F (see Fig. S1). As a control, to prove that the tyrosine replacement did not effect the protein secondary structure, CD spectra were measured for all mutants.
Flow-LD spectra of wild-type and modified HsRad51 protein complexes with double-stranded calf-thymus DNA in the presence of ATP as a cofactor were measured. These complexes represent the immediate-product state after the strand-exchange reaction has been finished but before DNA leaves the filamentous protein scaffold. The LD signal of the substrate complex of HsRad51 with single-stranded DNA and ATP (precursory complex) was too weak to allow for a quantitative analysis.
An LD spectrum of wild-type protein complex was measured first and last in each series of measurement; comparison of these spectra showed only minor structural differences, assuring that ATP hydrolysis was sufficiently slow to be negligible on the time scale of experiment. Assuming that the structure of nucleo-protein filaments formed by wild-type and modified proteins are effectively the same the differentials of LD spectra can be evaluated to determine orientation coordinates of the replaced residue chromophores according to the SSLD approach.
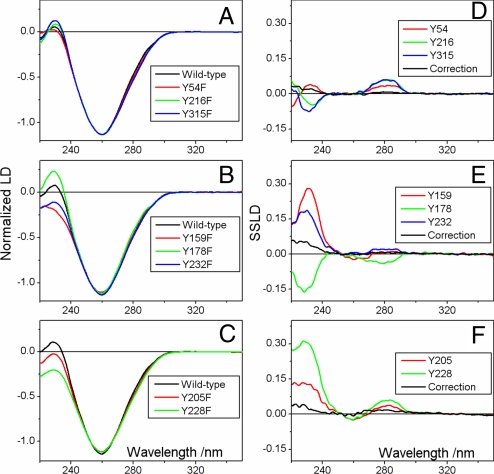
Fig. 3 A–C show the LD spectra of HsRad51-DNA-ATP filaments, normalized at the “magic wavelength” 250–255 nm (wavelengths where identical extinction coefficients of pure absorbers with identical specific LD allow for normalization of orientation factors) (see SI Text, Table S2, and Figs. S2–S4). There are clearly some distinct variations in the spectra of modified protein complexes, compared with the spectrum of the complex formed by wild-type protein. The most obvious differences are seen around 230 nm, but also around 280 nm the spectra differ significantly. These wavelength regions correspond to the La and Lb transition moments, respectively, of tyrosine. The spectral difference, computed as the differential spectrum between LD spectra of wild-type protein complex and modified protein complex, is the SSLD spectrum of the particular substituted residue (Fig. 3 D–F).
Fig. 3.
Experimental flow LD spectra (A–C) of wild-type and modified HsRad51 nucleo-protein complexes and corresponding SSLD spectra (D–F) for the tyrosine residues. For details about normalization and baseline corrections, see SI Text.
The angular orientation α of transition moments, for each substituted tyrosine residue relative to the helix axis of the nucleo-protein filament, can be determined from the reduced LD if the orientation factor S is known (Eq. 3 in SI Text). Assuming that the orientation of DNA in the complex with Rad51 is similar to that in the RecA-DNA complexes, which has been assessed from small-angle neutron scattering measurements to be 80° ± 10° for double-stranded DNA (20), an approximate absolute value of the S factor can be calculated from the normalized LD intensity at the “magic wavelength” 250–255 nm. Using this approach the orientation angles for the 8 tyrosine residues of HsRad51 included in this study were determined from the SSLD spectra (Table 1). The confidence intervals for the calculated α angles originate from the limits given for the DNA base orientation.
Discussion
We report a structural model for the filamentous complex of HsRad51 protein with double-stranded DNA in presence of ATP in solution based on molecular modeling of protein filament structure combined with angular information from SSLD spectra of mutant complexes. The model opens a possibility for systematic study of Rad51 function at different steps of strand-exchange reaction.
Protein–Protein Interactions.
