Abstract
Combinatorial genetics for conditional transgene activation allows studying gene function with temporal and tissue specific control like the Gal4-UAS system, which has enabled sophisticated genetic studies in Drosophila. Recently this system was adapted for zebrafish and promising applications have been introduced. Here, we report a systematic optimization of zebrafish Gal4-UAS genetics by establishing an optimized Gal4-activator (KalTA4). We provide quantitative data for KalTA4-mediated transgene activation in dependence of UAS copy numbers to allow for studying dosage effects of transgene expression. Employing a Tol2 transposon-mediated KalTA4 enhancer trap screen biased for central nervous system expression, we present a collection of self-reporting red fluorescent KalTA4 activator strains. These strains reliably transactivate UAS-dependent transgenes and can be rendered homozygous. Furthermore, we have characterized the transactivation kinetics of tissue-specific KalTA4 activation, which led to the development of a self-maintaining effector strain “Kaloop.” This strain relates transient KalTA4 expression during embryogenesis via a KalTA4-mediated autoregulatory mechanism to live adult structures. We demonstrate its use by showing that the secondary octaval nucleus in the adult hindbrain is likely derived from egr2b-expressing cells in rhombomere 5 during stages of early embryogenesis. These data demonstrate prolonged and maintained expression by Kalooping, a technique that can be used for permanent spatiotemporal genetic fate mapping and targeted transgene expression in zebrafish.
Keywords: enhancer trap, fate mapping, Gal4-UAS, secondary octaval nucleus
Zebrafish has received increasing attention as a model organism in the fields of developmental biology, cell biology, and behavioral analysis, and thus in recent years, combinatorial genetics has been established for this model organism as well. So far the adaptation of the Gal4 system used in Drosophila has received the most attention (1, 2). Here, variants of the yeast transcriptional activator Gal4 are expressed to drive the expression of effector transgenes under the control of Gal4-specific binding sites called upstream activating sequences (UAS). When the Gal4 activator and UAS effector are expressed by different stable transgenic strains, the offspring of activator/effector crosses express a Gal4-dependent transgene in a tissue-specific manner. As activator and effector strains can be freely combined, a multitude of targeted transgene expression studies can be performed from a limited number of transgenic strains. Such transgenic zebrafish strains have already proven their usefulness in cell type-specific ablation studies, the mapping of neuronal circuits and the inhibition of neuronal activity in distinct neuronal populations (3–5).
To fully exploit the potential of Gal4 genetics in zebrafish we have tested a series of activator and effector combinations and have successively optimized their transactivation potential in zebrafish. Subsequently, a Tol2 transposon mediated self-reporting Gal4 enhancer trap screen was performed to establish a collection of cell type-specific activator lines with characterized transactivation kinetics that will provide a useful resource for targeted transgene activation in zebrafish.
Recently, a combinatorial genetic fate mapping system was established in mouse to permanently label cells with reporter gene expression under inducible temporal and cell type-specific control (6). The use of this technique changed long-standing views about cerebellar and hindbrain development (7–9). Such a genetic fate mapping system is not yet available in zebrafish, but a first crucial step toward it is the possibility to continuously express transgenes in specific cell types by combinatorial genetics, thereby reporting the transient activity of an enhancer within the subsequently formed adult structures. We thus generated a stable transgenic UAS effector strain, 4xKaloop, which continuously self-maintains a period of transient embryonic Gal4 expression to adulthood via an autoregulatory Gal4-mediated expression loop. By such “Kalooping” we demonstrate that the secondary octaval nucleus in the adult zebrafish hindbrain is likely derived from rhombomere 5 during early embryogenesis. Thus, by establishing a combinatorial genetic system for continuously reporting transient gene activity, we have laid the foundation for non-invasive spatiotemporal fate mapping and life-long continuous transgene expression in zebrafish.
Results
Optimizing the Gal4 System for Zebrafish.
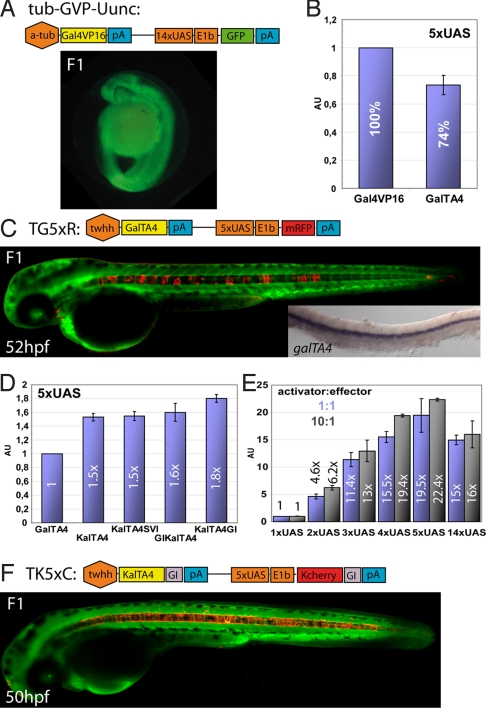
To establish transgenic zebrafish with neuronal-specific expression of the transcriptional activator Gal4VP16 we injected linearized, previously established tub-GVP-Uunc vector (10) into 1-cell stage embryos. Transgenic F1-founders showed strong ubiquitous expression (Fig. 1A) and failed to survive beyond embryonic day 10. Previously, attenuated repeats of the 13-amino acid core sequence of the VP16 transactivation domain have been shown to be similarly potent in transgene activation but displaying fewer promiscuous and likely toxic protein-protein interactions and are thus better tolerated by cells (11). In luciferase assays (n = 3) using a 5xUAS:luciferase construct as reporter GalTA2, TA3, and TA4 fusion proteins, produced the same order of activation potential (TA2>TA3>TA4) in zebrafish Pac2 fibroblasts than reported for human cells (11). As the reduction of the activation potential was less severe than in these human cells, we continued to work with the least active GalTA4 fusion protein (74% activity of full VP16 domain, Fig. 1B), reasoning that zebrafish would better tolerate further attenuations of this milder Gal4 activator.
