Abstract
Numerous protists and rare fungi have truncated Asn-linked glycan precursors and lack N-glycan-dependent quality control (QC) systems for glycoprotein folding in the endoplasmic reticulum. Here, we show that the abundance of sequons (NXT or NXS), which are sites for N-glycosylation of secreted and membrane proteins, varies by more than a factor of 4 among phylogenetically diverse eukaryotes, based on a few variables. There is positive correlation between the density of sequons and the AT content of coding regions, although no causality can be inferred. In contrast, there appears to be Darwinian selection for sequons containing Thr, but not Ser, in eukaryotes that have N-glycan-dependent QC systems. Selection for sequons with Thr, which nearly doubles the sequon density in human secreted and membrane proteins, occurs by an increased conditional probability that Asn and Thr are present in sequons rather than elsewhere. Increasing sequon densities of the hemagglutinin (HA) of influenza viruses A/H3N2 and A/H1N1 during the past few decades of human infection also result from an increased conditional probability that Asn, Thr, and Ser are present in sequons rather than elsewhere. In contrast, there is no selection on sequons by this mechanism in HA of A/H5N1 or 2009 A/H1N1 (Swine flu). Very strong selection for sequons with both Thr and Ser in glycoprotein of Mr 120,000 (gp120) of HIV and related retroviruses results from this same mechanism, as well as amino acid composition bias and increases in AT content. We conclude that there is Darwinian selection for sequons in phylogenetically disparate eukaryotes and viruses.
Keywords: Asn-linked glycan, evolution, influenza, sequon, N-glycan-dependent quality control
Asn-linked glycans (N-glycans) are built on lipid-linked precursors that contain 14 sugars (Glc3Man9GlcNAc2) in most animals, plants, and fungi, as well as Dictyostelium (1). In contrast, medically important protists make either no N-glycans (Theileria) or truncated N-glycan precursors composed of 2 sugars (Giardia and Plasmodium), 7 sugars (Entamoeba and Trichomonas), or 7–11 sugars (Leishmania, Trypanosoma, Toxoplasma, and Cryptosporidium) (2).
An oligosaccharyltransferase (OST) transfers N-glycans from the lipid-linked precursor to sequons (Asn-Xaa-Ser or Asn-Xaa-Thr, where Xaa cannot be Pro) of nascent peptides in the lumen of the endoplasmic reticulum (ER) (3). From Swiss-Prot data, Sharon and colleagues (4) estimated that two thirds of sequons are modified by N-glycans. Both in vitro and in vivo, sequons containing Thr are more often glycosylated than sequons containing Ser (5–8). Although the preference for sequons with Thr also occurs for Giardia, which has a single subunit OST and adds N-glycan composed of just 2 sugars (9), the sequon preferences of OSTs of the majority of eukaryotes have not been determined experimentally.
N-glycans of animals, plants, and fungi play important roles in the quality control (QC) of glycoprotein folding in the ER lumen (1, 10, 11). Important components of this QC system are a UDP-Glc-dependent glucosyltransferase (UGGT), which adds a Glc to a terminal Man of the D1 arm of the N-glycans of misfolded glycoproteins, and calnexin and/or calreticulin, which are lectins that bind the glucosylated glycoproteins and assist in their folding. In general, organisms with at least 5 Man residues in their N-glycans have this QC system for glycoprotein folding (all metazoans and plants, most fungi, and some protists, including Entamoeba, Trichomonas, Trypanosoma, and Leishmania; colored blue in Table S1) (12). However, some protists with 5 Mans in the N-glycans (Cryptosporidium and Toxoplasma), as well as those protists (Plasmodium, Theileria, and Giardia) and fungi (Encephalitozoon and Antonospora) that lack Man in their N-glycans, are missing this QC system (colored red in Table S1) (12).
Here, we studied the factors that affect the abundance of sequons (sites of N-linked glycosylation) in secreted proteins of metazoan, fungi, and protists, as well as envelope proteins of influenza virus and HIV. These studies attempt to answer the following questions:
First, how does N-glycan length, which varies from 0 to 14 sugars in diverse eukaryotes (1, 2), covary with the density of sequons in their secreted proteins?
Second, what is the relationship of AT content in protein-coding regions, which is known to vary widely among protists (13, 14), to sequon abundance, because Asn is encoded by AA(TC), whereas Pro is encoded by CC(AGCT)?
Third, is there Darwinian selection for sequons in secreted and membrane proteins (versus cytosolic control proteins) in eukaryotes, which have N-glycan-dependent QC of glycoprotein folding (1, 10–12)? If so, is the mechanism for selection for sequons due to (i) increased AT content, (ii) increased Asn, Ser, and Thr and decreased Pro in secreted proteins (amino acid composition bias), or (iii) increased conditional probability that Asn, Ser, and Thr will be present in sequons rather than elsewhere in secreted and membrane proteins?
Fourth, how are the very high densities of sequons in hemagglutinin (HA) of influenza viruses and glycoprotein of Mr 120,000 (gp120) of HIV achieved (15–18)?
Results and Discussion
The Density of Sequons, Which Varies >4-Fold Among Protists, Is Not Well-Correlated with N-Glycan Precursor Length.