The filament assembly following upon the monomer modeling was based on angular SSLD data, providing location of filament axis, and on tryptophan-scanning mutation analysis defining the monomer orientations relative to each other and the filamentous axis (18, 24, 25). A final constraint was the helical pitch and the number of monomers per turn of the HsRad51 filament that can be estimated from electron microscopy studies (10, 27). None of these data alone could determine the filament structure, and combined together they resulted in a few steric inconsistencies; therefore, further structural refinement was necessary. A molecular dynamics simulation of a 3 monomers long filament solved all clashes and inconsistencies resulted in a conformational change of flexible interdomain linkers at monomer-monomer interfaces. The resulting conformation of the interdomain joint together with the neighboring promoter's β-sheet is in good agreement (Fig. 4), a <2 Å rmsd over backbone atoms, with the corresponding region of the HsRad51 central domain fusion complex with a Rad51-binding motif of the BRCA2 protein (14).
Fig. 4.
Oligomerization of HsRad51. The Rad51 filament assembly is coordinated by the β-strand region of the interdomain linker (red) together with a β-strand (blue) of the adjacent monomer. Interaction between neighboring monomers follows a “key-lock” principle; Phe-86 is intercalated between Ala-190 and Ala-192.
When examining the monomer-monomer interface in the filament model structure, Phe-126 and Phe-129 of one subunit, and His-294 of a neighboring monomer were found inserted into a pocket at the interface between 2 monomers. These residues have been suggested to be close to the ATP binding site (18) and the pocket in our model is well suited to snugly fit the nucleotide cofactor. In the recently presented structure of the RecA-DNA complex filaments, the ATP analogue is located in a similar pocket between adjacent monomers (15). Moreover, in our model the residues lie in the proximity of the L2 loop, one of the proposed DNA binding sites. One could speculate that after completion of the strand-exchange reaction ATP to ADP hydrolysis could promote filament disassembly by destabilizations of both DNA-monomer and monomer-monomer interfaces. These speculations are consistent with the observation that Phe-129 and His-294 are important for the ATP-dependent DNA binding and the strand-exchange activity of HsRad51 (18) and with the observed rotation of protein lobes upon variation of nucleotide cofactor (10). The effects of ATP/ADP (not included explicitly in current modeling) on protein filament assembly and function will be the target of a separate study.
Protein–DNA Interactions.
Our computer modeling includes analysis of the proposed DNA binding loops L1 and L2. In the resulting structure, the short L1 loop is flexible and in a random coil conformation, whereas the longer L2 loop shows some ordered α-helical structure that is different from the conformation reported for the L2 loop of RecA (15). This is not surprising because the RecA and Rad51 have different sequences of the L2 loops, and possibly different modes of DNA interaction (24). The compact conformation of the L2 loop and the fact that mutations of a number of residues within the loop do not significantly affect HsRad51-DNA binding (24, 33), leads us to conclude that this loop provides additional, nonspecific stabilization to the nucleo-protein complex. In our structure both loops face the central cavity of the helical protein filament, which has the right dimensions to fit an extended double stranded DNA molecule (Figs. 2 and 5). The L2 loop is located in a position that enables direct contact with the DNA, in agreement with the claim that this loop is close to the DNA-binding site (24).
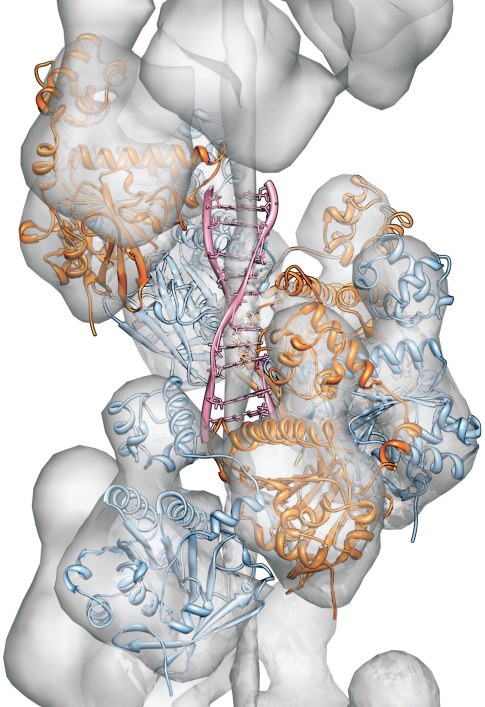
Fig. 5.