Fig. 1.
Optimization of the Gal4 system in zebrafish. (A) Schematic representation of construct tub-GVP-Uunc (10) and transgenic F1 zebrafish embryos. (B) Comparison of Gal4VP16 (set as 100%) and GalTA4 activity in luciferase assays (n = 3, Pac2 fibroblasts) using a 5xUAS:luciferase construct. (C) Schematic representation of the TG5xR construct and transgenic F1 embryo. Inset: mRNA in situ hybridization of GalTA4 expression throughout the notochord. (D) Activation potentials of differently modified Gal4 activators as determined by luciferase assays (n = 3). The activity of GalTA4 was set as 1. (E) Effects of different numbers of Gal4 DNA binding sites using pCSKalTA4GI and UAS-luciferase constructs (ratio 1:1 blue columns or 10:1 black columns) in luciferase assays (n = 3, 1xUAS:luciferase set as 1). (F) Schematic representation of the TK5xC construct and transgenic F1 embryo. Data are presented as mean ± SEM. Embryos in C and F are counterstained with green Bodipy Ceramide.
To establish stable transgenic fish, a construct was designed carrying the notochord specific regulatory element of the tiggy winkle hedgehog (twhh) promoter to drive expression of GalTA4 (12). As an internal reporter cassette, 5 Gal4-binding sites (5xUAS) followed by the EIb basal promoter and the red fluorescent protein mRFP1 were used (1, 13). Stable transgenic embryos derived from injections of this linearized expression construct, TG5xR, were viable, showed no malformations and could be raised to sexual maturity. Although they displayed notochord specific mRFP1 fluorescence, this expression was highly mosaic (Fig. 1C). Surprisingly, RNA in situ hybridizations revealed ubiquitous transcription of GalTA4 within the notochord (Fig. 1C, inset) suggesting a problem with GalTA4 translation. We thus altered the codon usage of GalTA4 ORF for efficient translation in zebrafish and added 5′ the Kozak sequence GCCGCCACC (KalTA4). In addition, we tested intron sequences either from SV40 placed 3′ to the ORF of KalTA4 (KalTA4SVI), or the rabbit β-globin intron used either in 5′ or 3′ position (GIKalTA4, KalTA4GI) (14, 15). Luciferase assays (n = 3 each) in zebrafish Pac2 fibroblasts revealed KalTA4GI as the most active variant, providing 1.8-fold luciferase activity compared to GalTA4 (Fig. 1D). Next, the effect of the number of Gal4-binding site repeats on KalTA4GI-mediated transgene activation was evaluated by luciferase assays. We found 2xUAS to be 4.6× stronger, 3xUAS 11.4×, 4xUAS 15.5×, and 5xUAS 19.5× stronger in transgene activation than the minimal 1xUAS effector cassette (Fig. 1E). Intriguingly, 14xUAS did not show the maximal activation level, but was comparable to 4xUAS (similar results were found with a 10-fold excess of transfected activator constructs, black columns). These findings show that adding Gal4-binding sites in effector constructs increases transgene expression in a roughly linear manner, but suggest a threshold for the optimal number of Gal4-binding repeats. In addition, our data provide a quantitative basis for dose-varying transgene expression in zebrafish.
Next, we cloned an expression construct, TK5xC, in which translation-optimized KalTA4GI activated by the twhh-enhancer was followed by 5xUAS sites controlling the expression of KCherryGI (Kozak sequence 5′ and rabbit β-globin intron 3′ of ORF encoding the red fluorescent protein mCherry). In addition, this construct was flanked by inverted repeats of the Tol2 transposon. Co-injection of TK5xC with Tol2 transposase mRNA into 1-cell stage embryos resulted in the generation of transgenic strains, of which F2 founder embryos displayed bright red fluorescence throughout the notochord (Fig. 1F, 3 independent lines). This demonstrates that optimization of translation can efficiently counteract mosaic Gal4-dependent transgene expression (compare Fig. 1 C with F) (16). Thus, for optimizing Gal4-dependent tissue-specific expression in zebrafish we have successfully balanced cellular concentrations of the activator (optimized translation) with its non-promiscuous activation strength (TA4 domain).
KalTA4 Enhancer Trapping.
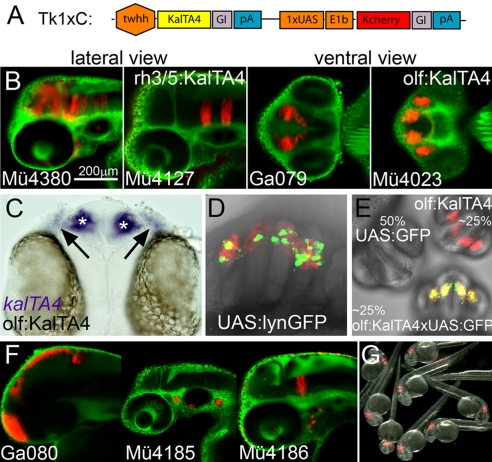
The twhh enhancer drives expression in notochord cells. The construction of TK5xC transgenic fish strains although revealed that this enhancer is sensitive to positional effects and does not result in any background expression (see database in SI Text). Based on this experimental observation we used this construct for systematic Tol2 transposon-based KalTA4 enhancer trapping. To bias our screen toward detecting only strong positional effects only 1 instead of 5 Gal4-binding sites was used (construct: TK1xC, Fig. 2A). Screening of the F1 generation was performed between 24 and 36 hpf.