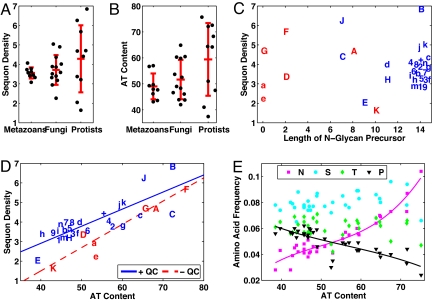
The sequon densities of the secreted and membrane proteins of 9 representative metazoans were very similar, averaging ≈3.6 sequons ± 0.3 per 500 aa (the approximate average length of a secreted or membrane protein) (Fig. 1A and Table S1). As discussed in Materials and Methods, we excluded from the set of secreted and membrane proteins those with multiple transmembrane helices where the ectoplasmic and cytoplasmic domains are difficult to predict. We also excluded those proteins with greater than 70% identity to reduce the impact of large families of recently duplicated genes and to exclude splice variants of the same protein. Examined from another point of view, ≈80% of the secreted and membrane proteins of metazoans had at least 1 sequon that may be N-glycosylated (Fig. S1A). The predicted secreted and membrane proteins of 13 fungi had sequon densities similar to those of metazoans, only there was somewhat greater variability among fungi.
Fig. 1.
The density of sequons is positively correlated with the AT richness of the coding regions of each eukaryote. (A) Sequon densities (average number of sequons per 500 aa plus SD) for the secreted and membrane proteins are similar among metazoans and fungi but are much more variant among protists. (B) AT contents of coding regions of predicted secreted and membrane proteins are also similar among metazoans and fungi but are much more variant among protists. (C) Sequon density is not directly related to length of N-glycan precursors, where metazoans are numbered [Anopheles gambiae (1), Caenorhabditis elegans (2), Canis familiaris (3), Ciona intestinalis (4), Danio rerio (6), Drosophila melanogaster (5), Homo sapiens (7), Muris muscularis (8), and Tetraodon nigroviridis (9). Fungi are in lowercase [Antonospora locustae (a), Aspergillus nidulans (b), Candida albicans (c), Cryptococcus neoformans (d), Encephalitozoon cuniculi (e), Gibberella zeae (f), Kluyveromyces lactis (g), Magnaporthe grisea (h), Neurospora crassa (i), Saccharomyces cerevisiae (j), Schizosaccharomyces pombe (k), Ustilago maydis (m), and Yarrowia lipolytica (n)]. Protists are in uppercase [Cryptosporidium parvum (A), Dictyostelium discoideum (B), Entamoeba histolytica (C), Giardia lamblia (D), Leishmania major (E), Plasmodium falciparum (F), Theileria anulata (G), Trypanosoma cruzi (H), and Trichomonas vaginalis (J)]. One plant (Arabidopsis thaliana) is marked with a plus sign. Eukaryotes that have N-glycan-dependent QC of glycoprotein folding are marked in blue. Eukaryotes that lack N-glycan-dependent QC of glycoprotein folding are marked in red. (D) Sequon density is positively correlated with AT content of secreted and membrane proteins of all eukaryotes (R2 values are 0.68 and 0.89 for blue and red lines, respectively). An analysis of variance shows AT content accounts for 63% of the variance, whereas N-glycan-dependent QC accounts for 11%. The percentage of predicted secreted and membrane proteins with at least 1 sequon is also correlated with the AT richness (Fig. S1). In addition, when AT content is ≤55%, the sequon densities of secreted proteins of eukaryotes with N-glycan-dependent QC (marked in blue) are significantly greater than those of eukaryotes without QC (marked in red) by using rank-sum test at α = 5%. (E) Sequon density is positively correlated with AT content, because Asn is encoded by AA(TC), whereas Pro, which cannot be in sequons, is encoded by CC(AGCT) (R2 values are 0.91 and 0.71 for Asn and Pro, respectively).
In contrast, the sequon densities of the predicted secreted and membrane proteins of 9 protists varied by >4-fold (from 1.6 sequons per 500 aa for Toxoplasma to 6.8 sequons per 500 aa in Dictyostelium; Fig. 1A and Table S1). In turn, these protists showed 5-fold differences in the percentage of secreted and membrane proteins without sequons (31% in Toxoplasma vs. 6% in Plasmodium; Fig. S1A).
We used Alg enzymes to predict the length of N-glycan precursors, many of which have been experimentally demonstrated (1, 2). On rare occasions, some of the N-glycans of protists may be made from N-glycan precursors that are shorter than those predicted by the Alg enzymes (19). Although N-glycan precursors of protists vary from 0 to 11 sugars in length, N-glycan length variation does not correlate well with sequon densities among secreted and membrane proteins of protists (Fig. 1C) (2). For example, Trichomonas and Leishmania have similar-length N-glycans but vary by a factor of 3 in their sequon density. In general, however, eukaryotes with longer N-glycans are more likely to employ N-glycan-dependent QC of glycoprotein folding (colored blue in Fig. 1C) (11), whereas shorter N-glycans predict the absence of N-glycan-dependent QC (colored red in Fig. 1C). As shown below, there is increased sequon density in secreted proteins of eukaryotes with N-glycan-dependent QC of protein folding, so the effect of N-glycan length on sequon density appears to be indirect.