Model structure of the HsRad51 filament (monomer units in blue and orange) is aligned with the surface map of a 3-dimensional filament reconstruction from an EM density map (27). A double-stranded DNA (pink), extended in a heterogeneous manner as proposed by Prévost and Takahashi (5) and observed by Chen et al. (15), was fitted into the protein filament structure.
The L1 loop points toward the central axis and is also in direct proximity to the DNA, which is consistent with results from tryptophan-scanning mutagenesis of the L1 loop concluding that Tyr-232 is involved in DNA binding and essential for the filament assembly. Interestingly, the distance between the L1 loops of neighboring monomers in our model is ≈16 Å, which corresponds to a 3 nucleobases long DNA fragment in its stretched conformation. We may thus speculate that the L1 loop may serve like a positioning pin for the monomer during the filament assembly, having Tyr-232 intercalated between DNA bases, thus DNA itself plays a role of a precise scaffold for filament assembly. This idea also provides a possible explanation for the stretched DNA conformation reported for the RecA nucleo-protein filament (15), where exactly 3 nucleotides correspond to a protein monomer and are followed by a spatial gap that in RecA is filled by Ile-199 of the L2 loop, and in HsRad51 is filled by Tyr-232 of the L1 loop. That Tyr-232 is indeed intercalated is supported by the observation that its replacement by a tryptophan, which is a more bulky residue, strongly reduces the binding affinity (26). Arg-235, also sitting on L1 and shown to facilitate the strand-exchange reaction, occupies a position enabling direct binding to the DNA phosphate backbone of the other strand. Other residues, Asp-231, Ser-233 and Gly-236, are close to the DNA binding site but not in direct contact with DNA (24, 25). The energetic aspects of HsRad51-DNA interactions for the multistep recombination reaction will be investigated further.
SSLD Spectroscopy.
The orientations of the 8 tyrosine residues of the HsRad51 determined from the SSLD data were compared with the orientations of the same residues in the model structure (Table 1). Two angular orientations, those of the La and Lb transition moments are reported for each tyrosine residue. The Lb transition, which could be significantly affected simply by rotation of the aromatic ring around the C1−4 axis, should be considered less robust in general compared with the La transition, which requires a larger conformational change to be affected. Comparing the angles from the experimental and modeled data shows a good agreement for most of the residues, verifying the quality of the model structure. By contrast, for 2 residues, Tyr-178 and Tyr-216, significant discrepancies are observed for the La transitions. Both residues are located on the surface of the protein filament, hence exposed to solvent water and could be regarded as flexible, adopting conformations that might vary between different monomers in the filament. In the SSLD technique the measured orientation of a particular residue will be an average over the orientations of that residue in all monomers along the filament, whereas the computer modeling yields 1 distinct conformation for each residue. This could explain the angular variations between model structure and the experimental data for these flexible residues.
Methods
Molecular Modeling.
The modeling of the monomer structure of HsRad51 protein was performed on the basis of 2 known domains of the structure. The central domain (amino acid residues 98–339) (14) and the N-terminal domain (amino acid residues 16–85) (12), both structures are available in the protein data bank (34) (PDB entries 1N0W and 1B22, respectively). The sequences of the 2 domains do not overlap and the assembly of the domain structures, and the addition of other missing parts, was accomplished by homology modeling with the help of Swiss-PDB Viewer (35). As a homologous structure a monomer structure of the Rad51 protein from the yeast S. cerevisiae, ScRad51, was used (PDB entry 1SZP) (13). The ScRad51 protein filament shows a pseudo 6-fold helical symmetry, with alternating protein–protein interfaces being slightly different. However, the structural deviation between the 2 independent monomers is small enough to be negligible when using the ScRad51 structure as a scaffold for the starting model of HsRad51 and, thus, one of the ScRad51 monomers was arbitrarily chosen for the homology modeling. The assembled model, obtained from SWISS-MODEL Homology modeling server (36), including the merged domain structures and the missing regions taken from the ScRad51 structure was then subjected to further structure optimization consisting of 600 steps of steepest descent followed by 400 steps of conjugate gradient performed in explicit water, with the help of the molecular dynamics software package AMBER10 (37).