Fig. 2.
KalTA4GI enhancer trapping. (A) Schematic representation of the TK1xC enhancer trapping construct. (B) Examples of transgenic Gal4 enhancer trap lines at 50 hpf (C) kalTA4 mRNA expression (asterisk: olfactory bulb, arrow olfactory epithelium) in line olf:KalTA4. (D) Transactivation of lynGFP in Ulyn (10) injected olf:KalTA4 embryo. (E) KalTA4-mediated transactivation of GFP expression in offspring (26 hpf) from crosses of heterozygous olf:KalTA4 and 4xUAS-KGFPGI carriers. (F) Different expression patterns in F1 embryos (50 hpf, lateral view) derived from the same P0 founder fish. (G) Offspring of a rh3/5:KalTA4 homozygous carrier crossed to a wild-type fish. Note that all embryos display the characteristic mCherry fluorescence in rhombomeres 3 and 5. Embryos in B and F are counterstained with green Bodipy Ceramide.
Out of 94 injected P0 embryos that were raised to adulthood, 38 (40%) transmitted the transgene cassette to the next generation and gave stable transgenic strains with tissue specific mCherry fluorescence. These isolated enhancer trap strains showed no signs of malformation, retarded development or reduced viability.
For example, fish were recovered with expression in the midbrain and anterior hindbrain (Mü4380), specific rhombomeres (Mü4127), the diencephalon (Ga079), or the olfactory system (Mü4023, Fig. 2B). The latter (olf:KalTA4) was used for in situ hybridization, which showed that kalTA4 mRNA was expressed bilaterally in both the olfactory epithelia and olfactory bulbs (Fig. 2C, black arrows and white asterisks). When the 14xUAS:lynGFP reporter construct (10) was injected into 1-cell stage embryos of this strain, mosaic membrane-targeted GFP expression confined only to KalTA4/mCherry expressing cells was observed (Fig. 2D). To further confirm that the isolated enhancer trap lines are capable of transactivating tissue specific transgene expression by combinatorial genetics, a 4xUAS-KGFPGI (Kozak-GFP-globin intron) transgenic strain was established and F1 heterozygous carriers were crossed with heterozygous KalTA4 enhancer trap carriers. All strains tested so far (n = 15) reliably activated GFP expression in a Mendelian ratio, but strictly confined to mCherry expressing cells (Fig. 2E), indicating that the isolated KalTA4 enhancer trap strains activate robust UAS-dependent expression in trans. Comparison of carrier crosses of 4xUAS-KGFPGI with either TK1xC (KalTA4, almost non-mosaic GFP) or TG5xR (GalTA4, highly mosaic GFP) fish revealed that the effects of KalTA4 optimization are also effective in trans. Moreover, homozygous carriers were established for several strains, which reliably passed on mCherry expression to 100% of the progeny when crossed to wild-type fish (Fig. 2G). This will be very useful for future studies where large numbers of transgene expressing offspring can be recovered without laborious screening for positive fish.
A number of P0 or F1 founders gave rise to fluorescent F1 or F2 offspring (F2 at high non-Mendelian ratios) displaying different tissue specific expression patterns (for example, all 3 patterns in Fig. 2F were derived from the same P0 founder), suggesting multiple independent transposon integrations and enhancer trap events in the germline of a single P0 founder, as revealed by Southern blot analysis of F1 offspring. In total, we have established 60 independent stable KalTA4 activator lines with tissue specific expression that remained stable over several generations (Figs. S1 and S2A). Most of their fluorescent expression patterns were recorded by laser scanning confocal microscopy and incorporated into a searchable database (Fig. S2B), providing a valuable resource for future studies by the zebrafish community. This systematic analysis indicated a bias of our enhancer trap approach for isolating central nervous system activator lines as 70% (42/60) of the identified transgenic strains showed expression in this tissue (Fig. S2C). In addition, when all analyzed fluorescent expression patterns were classified, 66% of these patterns could be clearly attributed to a neuroanatomically or neurodevelopmentally distinct cellular entity of the CNS (67/102, Fig. S2D). Currently the molecular reason for this bias is unclear and requires a detailed dissection of the regulatory twhh fragment. Intriguingly although a first bioinformatic analysis of all predicted transcription factor binding sites (MatInspector, Genomatix Software) in the twhh regulatory element revealed that more than 40% of these sites can be attributed to transcription factors that are expressed in neural tissue. Most expression patterns in the isolated enhancer trap lines were restricted to defined cell populations rather than being broadly distributed (Figs. S1 and S2D). This suggests that using a larger promoter element instead of a basal promoter to drive the Gal4 activator in the trapping construct allows one to suppress unwanted background expression and to restrict isolated trap lines to certain tissues of interest.