Sequon Abundance Increases with the AT Richness of the Coding Regions of All Eukaryotes.
The percentage of AT in protein-coding sequences, which varies dramatically in protists, increases with sequon abundance in secreted membrane proteins of all eukaryotes (Fig. 1 B and D and Table S1). For each increase of ≈10% in the AT content of coding regions, there is an additional sequon per 500-aa protein. Similarly, the percentage of secreted proteins with at least 1 sequon increases with the AT richness (Fig. S1B).
The density of sequons in secreted proteins correlates with AT percentage, because in the presence of high AT, the concentration of Asn, which is encoded by an AT-rich codon, increases 4-fold when comparing extremes (Fig. 1E). Conversely, in the presence of high AT, the concentration of Pro, which is encoded by a GC-rich codon and cannot be in sequons, decreases 2-fold when comparing extremes (Fig. 1E). In contrast, there is no systematic change with AT content in concentrations of Ser and Thr, which are encoded by AT-neutral codons.
We used the presence of a UDP-Glc-dependent glucosyltransferase, glucosidase II, calnexin or calreticulin, and ERGIC-53 to infer that a particular organism has an N-glycan-dependent QC system for glycoprotein folding (10–12). Eukaryotes with N-glycan-dependent QC of protein folding, which are colored blue in Fig. 1D, have significantly greater densities of sequons (P = 0.026) in secreted proteins (for the same AT percentage) than eukaryotes without N-glycan-dependent QC of protein, which are colored red in Fig. 1D. The reasons for this difference are explored in the next section.
An analysis of variance on the sequon data in Fig. 1D and Table S1 shows AT content accounts for 63% of the variance, whereas N-glycan-dependent QC accounts for 11%. When sequons with Thr are examined separately from those with Ser, N-glycan-dependent QC accounts for 28% of the variance (see below for an explanation).
Apparent Darwinian Selection for Sequons Containing Thr in Secreted Proteins of Eukaryotes with N-Glycan-Associated QC of Glycoprotein Folding.
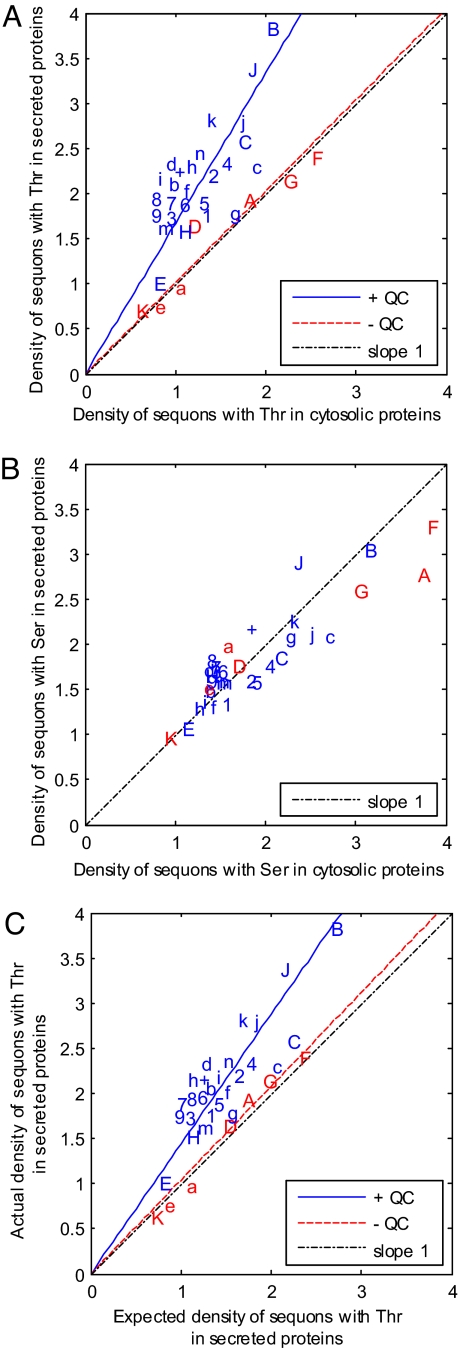
The density of sequons containing Thr, which are more likely to be N-glycosylated than sequons containing Ser (4–9), is increased by ≈70% in secreted and membrane proteins versus nucleocytosolic proteins for metazoans, fungi, and protists with N-glycan-dependent QC of glycoprotein folding (Fig. 2A, blue, and Table S1). In humans, the density of sequons with Thr in secreted and membrane proteins is nearly double that of nucleocytosolic controls, which are not N-glycosylated.
Fig. 2.