A protein filament structure was designed in 2 steps: first, determination of a filament axis and filament assembly, and second, determination of monomer interfaces and their orientations in the protein filament relative to the filament axis. To define the filament axis the monomer model structure was aligned with respect to a trial axis. The angular orientations of 8 tyrosine residues, relative to this axis, were calculated and compared with the transition moment angles of these residues as determined experimentally by SSLD. A rotation of the trial axis, to obtain the best angular fit, was determined by minimizing the functional that returns the root mean square deviation between angles measured experimentally and calculated from the model structure,
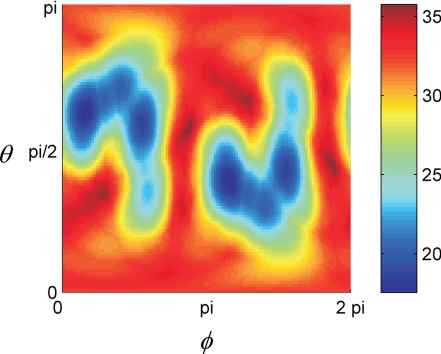
where (θ, φ) are zenith and azimuth angles from the spherical coordinate system that defines the orientation of the trial axis and α is the vector of angles between a transition moment in a tyrosine residue and the filament axis. The minimization of the functional was performed in 2 steps. In the first step the search was made for the values of zenith (0 ≤ θ < π) and of azimuth (0 ≤ φ < 2π) angles. The pair (θ,φ) giving the minimum value of the functional was taken as the starting point for the second step, in which the functional was subjected to minimization with the help of the conjugate gradient method. The functional has 2 local minima that correspond to the same orientation of the trial axes but with mutually inverse axis directions (Fig. 6). The trial axis that gave the best fit to the experimentally determined angles was set as the filament axis:
Once the protein filament axis was found the 6 monomer model structures were then assembled into a filamentous structure with a helix pitch distance of 99 Å taking into account that 6.39 monomers form 1 turn of a helical filament (10, 27). Orientation of the monomers in the filamentous structure was set to be consistent with the results from a tryptophan-scanning mutagenesis of informatively positioned aromatic residues across HsRad51: H294, Y315, and Y191 at the subunit-subunit interface (18, 26), Y232 and Y301 in the DNA binding regions and thus located in the nearest proximity of the filament center (18, 24, 25). Loop modeling was performed according to a protocol described by Simmerling et al. (30) for a loop of similar length as the L2 loop, using the molecular dynamics software package AMBER10. For the shorter L1 the conformation search was performed with the help of the loop modeling program MODELLER (31). AMBER10 was used for extra refinement of the structure, including modeling of a filament fragment of 3 monomers to include contact regions between subunits, which was subjected to a 300 ns run at 300 K and in explicit water.
Fig. 6.
Dependence of the functional (Eq. 1) on spherical (θ,φ) angles. The numbers to the right of the color bar represent the values of the functional given in degrees.
As a verification of the model the nucleo-protein filament structure was aligned to the density map from a 3-dimensional electron micrograph reconstruction of a HsRad51-DNA complex filament (27). The alignment was performed using UCSF Chimera computer modeling tool (38), result shown in (Fig. 5).
Preparation of Wild-Type and Modified HsRad51 Proteins.
Wild-type HsRad51 protein was expressed and purified as described in ref. 39, with minor modifications; the second column was a Sepharyl S-200 HR column (Amersham Biosciences) and peak fractions from DEAE-5PW column (Tosoh) were pooled and directly dialyzed against DEAE buffer. To prepare modified HsRad51 proteins site-directed mutagenesis was performed on M13-KM40, an M13 phage containing the HsRad51 gene under the T7 promoter control (oligonucleotide sequences used for mutagenesis are given in Table S1). The modified HsRad51 proteins were expressed by infecting E. coli strain cells XL1 Blue (40) carrying a plasmid pTI1219 that expresses T7 RNA polymerase, with the modified M13-KM40 phage in LB-medium containing 50 mg/L isopropyl-β-d-thiogalactopyranoside as described in ref. 16. Proteins were then purified as wild-type protein.
LD Measurements.