Based on the known flanking sequence of the Tol2 transposon, we established nested inverse PCR for mapping integration sites (17). For several enhancer trap lines, the inserted KalTA4 expression cassette could be located near a zebrafish gene for which KalTA4 and mCherry expression closely reflected the endogenous expression (Table S1). Among them we identified an inserted trap cassette about 1.5 kb downstream of the zebrafish egr2b gene known as krox20 in other vertebrates. Its prominent expression in rhombomere 3 and 5 during developmental stages of the hindbrain was reflected by mCherry fluorescence. We subsequently used this insertion to investigate the dynamic regulation of kalTA4 expression and of UAS:transgenes as direct KalTA4 downstream genes by mRNA in situ analysis.
Like endogenous egr2b (Fig. S3A–C), kalTA4 expression was initiated first in rhombomere 3 and next turned on in rhombomere 5 about 30 min later (Fig. S3D–F). Similarly, expression of both egr2b and kalTA4 ceased during embryogenesis first in rhombomere 3 and about 2 h later in rhombomere 5 (18) (Fig. S3J–L) and was never detected again in these rhombomeres until adulthood (60, 84, 132 hpf, 4 months). Thus kalTA4 expression in this strain closely reflects the endogenous expression dynamics of egr2b. Interestingly, we observed that kalTA4 expression is initiated about 90 min after egr2b (compare Fig. S3A and D) and turns off approximately 4–6 h earlier compared to egr2b (18). This is probably due to the different distance of the inserted TK1C cassette to key regulatory elements. When heterozygous carriers of the rh3/5:KalTA4 and the 4xUAS-KGFPGI transgenic strains were crossed, the first transactivated gfp mRNA expression could be observed about 30–60 min later, closely reflecting kalTA4 expression (Fig. S3G–I, note first visible gfp mRNA expression at 12 hpf). Thus, transactivation of a direct transcriptional target in zebrafish takes approximately 30–60 min. Termination of kalTA4 mRNA expression between 21 and 23 hpf (Fig. S3J–L) resulted in a decrease of transactivated gfp mRNA transcription at about 27 to 29 hpf (Fig. S3M–O), reflecting the time needed for the degradation of the KalTA4 protein. This knowledge is important for planning transactivation experiments in which the lifetime of the transactivated protein (several days in the case of GFP) affects the full duration of ectopic expression.
KalTA4-Mediated Self-Maintenance of Cell Labeling.
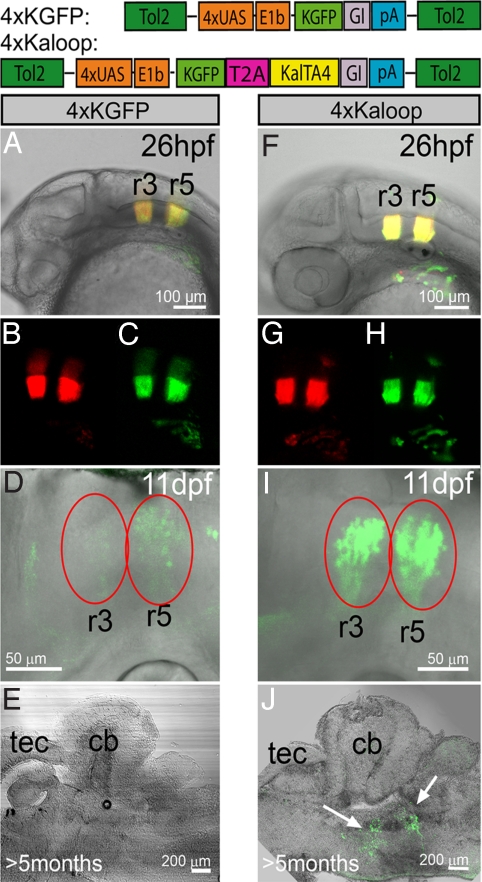
The detailed characterization of kalTA4 expression and its restriction to rhombomere 3 and 5 during a short time window of embryonic development in the rh3/5:KalTA4 strain offered the chance for developing a self-maintaining KalTA4-mediated cell labeling technique that reports the spatiotemporal expression of KalTA4 to adult stages. For this, we generated a stable transgenic effector strain, 4xKaloop, that carries a bicistronic 4xUAS effector construct driving GFP expression as a reporter followed by a peptide backbone breaking T2A sequence, which mediates stochiometric expression of KalTA4 (Fig. 3) (19). Once activated, this effector should continuously maintain its own expression by constantly providing the KalTA4 activator in a feedback loop. When heterozygous rh3/5:KalTA4 activator fish were crossed to carriers of the 4xUAS-KGFPGI and 4xKaloop effectors respectively, embryos displayed red and green fluorescence (cis- and trans-activation) strictly localized to rhombomeres 3 and 5 at 26 hpf (Fig. 3 A–C and F–H). Fluorescence was maintained in both embryos by 3 dpf due to the stability of the fluorescent proteins. After 11 days, fluorescence in 4xUAS-KGFPGI control fish was significantly decreased and later vanished completely (Fig. 3 D and E, n = 7). Strikingly, in the 4xKaloop carriers GFP continued to be expressed in 2 characteristic spatially separated stripes in the hindbrain through to adulthood (Fig. 3 I and J, n = 10). This indicates that fluorescent protein expression in 4xKaloop fish is continuously maintained.
Fig. 3.
Maintenance of rhombomeres 3/5 labeling in Kaloop fish. (A–E) Expression of GFP in rhombomeres 3/5 in offspring from a cross between rh3/5:KalTA4 and 4xUAS-KGFPGI or (F-J) rh3/5:KalTA4 and 4xKaloop carriers (schematic representation of effector constructs shown). (A–C, F–H) Expression of mCherry and transactivated GFP at 26 hpf. (D) At 11 hpf, GFP fluorescence (red ovals) is diminished in 4xUAS-KGFP carriers and (E) lost in the adult brain, while in 4xKaloop carriers (I, J) it is maintained in 2 clusters in the hindbrain until adulthood (white arrows). (E, J) Sagittal sections at 100 μm, abbr.: see Fig. 4.