Darwinian selection for sequons with Thr in the secreted and membrane proteins of eukaryotes with N-glycan-dependent QC of proteins folding is based for the most part on an increased conditional probability that Asn and Thr will be present in sequons rather than elsewhere in these proteins. (A) Density of sequons with Thr (number per 500 aa) in secreted and membrane proteins versus nucleocytosolic proteins (negative controls) for each organism, which are abbreviated as in Fig. 1. In the case of eukaryotes that have N-glycan-dependent QC of glycoprotein folding and are marked in blue, there is moderately strong selection for sequons with Thr, so that the slope of the blue line is 1.7 (R2 = 0.46) rather than 1. In contrast, eukaryotes that lack N-glycan-dependent QC of glycoprotein folding and are marked in red show no selection, so the slope of the red line is 1.1 (R2 = 0.91). (B) Density of sequons with Ser (number per 500 aa) in secreted and membrane proteins versus nucleocytosolic proteins for each organism. There is no selection for or against sequons with Ser in eukaryotes with or without N-glycan-dependent QC, so that all points fall on the dotted line with the slope of 1 and intercept of 0. (C) The mechanisms for positive selection for sequons with Thr is shown by plotting the actual density of sequons with Thr in secreted and membrane proteins versus that calculated by the Asn, Thr, and Pro frequencies for each organism (i.e., the expected density). In eukaryotes with N-glycan-dependent QC that are marked in blue, there is an increased conditional probability that Asn and Thr will be in sequons rather than elsewhere in secreted proteins, so that the slope of the blue line is 1.5 (R2 = 0.74). In contrast, there is no increased conditional probability that Asn and Thr will be in sequons rather than elsewhere in secreted proteins of eukaryotes without N-glycan-dependent QC, so that the slope of the red line is 1.0 (R2 = 0.98). In Fig. S2, a small positive selection for amino acid composition bias and negative results for AT-content bias are shown.
For multiple reasons, the increase in the density of sequons containing Thr appears to be the result of Darwinian selection. First, the data do not support selection for sequons with Thr in protists and fungi that lack N-glycan-dependent QC of glycoprotein folding (Fig. 2A, red, and Table S1). Second, there is also no support for selection for sequons with Ser, which are less likely to contain N-glycans than sequons with Thr (4–9), in eukaryotes with N-glycan-dependent QC (Fig. 2B).
Third, the major mechanism for the increased density of sequons with Thr in secreted and membrane proteins of eukaryotes, which have N-glycan-dependent QC of glycoprotein folding, is an increased likelihood that Asn and Thr will be present in sequons rather than elsewhere in secreted and membrane proteins. Because of this increased likelihood that Asn and Thr will be in sequons, the actual density of sequons with Thr (plotted on the y axis) was greater than the expected density of sequons with Thr (plotted on the x axis), which is calculated from the frequencies of Asn, Thr, and Pro for secreted and membrane proteins of these eukaryotes with N-glycan dependent QC (colored blue in Fig. 2C). In contrast, the actual and the expected densities of sequons with Thr were the same for secreted and membrane proteins of eukaryotes that lack N-glycan-dependent QC (colored red in Fig. 2C). The actual and the expected densities of sequons with Thr were also the same for cytosolic proteins of eukaryotes that have N-glycan-dependent QC (Fig. S2C).
A minor contributor to the increase in sequon density of secreted and membrane proteins of eukaryotes, which have N-glycan-dependent QC, was amino acid composition bias (Fig. S2A). Amino acid composition bias is shown by difference for each organism between the expected densities of sequons in its secreted and membrane proteins versus its nucleocytosolic proteins (negative controls). In contrast, secreted, membrane, and nucleocytosolic proteins were equally AT-rich in all eukaryotes (Fig. S2B).
Together, these results show that the apparent Darwinian selection for sequons is extraordinarily specific: it occurs only for sequons with Thr in eukaryotes with N-glycan-dependent QC of glycoprotein folding and occurs primarily by an increased conditional probability that Asn and Thr will be present in sequons rather than elsewhere in secreted and membrane proteins.
Increasing Sequon Densities of HA of Influenza Viruses A/H1N1 and A/H3N2 with Time in the Human Host Are Caused by an Increased Conditional Probability That Asn, Thr, and Ser Will Be Present in Sequons Rather Than Elsewhere.
When influenza virus A/H1N1 (also known as Spanish flu) first appeared and caused the great pandemic of 1918 (16), there was a modest amino acid composition bias in HA, which increased the density of sequons (white arrow in Fig. 3). Similarly, when influenza virus A/H3N2 (also known as the Hong Kong flu) appeared in 1968, there was a modest amino acid composition bias in HA, which increased the density of sequons. This is also the case for sequons in HA of A/H5N1, which presently infects poultry and threatens human populations, and for the 2009 A/H1N1, which has also been called “Swine flu” (20–23).
Fig. 3.
Increasing sequon densities of HA of A/H3N2 (Left) and A/H1N1 (Center) strains of influenza virus with antigenic drift results from an increased conditional probability that Asn, Thr, and Ser will be present in sequons rather than elsewhere in HA. Selection for sequons (solid arrow) based on this mechanism, which is determined by comparing actual (solid pink triangles) versus calculated or expected (open pink triangles) sequon densities for HA, increases with time. As a control, there is no selection for sequons in viral capsid and polymerases (capsidic proteins), where the observed density of sequons (solid blue circles) equals the expected density (open blue circles). In contrast, amino acid composition bias (white arrow), which is determined by comparing the expected sequon density of HA (open pink triangles) with that of capsid and polymerases of influenza viruses (open blue circles), remains the same with time. The HA proteins of A/H5N1 and 2009 A/H1N1 (Right) show modest selection based on amino acid composition bias but do not show selection based on an increased conditional probability of Asn, Thr, and Ser being present in sequons rather than elsewhere in HA (20, 25). Changes in the amino acid sequences of A/H3N2 influenza proteins with time is shown in Fig. S3.