Linear dichroism (LD) spectra were measured in a Couette flow device as described in ref. 41. The samples contained 4 μM HsRad51 protein, 300 μM ATP and 12 μM (base pair) calf-thymus DNA. The buffer (pH 7.1) contained 10 mM K2HPO4, 10 mM KH2PO4, 20 mM Tris·HCl, 0.4 mM EDTA, 1.4 mM MgCl2, 0.4 mM DTT, 80 mM NaCl and 30% glycerol. Samples were preincubated at 22 °C for 2 h before loading into the system. The spectra were averaged over 4 scans (scan speed 50 nm·min−1; bandwidth 2 nm; shear gradient 1,200 s−1). As a baseline, the signal obtained with very slow rotation of the cell (shear gradient 90 s−1) was used to minimize influence of optical inhomogeneities of the cell. For principles and technical details of LD measurements, see SI Text.
Supplementary Material
Acknowledgments.
We thank Professor Edward H. Egelman (University of Virginia, Charlottesville, VA) for kindly providing us with the surface map of a 3D reconstruction of a HsRad51 filament electron micrograph and Dr. Axelle Renodon-Cornière for performing the recombinase activity measurements. This work was supported by King Abdullah University of Science and Technology Grant KUK-11-008-23 and the Association pour la Recherche sur le Cancer (M.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 13147.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902723106/DCSupplemental.
References
- 1.Baumann P, West S. Role of the human Rad51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 2.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 3.Bianco P, Tracy R, Kowalczykowski S. DNA strand exchange proteins: A biochemical and physical comparison. Front Biosci. 1998;3:570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 4.Cox MM. Recombinational DNA repair in bacteria and the RecA protein. Prog Nucleic Acid Res Mol Biol. 1999;63:311–366. doi: 10.1016/s0079-6603(08)60726-6. [DOI] [PubMed] [Google Scholar]
- 5.Prévost C, Takahashi M. Geometry of the DNA strands within the RecA nucleofilament: Role in homologous recombination. Q Rev Biophys. 2003;36:429–453. doi: 10.1017/s0033583504003956. [DOI] [PubMed] [Google Scholar]
- 6.Radding CM. Helical interactions in homologous pairing and strand exchange driven by RecA protein. J Biol Chem. 1991;266:5355–5358. [PubMed] [Google Scholar]
- 7.Roca AI, Cox MM. RecA protein: Structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi M, Nordén B. Structure of RecA-DNA complex and mechanism of DNA strand exchange reaction in homologous recombination. Adv Biophys. 1994;30:1–35. doi: 10.1016/0065-227x(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 9.Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast Rad51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 10.Yu X, Jacobs S, West S, Ogawa T, Egelman EH. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc Natl Acad Sci USA. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Story RM, Weber IT, Steitz TA. The structure of the E. coli RecA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 12.Aihara H, Ito Y, Kurumizaka H, Yokoyama S, Shibata T. The N-terminal domain of the human Rad51 protein binds DNA: Structure and a DNA binding surface as revealed by NMR. J Mol Biol. 1999;290:495–504. doi: 10.1006/jmbi.1999.2904. [DOI] [PubMed] [Google Scholar]
- 13.Conway AB, et al. Crystal structure of a Rad51 filament. Nat Struct Mol Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 14.Pellegrini L, et al. Insights into DNA recombination from the structure of a Rad51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 16.Morimatsu K, Takahashi M, Nordén B. Arrangement of RecA protein in its active filament determined by polarized-light spectroscopy. Proc Natl Acad Sci USA. 2002;99:11688–11693. doi: 10.1073/pnas.142404499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.VanLoock MS, et al. ATP-mediated conformational changes in the RecA filament. Structure. 2003;11:187–196. doi: 10.1016/s0969-2126(03)00003-0. [DOI] [PubMed] [Google Scholar]