The Secondary Octaval Nucleus Is Likely Derived from Rhombomere 5.
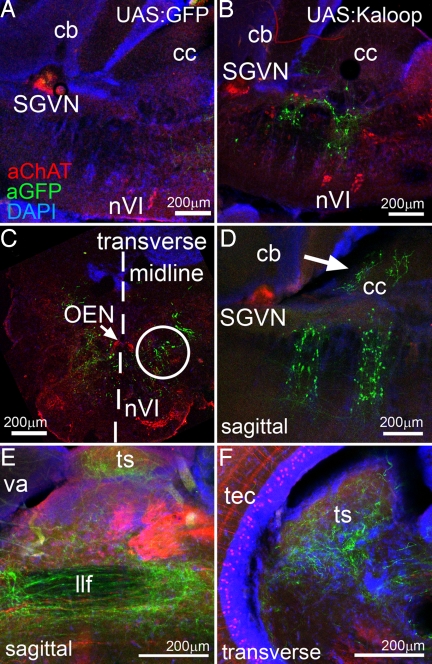
Vibratome sections of adult rh3/5:KalTA4 × 4xKaloop fish revealed that the stripes of GFP-expressing cell clusters in the hindbrain were not continuous, but displayed a mosaic arrangement of fluorescent cells (Figs. 3J and 4B, n = 10). Each time although the fluorescent cells labeled the same structures in the hindbrain but to a different extent. While this may be due to an inactivation of the 4xKaloop cassette in some cells of the total marked cell population, it serves to better reveal the detailed axonal projections of individual GFP-expressing neurons. However, it has to be kept in mind that a number of adults have to be analyzed to judge the extent of the cell population derived from the initial embryonic KalTA4-expression domain. First, to address the localization of the GFP-expressing cell clusters, we performed immunohistochemistry against GFP and choline acetyltransferase (ChAT) on sagittal sections through the hindbrain. These sections showed that GFP-expressing neurons in the anterior cluster were positioned caudally to the cholinergic secondary gustatory/viscerosensory nucleus (SGVN), which is derived from the embryonic cerebellar upper rhombic lip and positioned in the tegmental area of rhombomere 1 underneath the cerebellum. The posterior cluster of GFP-expressing neurons was positioned dorsally to the cholinergic abducens nuclei (Fig. 4B, nVI), which are localized in the ventral area of rhombomere 5-derived hindbrain tissue (20). Thus, the spatial arrangement of the GFP-expressing neurons in the adult hindbrain corresponds well to the embryonic expression pattern of egr2b. Transverse hindbrain sections revealed that the GFP-expressing neurons from rhombomere 5 were localized in a dorso-medial position with respect to the abducens nucleus (Fig. 4C, white circle) suggesting an identity as neurons of the secondary octaval nucleus [SON, (21)]. Neurons of this nucleus send dendrites into the crista cerebellaris and participate in transmitting directional acoustic information into the torus semicircularis of the midbrain, where this information converges at least in some species with sensory input from second-order projections of the lateral line (22, 23). Indeed, confocal sectioning revealed GFP-containing dendrites in the crista cerebellaris (Fig. 4D, white arrow) and axons leaving GFP-expressing neurons of the caudal GFP cell cluster in ventral directions and turning rostrally. These axonal projections passed ventrally to the tectum (Fig. 4E) into the midbrain, where they turned dorsally to terminate in the central nucleus of the torus semicircularis just underneath the optic tectum (Fig. 4F).
Fig. 4.
Rh3/5-Kalooping identifies the secondary octaval nucleus (SON) as rhombomere 5-derived. Immunhistochemistry for GFP (green) and Cholinacetyltransferase (ChAT; red) on brain vibratome sections of adult zebrafish derived from crosses of (A) rh3/5:KalTA4 × 4xKGFP and (B–F) rh3/5:KalTA4 × 4xKaloop. All nuclei were counterstained with DAPI (blue). (A and B) Sagittal sections of the hindbrain. (C) Transverse section through the caudal GFP-expressing hindbrain region (r5) in B. (midline marked by dashed line). (D) Sagittal section (rostral is left) showing GFP-positive dendrites in the crista cerebellaris (white arrow). (E) Axons of GFP-positive neurons project through the midbrain into the (F) torus semicircularis. Note typical periventricular cholinergic cells in optic tectum. Abbr.: cb: corpus cerebelli, cc: crista cerebellaris, llf: lateral longitudinal fascicle, nVI: rostral and caudal abducens nuclei, OEN: octavolateralis efferent neurons, r: rhombomere, SGVN: secondary gustatory/viscerosensory nucleus, ts: torus semicircularis, tec: optic tectum, va: valvula cerebelli.
To independently confirm the torus semicircularis as a target for axons of GFP-expressing neurons we performed retrograde tracing studies in adult rh3/5:KalTA4 × 4xKaloop fish. When axon terminals in the central nucleus of the torus semicircularis were labeled by focal rhodamine dextrane injections (Fig. S4A), the visualized afferents (Fig. S4B) were derived from the lateral longitudinal fascicle (llf), which descends caudally into the tegmentum and co-localized there with GFP-fluorescent axons (Fig. S4C, white arrow). At the level of both neuronal GFP clusters these retrogradely labeled axons traveled through ventral hindbrain regions (Fig. S4D), crossed the midline (Fig. S4E) and then turned dorsally (Fig. S4F) to eventually connect to their GFP-expressing somata (Fig. S4G–I) in the caudal cluster of rh3/5:KalTA4 × 4xKaloop fish.