With genetic drift of A/H1N1 from 1918 to 1958 and of A/H3N2 from 1968 to the present, the sequon densities of each HA doubled, adding numerous N-glycans to the head group (15, 16). The mechanism of positive selection for sequons in HA proteins of both viruses is an increased likelihood that Asn, Ser, and Thr are in sequons rather than elsewhere in the HA proteins (black arrows in Fig. 3). As is the case for secreted and membrane proteins of the host (Fig. 2C), this increased likelihood of Asn, Ser, and Thr in sequons rather than elsewhere in HA is shown by an increasing difference between the actual sequon density of HA and the expected or calculated sequon density. In contrast, there was no change with time in the amino acid composition bias of HA proteins (white arrows in Fig. 3). The remarkable linearity of the increase in sequon density of HA of A/H3N2 superficially resembles previous demonstrations of linear rates of change in the amino acid sequence of HA and other viral proteins with time (Fig. S3) (24, 25). Rather than selection for sequons, as described here, most changes in amino acid sequence of HA are due to diversifying selection.
Very Strong Selection for Sequons with Both Thr and Ser in gp120 of HIV Results from an Increased Likelihood That Asn, Ser, and Thr Are Present in Sequons, Amino Acid Composition Bias, and Increases in AT Richness.
The very strong selection for sequons with both Thr and Ser in gp120 of HIV-1, HIV-2, and related retroviruses resulted from all possible mechanisms for positively selecting sequons (Fig. 4). There was a marked increase in the probabilities of Asn, Ser, and Thr in sequons rather than elsewhere in gp120, which resulted in a ≈4-fold increase in the density of sequons of gp120 (Fig. 4B).
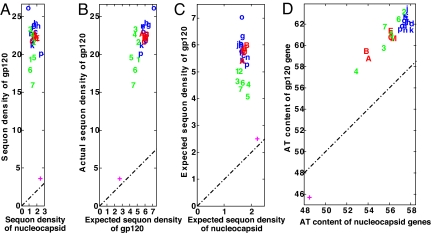
Fig. 4.
Very strong positive selection for sequons with Thr and Ser in gp120 of HIV and other retroviruses results from an increased conditional probability that Asn, Thr, and Ser will be present in sequons rather than elsewhere for selection in gp120, amino acid composition bias, and changes in AT content. (A) Density of sequons per 500 aa in gp120 versus capsid proteins and enzymes (negative controls) for various retroviruses, which are abbreviated and color-coded as follows: HIV-1 strains (A–O) are marked with lowercase blue letters. HIV-2 strains are marked with uppercase red letters, whereas other lentiviruses are marked with green numbers. Very strong selection for sequons in gp120 of all of the retroviruses dwarfs the relatively modest selection for sequons in host-secreted and membrane proteins (marked with a red plus sign). (B) An important mechanism for positive selection for sequons in gp120 of retroviruses, which is based on an increased conditional probability that Asn, Thr, and Ser will be present in sequons rather than elsewhere in gp120, is shown by plotting the counted density of sequons in gp120 versus that calculated (expected value) by the Asn, Thr, Ser, and Pro content of gp120. (C) Amino acid composition bias, which increases the number of sequons in gp120, is shown by plotting the calculated sequon density of gp120 versus that of capsid and enzymes. Although Asn, Ser, and Thr are relatively increased in gp120 versus retroviral capsid and enzymes, Pro is decreased (Fig. S4). (D) AT content of gp120 coding sequence versus the rest of the coding sequence of the retrovirus shows there is moderate positive selection for AT in gp120s of all retroviruses examined. Fig. S5 shows there is no change in sequon densities of gp120 of HIV strains A1, B, C, and D with time.
Amino acid composition bias increased by ≈3-fold the density of sequons in gp120 of HIV-1, HIV-2, and related retroviruses (Fig. 4C and Fig. S4). There was a modest increase in AT in the coding region of gp120 versus that for structural proteins and enzymes of HIV and other retroviruses (Fig. 4D). In contrast to influenza viruses and despite the fact that there is a very high mutation rate in gp120, there has been little change in the density of sequons of gp120 of HIV-1 over time in the human host (Fig. S5). It is likely that the very high sequon density of gp120 (≈25 sequons per protein) is the upper limit for the number of sequons for this protein.
Other viral pathogens showed a broad range of sequon densities in their envelope glycoproteins and multiple mechanisms for selecting sequons (Fig. S6) (26–28). Envelope proteins of hepatitis C virus (HCV), Severe Acute Respiratory Syndrome (SARS) virus, and Ebola virus all have very high sequon densities. An important contributor to these high sequon densities is an increased likelihood that Asn, Ser, and Thr will be in sequons rather than elsewhere in viral envelope proteins, as described for host, influenza, and HIV glycoproteins.
Major Conclusions and Inferences.