- 18.Renodon-Corniere A, et al. Structural analysis of the human Rad51 protein–DNA complex filament by tryptophan fluorescence scanning analysis: Transmission of allosteric effects between ATP binding and DNA binding. J Mol Biol. 2008;383:575–587. doi: 10.1016/j.jmb.2008.08.030. [DOI] [PubMed] [Google Scholar]
- 19.Yu X, VanLoock MS, Yang S, Reese JT, Egelman EH. What is the structure of the RecA-DNA filament? Curr Protein Pept Sci. 2004;5:73–79. doi: 10.2174/1389203043486883. [DOI] [PubMed] [Google Scholar]
- 20.Nordén B, et al. Structure of RecA-DNA complexes studied by combination of linear dichroism and small-angle neutron scattering measurements on flow-oriented samples. J Mol Biol. 1992;226:1175–1191. doi: 10.1016/0022-2836(92)91060-3. [DOI] [PubMed] [Google Scholar]
- 21.Frykholm K, Morimatsu K, Nordén B. Conserved conformation of RecA protein after executing the DNA strand-exchange reaction. a site-specific linear dichroism structure study. Biochemistry. 2006;45:11172–11178. doi: 10.1021/bi060621q. [DOI] [PubMed] [Google Scholar]
- 22.Larkin MA, et al. ClustalW and ClustalX version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 23.Shinohara A, et al. Cloning of human, mouse and fission yeast recombination genes homologous to Rad51 and RecA. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 24.Matsuo Y, Sakane I, Takizawa Y, Takahashi M, Kurumizaka H. Roles of the human Rad51 L1 and L2 loops in DNA binding. FEBS J. 2006;273:3148–3159. doi: 10.1111/j.1742-4658.2006.05323.x. [DOI] [PubMed] [Google Scholar]
- 25.Prasad T, Yeykal C, Greene E. Visualizing the assembly of human Rad51 filaments on double-stranded DNA. J Mol Biol. 2006;363:713–728. doi: 10.1016/j.jmb.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 26.Selmane T, et al. Identification of the subunit-subunit interface of Xenopus Rad51.1 protein: Similarity to RecA. J Mol Biol. 2004;335:895–904. doi: 10.1016/j.jmb.2003.11.045. [DOI] [PubMed] [Google Scholar]
- 27.Galkin V, et al. BRCA2 BRC motifs bind Rad51-DNA filaments. Proc Natl Acad Sci USA. 2005;102:8537–8542. doi: 10.1073/pnas.0407266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinebuchi T, et al. Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein DMC1. Mol Cell. 2004;14:363–374. doi: 10.1016/s1097-2765(04)00218-7. [DOI] [PubMed] [Google Scholar]
- 29.Shin DS, et al. Full-length archaeal Rad51 structure and mutants: Mechanisms for Rad51 assembly and control by BRCA2. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simmerling C, Strockbine B, Roitberg AE. All-atom structure prediction and folding simulations of a stable protein. J Am Chem Soc. 2002;124:11258–11259. doi: 10.1021/ja0273851. [DOI] [PubMed] [Google Scholar]
- 31.Fiser A, Do R, Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benson F, Stasiak A, West S. Purification and characterization of the human Rad51 protein, an analog of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurumizaka H, Aihara H, Kagawa W, Shibata T, Yokoyama S. Human Rad51 amino acid residues required for Rad52 binding. J Mol Biol. 1999;291:537–548. doi: 10.1006/jmbi.1999.2950. [DOI] [PubMed] [Google Scholar]
- 34.Berman H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaplan W, Littlejohn TG. Swiss-PDB Viewer (Deep View) Brief Bioinform. 2001;2:195–197. doi: 10.1093/bib/2.2.195. [DOI] [PubMed] [Google Scholar]
- 36.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Case D, et al. San Francisco: University of California; 2008. AMBER 10. [Google Scholar]
- 38.Pettersen E, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Kim HK, Morimatsu K, Nordén B, Ardhammar M, Takahashi M. ADP stabilizes the human Rad51-single stranded DNA complex and promotes its DNA annealing activity. Genes Cells. 2002;7:1125–1134. doi: 10.1046/j.1365-2443.2002.00588.x. [DOI] [PubMed] [Google Scholar]
- 40.Bullock WO, Fernandez JM, Short JM. XL1-Blue—a high-efficiency plasmid transforming RecA Escherichia-coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 41.Nordén B, Kubista M, Kurucsev T. Linear dichroism spectroscopy of nucleic acids. Q Rev Biophys. 1992;25:51–170. doi: 10.1017/s0033583500004728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.