The position and characteristic projection of GFP-expressing neurons in the caudal hindbrain cluster in each of the analyzed adult rh3/5:KalTA4 × 4xKaloop fish confirm their identity as neurons of the SON. Furthermore, these findings suggest that SON neurons in the adult hindbrain are derived from egr2b-expressing progenitors in rhombomere 5 during early stages of embryonic hindbrain development (before 29 hpf). Thus, genetic approaches based on the 4xKaloop effector fish may serve to permanently mark a period of transient transgene expression and provide a promising entry point for the development of genetic fate mapping technology with precise tissue specific and temporal control based on combinatorial KalTA4-UAS genetics.
Discussion
Systematic Optimization of Gal4-UAS Genetics in Zebrafish.
Based on our previous experience with Gal4 expression in zebrafish (10) we have aimed to systematically optimize this system for stable transgenic combinatorial genetics. By choosing the minimal but potent TA4 core region from the herpes simplex virus VP16 transactivation domain, optimizing the codon usage of the Gal4 DNA-binding domain for use in zebrafish, adding a Kozak sequence and identifying the 3 prime region of the KalTA4 coding sequence as the best position for a globin intron, we optimized the expression and function of a Gal4-derived transactivator. KalTA4GI produced robust non-variegated and non-deleterious transactivation, which could be further enhanced by using the more active TA3 or TA2 transactivation domains (11). As the established KalTA4-UAS vectors were adapted for a vertebrate model organism, they should provide similar results in other vertebrates including mammals, where Gal4-genetics have not gained much attention so far (24). In addition, we showed that the expression levels of transactivated transgenes could be varied stepwise from minute (1xUAS) to intense overexpression concentrations (5xUAS) by increasing the numbers of UAS sites. This finding offers the chance for studying dose-dependent effects of transgene expression and for adjusting transgene expression to physiological levels. For example, it should be possible to tailor disease models for either slow or fast progressing phenotype development.
Enhancer Trapping with Self-Reporting Optimized KalTA4 Activator.
We have used the optimized KalTA4-UAS genetic system to provide a collection of cell type-specific self-reporting KalTA4 activator lines with the help of a Tol2 transposon enhancer trap screen. These lines themselves provide a valuable resource for characterizing the dynamics of the respective fluorescent tissues. Moreover, these red fluorescent strains can be readily combined with the numerous existing GFP-expressing zebrafish lines. Further, the red fluorescent cell population simultaneously expresses KalTA4, and so can be genetically manipulated either through crossing to fish stably expressing transgenes under UAS control or through the injection of embryos with UAS constructs for mosaic analysis within a few days (10, 19).
All tissue-specific KalTA4 activator lines tested provided reliable robust transactivation when crossed to a 4xUAS-KGFPGI reporter strain and showed no expression in other tissues beside the intrinsic notochord activity of the twhh promoter. This notochord expression was weak and often appeared only at 2–3 dpf. As we have used only a single UAS site for reporting an enhancer trap event and as the trapped expression was usually much stronger than the weak notochord expression in these strains, we likely selected for strong position effects that provide sufficient levels of KalTA4 expression for robust transactivation.
Our thorough characterization of KalTA4-mediated transactivation kinetics will allow for better planning of combinatorial genetic experiments. We demonstrated that kalTA4 mRNA expression was initiated with a lag time as compared to the endogenous expression of a gene nearby the insertion site. Of note however is our observation that the positional effect of the trapped enhancer acts not only on KalTA4 expression, but it also directly activates basal promoter driven mcherry transcription. Due to this additive effect of the positional effect and KalTA4-activation, mCherry expression is higher when activated in cis from the self-reporting cassette as compared to activation in trans. Thus fluorescence from 1xUAS-mCherry (cis) and GFP (4xUAS-KGFPGI reporter strain, trans) becomes visible about the same time. First mCherry fluorescence in our transgenic strains can therefore serve as a rough indicator for the onset of activity from a 4xUAS-transgene cassette activated by KalTA4 in trans. The collection of KalTA4 activator strains annotated in an image database searchable for tissue-specific mCherry expression patterns should provide a helpful resource for targeted gene expression studies in zebrafish.
We demonstrated that our KalT4 activator strains could be bred to homozygosity. When these activator fish are crossed with homozygous UAS-effector strains, all of the offspring express the transgene of interest. This makes KalTA4-UAS genetics suitable for high-throughput in vivo approaches such as small compound screening in neurodegenerative disease models (25).
Self-Maintenance of Transgene Expression by a KalTA4-Dependent Feedback Loop.