Although AT content has previously been shown to modify the amino acid composition of host proteins (14, 15), we show here that AT content has profound effects on sequon density (>4-fold effect), and therefore on the N-glycosylation of glycoproteins. We can think of no other example where the genetic code has such profound effects on protein phenotype. However, because the AT content was the same for nucleocytosolic, secreted, and membrane proteins of all eukaryotes examined here (Fig. S2B), we cannot infer any causality with regard to AT content and sequon density. In addition, we recognize that there may be other determinants (growth at high temperature, tRNA abundance, rRNA operon abundance) that affect AT content of an organism (29, 30).
The “smoking gun” for Darwinian selection in eukaryotes with N-glycan-dependent QC of glycoprotein folding is an increased likelihood that Asn and Thr will be present in sequons rather than elsewhere in secreted and membrane proteins. This positive selection for sequons with Thr increases the actual density of sequons in secreted and membrane proteins of each organism versus that calculated from the frequencies of Asn, Thr, and Pro. Also arguing for Darwinian selection are the absence of selection for sequons with Ser in these eukaryotes and the absence of selection for sequons with Thr in secreted and membrane proteins of eukaryotes without N-glycan-dependent QC. Although the role of N-glycans in the QC of glycoprotein folding is emphasized here, we recognize that N-glycans also play roles in cell signaling by notch, integrins, and cadherins, as well as in cancer cell progression (31–33).
Although the importance of N-glycan-dependent QC has been shown for folding of a small number of glycoproteins in vitro, including HA of influenza virus (10, 11, 34), our results here suggest N-glycan-dependent QC of glycoprotein folding has nearly doubled the sequon densities of tens of thousands of secreted and membrane proteins of phylogenetically diverse protists, fungi, and metazoans.
Although the abundance of sequons in gp120 of HIV, HA of influenza viruses, and envelope proteins of HCV and Ebola viruses has been noted before (15–18, 26–28), to our knowledge the mechanisms for generating the high sequon densities in these viral glycoproteins have not previously been described. In addition to the increased conditional probability that Asn, Ser, and Thr will be in sequons rather than elsewhere in envelope protein, some of these viruses change the AT and amino acid composition of envelope proteins. Although there has been no change in the sequon density of gp120 as HIV has moved from primates to humans, there is a marked increase in sequon densities of HA as influenza has evolved with time in humans. If it is the case that either of the A/H5N1 or 2009 A/H1N1 viruses cause pandemic infections in people, we expect that the density of sequons in the HA proteins would increase over time by the same mechanisms shown for increasing density of sequons in HA proteins of Spanish and Hong Kong influenza viruses.
In influenza virus, HIV, HCV, and Ebola virus, the sequon densities of the envelope proteins appear to be far greater than that required to fold the vast majority of host proteins. It is not surprising then that viral N-glycans have important roles in pathogenesis, including masking host antigens, stimulating host cytokine production, and viral entry into host cells (17–18, 26–28, 35). Although increases in HA sequon density are associated with decreased virulence of influenza A/H3N2 in a mouse model system (36), this may not be the case for people (16). In addition, many parameters other than sequon density contribute to the virulence of influenza viruses (16, 20, 22–25). Finally, there has been speculation that the high frequency of hypomorphic alleles in N-glycan synthesis in human populations may be an adaptive response against viruses, which depend on N-linked glycosylation for their pathogenesis (37).
Materials and Methods
Identification of Secreted and Cytosolic Proteins of 33 Representative Eukaryotes.
Predicted proteins, as well as the cDNA sequences, from the complete or nearly complete genome sequences of 9 protists, 9 metazoans, 13 fungi, and a single plant were downloaded from databases at GenBank, Ensembl, and The Institute for Genomic Research. Proteins shorter than 100 aa and those with >70% identity with other proteins were removed from each set by using the cdhit program (38) to reduce the effect of large protein families and to exclude splice variants of the same protein. The low-complexity regions, which cause misleading amino acid frequencies, were identified by using the SEG algorithm (39) and were excluded from the analyses.
Secreted and membrane proteins were identified by the presence of an N-terminal secretory signal, a signal anchor, or at least 1 transmembrane helix (TMH) (40, 41). Because it can be difficult to accurately predict ectoplasmic domains (where N-glycans may be added to sequons) and cytoplasmic domains (where N-glycans cannot be added to sequons) in proteins with multiple TMHs, proteins with multiple TMHs were excluded from the analysis of secreted proteins. Nuclear and cytosolic proteins (referred to as “nucleocytosolic” for brevity) were defined as those without a secretory signal, signal anchor, or TMH.
Identification of Viral Sequences.
Whole-genome sequences of representative human HIV strains and primate lentiviruses were downloaded from the HIV database at Los Alamos National Laboratory (Los Alamos, NM). Influenza sequences were downloaded from the Influenza Virus Resource at the National Center for Biotechnology Information (NCBI), whereas other representative viruses infecting humans (e.g., HCV and Herpes virus) were downloaded from NCBI. For analysis of sequons, viral envelope proteins were compared with viral “capsidic proteins,” which include capsid and other structural proteins, as well as polymerases and other enzymes.
Analysis of Sequon Evolution.