A challenge in developmental biology is to relate embryonic gene expression to adult functional structures. We have developed the 4xKaloop effector strain, which provides a self-maintaining KalTA4-mediated feedback expression loop for propagating activator activity and reporter fluorescence until adulthood. When combined with the collection of available activator strains, this single effector line could serve to map the expression activity of many genetic loci to adult tissues and organs. We have demonstrated this reporting ability of the Kaloop strain for the krox20/egr2b locus, which in zebrafish shows highly conserved expression restricted to rhombomeres 3 and 5 during early embryogenesis (18). The 2 stripes of GFP-expressing cells in adult rh3/5:KalTA4 × 4xKaloop fish, spatially distant from one another and to the rhombomere 1 derived SGVN, resemble the organization of the egr2b expression in the embryonic brain. This suggests a preservation of segmental hindbrain organization in sequential rostro-caudal neuromeres in zebrafish, similar to the frog and chick brain (26, 27). In addition, histological analysis, tracking of axonal projections and retrograde tracing of axons derived from neurons of the posterior GFP-expressing hindbrain stripe revealed that neurons of the secondary octaval nucleus are likely derived from cells of rhombomere 5 expressing egr2b during the first day of embryonic development. This nucleus is part of the ascending acoustic neuronal network in teleosts (22), but its developmental origin has remained unclear so far.
Expression of GFP in both adult hindbrain stripes was found to be mosaic although during the first 2 weeks of development GFP expression was continuous and hindbrain organization is rather manifest by this time. This makes a migration of cells into or out of the expression stripes resulting in a dilution of the GFP expression pattern unlikely. As the degree of mosaic labeling of SON neurons varied in each analyzed specimen we do not think that only a specific subset of SON neurons is derived from the embryonic egr2b expression domain. We rather favor the explanation that GFP expression is prolonged but not maintained to the same degree in all cells derived from rhombomeres 3 and 5. In many cases, such a prolongation of reporter expression up to at least 4 weeks may be sufficient to identify the final fate of cells in the juvenile brain. It is important although to analyze several adult double transgenic carriers to fate map all adult structures that are derived from a transient tissue-specific embryonic KalTA4 expression domain in the activator strain.
At the moment, the cause for the mosaic inactivation of the 4xKaloop cassette is unclear, but it has been suggested that epigenetic events could inactivate oligomeric UAS repeats of multiple UAS sites (16). Alternatively, fewer amounts of KalTA4 from the Kaloop cassette may be produced than are needed to achieve a steady state level of KalTA4 activator concentration. Solutions to either explanation could potentially be achieved by further optimization of the Kaloop effector through the substitution of the 4xUAS with fewer UAS-sites (to reduce potential epigenetic inactivation) and of the KalTA4 with the stronger KalTA3 or KalTA2 activators (to counterbalance reduced expression from fewer UAS-sites). Our quantification of the effect of UAS numbers and different activation domains on KalTA-dependent transactivation will help to guide this optimization process. Given that GFP expression in the secondary octaval nucleus lasted for more than half a year although the initial enhancer-driven KalTA4 in the rh3/5:KalTA4 line is only active until 29 hpf (Fig. S3L and O), such further optimization promises to be feasible.
To further exploit continuous expression mediated by Kalooping and to allow for spatiotemporal fate mapping of KalTA4-expressing cells into adulthood, temporal control over KalTA4 feedback loop activation should be developed as a next step. Recent progress in establishing inducible Gal4 or recombinase activity in zebrafish (28, 29) promises that spatiotemporal Kalooping could be established in a straight-forward manner, which will provide a powerful system for precise genetic control for functional in vivo analysis in zebrafish.
Materials and Methods
Microscopy.
Counterstaining of cellular membranes in living zebrafish embryos was performed using 0,001% BODIPY FL C5ceramide (Invitrogen)/30% Danieau over night before image acquisition. For image recording embryos were dechorionated and embedded in 1.2% ultra low melting agarose/30% Danieau (30). Images of living embryos were recorded using a Zeiss LSM510 confocal microscope. Images of in situ hybridizations were recorded using an Axioplan2 microscope equipped with an AxioCam HRc and Axiovision 4.5 software (all Zeiss).
Microinjection and Generation of Transgenic Zebrafish Lines.
Zebrafish embryos were injected with Tol2 mRNA and the respective Tol2 based construct (25 ng/μL each, 1.5 nL) at 1-cell stage. Fluorescent embryos were raised to adulthood and crossed for analyzing germline transmission by screening the offspring for tissue-specific expression.
Immunohistochemistry.
For immunohistochemistry brains were dissected from 3 month or older adult zebrafish and fixed in 4% PFA over night. After washing with PBS, brains were embedded in 2% agarose/PBS for vibratome sectioning. One hundred-micrometer brain slices were cut and fixed on slides for immunohistochemical analysis. After blocking in 5%BSA in PBS/0.2% Triton X-100 for 1 h at room temperature slides were incubated with the primary chicken α-GFP (1:500) (Jackson ImmunoResearch) and goat α-ChAT (1:100) (Chemicon) antibodies in 5% BSA in PBS/0.2% Triton X-100 at 4 °C over night. After several washes in PBS/0.2% Triton X-100 slides were incubated with the secondary α-goat Alexa546 (1:100, Invitrogen) and α-chicken FITC (1:100, Jackson ImmunoResearch) antibodies in 5% BSA in PBS/0.2% Triton X-100 at 4 °C over night. Nuclei were stained using DAPI (10236276001, 1 μg/μL, Roche) and slides were mounted in Aqua Polymount (Polysciences Inc.).
Supplementary Material
Acknowledgments.
We thank Olga Lositsky, Vanessa Bednarz, Benjamin Wolf, Yuanyuan Chu, Mark Schibler, Nina Dedic, Enrico Kühn, and Petra Hammerl for excellent technical assistance and animal care. We are grateful for obtaining vectors from Elwood Linney, Christoph Winkler, Adam Amsterdam, Atsushi Miyawaki, and Roger Tsien. We thank Ralf Kühn, Jan Deussing, and Jennifer Hocking for carefully commenting on the manuscript. We acknowledge the financial support of this study by a BioFuture Award Grant (R.W.K.) of the German Ministry of Education and Research (BMBF 0311889), and a Fellowship of the Studienstiftung des deutschen Volkes (M.D.), and the Graduate School for Systemic Neurosciences (L.M.U.-Munich).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903060106/DCSupplemental.