The cDNA sequence for each protein was used to calculate the percentage of AT in the secreted and cytosolic proteins of each organism. Sequons were identified as NxS or NxT, where “x” cannot be Pro (3), and the sequon densities per 500 aa for secreted proteins and cytosolic proteins (control) were determined for each organism in 2 ways. First, sets of secreted and nucleocytosolic proteins were concatenated into a single, very long sequence, and average densities of sequons; densities of sequons with Thr or Ser; densities of Thr, Ser, Asn, and Pro; and percentage of sequons with Thr were determined. Second, average sequon densities, densities of amino acids, and percentage of sequons with Thr were determined for each protein and then averaged. These 2 methods gave similar results, so that numbers in the figures and text refer to the first method.
Possible Darwinian selection for sequons in each organism was suspected when the density of sequons in secreted and membrane proteins (which are N-glycosylated) was greater than that of nucleocytosolic proteins (which are not N-glycosylated). Differences between sequon densities of secreted proteins and cytosolic proteins, which were computed by the second method, were compared for each organism by using a Mann–Whitney rank-sum test.
Mechanisms for selection of sequons in each organism were determined in 3 ways. First, the AT content of coding regions of secreted and membrane proteins was compared with that of nucleocytosolic proteins. Second, amino acid composition bias was determined by comparing the expected sequon densities of secreted and membrane proteins, which was calculated from their frequencies of Asn, Ser, Thr, and Pro, with the expected sequon densities of nucleocytosolic proteins (negative controls that do not have N-glycans). Third, selection for sequons in secreted and membrane proteins was also determined by comparing the actual sequon density with the expected sequon density, which was calculated based on the frequencies of Asn, Ser, Thr, and Pro in the set of secreted proteins for each organism. This method determines whether there is an increase, decrease, or no change in the conditional probability that Asn, Thr, and Ser will be present in sequons rather than elsewhere in secreted and membrane proteins. A negative control for sequon selection was the nucleocytosolic proteins, which are not N-glycosylated. The statistical significance of the difference between the actual and expected sequon densities for each organism was determined with a Wilcoxon matched-pairs test.
Prediction of N-Glycan Length and Prediction of N-Glycan-Associated Quality Control of Protein Folding.
N-glycan length was predicted by probing with PSI-BLAST the proteins of each organism with the Asn-linked glycosyltransferases of Saccharomyces cerevisiae (1, 2). The absence of N-glycans was predicted when there were no Asn-linked glycosyltransferases and no OST present.
The following Schizosaccharomyces proteins were used to infer the presence of an N-glycan-associated QC system for protein folding: UDP-Glc-dependent glucosyltransferase, glucosidase II, calnexin or calreticulin, and ERGIC-53 (10–12, 42). To determine the effect of N-glycan-dependent QC of protein folding on sequon density, numerous plots were made that distinguished eukaryotes with N-glycan-dependent QC from eukaryotes without N-glycan-dependent QC.
Supplementary Material
Acknowledgments.
We thank Ben Rosenthal and Mark Kon for comments on the manuscript. This work was supported in part by National Institutes of Health Grants AI48082 (to J.S.) and GM31318 (to P.W.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905818106/DCSupplemental.
References
- 1.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 2.Samuelson J, et al. The diversity of protist and fungal dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc Natl Acad Sci USA. 2005;102:1548–1553. doi: 10.1073/pnas.0409460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 4.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Dor S, Esterman N, Rubin E, Sharon N. Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiology. 2004;14:95–101. doi: 10.1093/glycob/cwh004. [DOI] [PubMed] [Google Scholar]
- 6.Petrescu AJ, Milac AL, Petrescu SM, Dwek RA, Wormald MR. Statistical analysis of the protein environment of N-glycosylation sites: Implications for occupancy, structure, and folding. Glycobiology. 2004;14:103–114. doi: 10.1093/glycob/cwh008. [DOI] [PubMed] [Google Scholar]
- 7.Breuer W, Klein RA, Hardt B, Bartoschek A, Bause E. Oligosaccharyltransferase is highly specific for the hydroxy amino acid in Asn-Xaa-Thr/Ser. FEBS Lett. 2001;501:106–110. doi: 10.1016/s0014-5793(01)02641-2. [DOI] [PubMed] [Google Scholar]
- 8.Kasturi L, Eshleman JR, Wunner WH, Shakin-Eshleman SH. The hydroxy amino acid in an Asn-X-Ser/Thr sequon can influence N-linked core glycosylation efficiency and the level of expression of a cell surface glycoprotein. J Biol Chem. 1995;270:14756–14761. doi: 10.1074/jbc.270.24.14756. [DOI] [PubMed] [Google Scholar]
- 9.Ratner DM, et al. Changes in the N-glycome, glycoproteins with Asn-linked glycans, of Giardia lamblia with differentiation from trophozoites to cysts. Eukaryot Cell. 2008;7:1930–1940. doi: 10.1128/EC.00268-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trombetta ES, Parodi AJ. Quality control and protein folding in the secretory pathway. Annu Rev Cell Dev Biol. 2003;19:649–676. doi: 10.1146/annurev.cellbio.19.110701.153949. [DOI] [PubMed] [Google Scholar]