References
- 1.Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev. 1999;80:153–158. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- 2.Deiters A, Yoder JA. Conditional transgene and gene targeting methodologies in zebrafish. Zebrafish. 2006;3:415–429. doi: 10.1089/zeb.2006.3.415. [DOI] [PubMed] [Google Scholar]
- 3.Davison JM, et al. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol. 2007;304:811–824. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott EK, et al. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods. 2007;4:323–326. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- 5.Asakawa K, et al. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci USA. 2008;105:1255–1260. doi: 10.1073/pnas.0704963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joyner AL, Zervas M. Genetic inducible fate mapping in mouse: Establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev Dyn. 2006;235:2376–2385. doi: 10.1002/dvdy.20884. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez CI, Dymecki SM. Origin of the precerebellar system. Neuron. 2000;27:475–486. doi: 10.1016/s0896-6273(00)00059-3. [DOI] [PubMed] [Google Scholar]
- 8.Landsberg RL, et al. Hindbrain rhombic lip is comprised of discrete progenitor cell populations allocated by Pax6. Neuron. 2005;48:933–947. doi: 10.1016/j.neuron.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 9.Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 10.Köster RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001;233:329–346. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- 11.Baron U, Gossen M, Bujard H. Tetracycline-controlled transcription in eukaryotes: Novel transactivators with graded transactivation potential. Nucleic Acids Res. 1997;25:2723–2729. doi: 10.1093/nar/25.14.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du SJ, Devoto SH, Westerfield M, Moon RT. Positivie and negative regulation of muscle cell identity by members of the hedgehog and TGF-ss gene families. J Cell Biol. 1997;139:145–156. doi: 10.1083/jcb.139.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell RE, et al. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:377–382. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amsterdam A, Lin S, Hopkins N. The Aequorea victoria green fluorescent protein can be used as a reporter in live zebrafish embryos. Dev Biol. 1995;171:123–129. doi: 10.1006/dbio.1995.1265. [DOI] [PubMed] [Google Scholar]
- 15.Perz-Edwards A, Hardison NL, Linney E. Retinoic acid-mediated gene expression in transgenic reporter zebrafish. Dev Biol. 2001;229:89–101. doi: 10.1006/dbio.2000.9979. [DOI] [PubMed] [Google Scholar]
- 16.Halpern ME, et al. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish. 2008;5:97–110. doi: 10.1089/zeb.2008.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawakami K, et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Provost E, Rhee J, Leach SD. Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis. 2007;45:625–629. doi: 10.1002/dvg.20338. [DOI] [PubMed] [Google Scholar]
- 20.Mueller T, Vernier P, Wullimann MF. The adult central nervous cholinergic system of a neurogenetic model animal, the zebrafish Danio rerio. Brain Res. 2004;1011:156–169. doi: 10.1016/j.brainres.2004.02.073. [DOI] [PubMed] [Google Scholar]
- 21.Wullimann MF, Rupp B, Reichert H. Neuroanatomy of the Zebrafish Brain: A Topological Atlas. Basel: Birkhäuser; 1996. [Google Scholar]
- 22.McCormick CA, Hernandez CV. Connections of the octaval and lateral line nuclei of the medulla in the goldfish, including the cytoarchitecture of the secondary octaval population in goldfish and catfish. Brain Behav Evol. 1996;47:113–138. doi: 10.1159/000113232. [DOI] [PubMed] [Google Scholar]
- 23.Fame RM, Brajon C, Ghysen A second-order projection from the posterior lateral line in the early zebrafish brain. Neural Dev. 2006;1:4. doi: 10.1186/1749-8104-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ray P, et al. Noninvasive quantitative imaging of protein-protein interactions in living subjects. Proc Natl Acad Sci USA. 2002;99:3105–3110. doi: 10.1073/pnas.052710999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paquet D, et al. A novel Tau transgenic zebrafish model for in-vivo research and drug discovery. J Clin Invest. 2009 [Google Scholar]
- 26.Wingate RJ, Lumsden A. Persistence of rhombomeric organisation in the postsegmental hindbrain. Development. 1996;122:2143–2152. doi: 10.1242/dev.122.7.2143. [DOI] [PubMed] [Google Scholar]
- 27.Straka H, Baker R, Gilland E. Preservation of segmental hindbrain organization in the adult frogs. J Comp Neurol. 2006;494:228–245. doi: 10.1002/cne.20801. [DOI] [PubMed] [Google Scholar]
- 28.Esengil H, Chang V, Mich JK, Chen JK. Small-molecule regulation of zebrafish gene expression. Nat Chem Biol. 2007;3:154–155. doi: 10.1038/nchembio858. [DOI] [PubMed] [Google Scholar]
- 29.Hans S, Kaslin J, Freudenreich D, Brand M. Temporally-controlled site-specific recombination in zebrafish. PLoS ONE. 2009;4:e4640. doi: 10.1371/journal.pone.0004640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Distel M, Köster RW. In vivo time-lapse imaging of zebrafish embryonic development. CSH Protocols. 2007;2 doi: 10.1101/pdb.prot4816. doi: 10.1101/pdb.prot4816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.