- 11.Hebert DN, Molinari M. In and out of the ER: Protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 12.Banerjee S, et al. Evolution of quality control of protein-folding in the ER lumen. Proc Natl Acad Sci USA. 2007;104:11676–11681. doi: 10.1073/pnas.0704862104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bastien O, et al. Analysis of the compositional biases in Plasmodium falciparum genome and proteome using Arabidopsis thaliana as a reference. Gene. 2004;336:163–173. doi: 10.1016/j.gene.2004.04.029. [DOI] [PubMed] [Google Scholar]
- 14.Singer GA, Hickey DA. Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Mol Biol Evol. 2000;17:1581–1588. doi: 10.1093/oxfordjournals.molbev.a026257. [DOI] [PubMed] [Google Scholar]
- 15.Zhang M, et al. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- 16.Nelson MI, Holmes EC. The evolution of epidemic influenza. Nat Rev Genet. 2007;8:196–205. doi: 10.1038/nrg2053. [DOI] [PubMed] [Google Scholar]
- 17.Kwong PD, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 18.Poon AF, Lewis FI, Pond SL, Frost SD. Evolutionary interactions between N-linked glycosylation sites in the HIV-1 envelope. PLoS Comput Biol. 2007;3:e11. doi: 10.1371/journal.pcbi.0030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones DC, Mehlert A, Güther ML, Ferguson MA. Deletion of the glucosidase II gene in Trypanosoma brucei reveals novel N-glycosylation mechanisms in the biosynthesis of variant surface glycoprotein. J Biol Chem. 2005;280:35929–35942. doi: 10.1074/jbc.M509130200. [DOI] [PubMed] [Google Scholar]
- 20.Peiris JS, de Jong MD. Guan Y Avian influenza virus (H5N1): A threat to human health. Clin Microbiol Rev. 2007;20:243–267. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallace RG, Hodac H, Lathrop RH, Fitch WM. A statistical phylogeography of influenza A H5N1. Proc Natl Acad Sci USA. 2007;104:4473–4478. doi: 10.1073/pnas.0700435104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garten RJ, et al. Antigenic and genetic characteristics of Swine-origin 2009 A(H1N1) Influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinde V, et al. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N Engl J Med. 2009;360:2616–2625. doi: 10.1056/NEJMoa0903812. [DOI] [PubMed] [Google Scholar]
- 24.Bush RM, Bender CA, Subbarao K, Cox NJ, Fitch WM. Predicting the evolution of human influenza A. Science. 1999;286:1921–1925. doi: 10.1126/science.286.5446.1921. [DOI] [PubMed] [Google Scholar]
- 25.Fitch WM, Leiter JM, Li XQ, Palese P. Positive Darwinian evolution in human influenza A viruses. Proc Natl Acad Sci USA. 1991;88:4270–4274. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goffard A, et al. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J Virol. 2005;79:8400–8409. doi: 10.1128/JVI.79.13.8400-8409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helle F, et al. The neutralizing activity of anti-hepatitis C virus antibodies is modulated by specific glycans on the E2 envelope protein. J Virol. 2007;81:8101–8111. doi: 10.1128/JVI.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dowling W, et al. Influences of glycosylation on antigenicity, immunogenicity, and protective efficacy of Ebola virus GP DNA vaccines. J Virol. 2007;81:1821–1837. doi: 10.1128/JVI.02098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharp PM, Bailes E, Grocock RJ, Peden JF, Sockett RE. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 2005;33:1141–1153. doi: 10.1093/nar/gki242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paz A, Mester D, Baca I, Nevo E, Korol A. Adaptive role of increased frequency of polypurine tracts in mRNA sequences of thermophilic prokaryotes. Proc Natl Acad Sci USA. 2004;101:2951–2956. doi: 10.1073/pnas.0308594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanley P. Regulation of Notch signaling by glycosylation. Curr Opin Struct Biol. 2007;17:530–535. doi: 10.1016/j.sbi.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y, et al. Branched N-glycans regulate the biological functions of integrins and cadherins. FEBS J. 2008;275:1939–1948. doi: 10.1111/j.1742-4658.2008.06346.x. [DOI] [PubMed] [Google Scholar]
- 33.Lau KS, Dennis JW. N-Glycans in cancer progression. Glycobiology. 2008;18:750–760. doi: 10.1093/glycob/cwn071. [DOI] [PubMed] [Google Scholar]
- 34.Daniels R, Kurowski B, Johnson AE, Hebert DN. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol Cell. 2003;11:79–90. doi: 10.1016/s1097-2765(02)00821-3. [DOI] [PubMed] [Google Scholar]
- 35.Hong PW, Nguyen S, Young S, Su SV, Lee B. Identification of the optimal DC-SIGN binding site on human immunodeficiencyvirus type 1 gp120. J Virol. 2007;81:8325–8336. doi: 10.1128/JVI.01765-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vigerust DJ, et al. N-linked glycosylation attenuates H3N2 influenza viruses. J Virol. 2007;81:8593–8600. doi: 10.1128/JVI.00769-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freeze HH, Westphal V. Balancing N-linked glycosylation to avoid disease. Biochimie. 2001;83:791–799. doi: 10.1016/s0300-9084(01)01292-5. [DOI] [PubMed] [Google Scholar]
- 38.Li W, Jaroszewski L, Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17:282–283. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- 39.Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 40.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 41.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 42.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.