Abstract
Formins are present in all eukaryotes and are essential for the creation of actin-based structures responsible for diverse cellular processes. Because multicellular organisms contain large formin gene families, establishing the physiological functions of formin isoforms has been difficult. Using RNAi, we analyzed the function of all 9 formin genes within the moss Physcomitrella patens. We show that plants lacking class II formins (For2) are severely stunted and composed of spherical cells with disrupted actin organization. In contrast, silencing of all other formins results in normal elongated cell morphology and actin organization. Consistent with a role in polarized growth, For2 are apically localized in growing cells. We show that an N-terminal phosphatase tensin (PTEN)-like domain mediates apical localization. The PTEN-like domain is followed by a conserved formin homology (FH)1-FH2 domain, known to promote actin polymerization. To determine whether apical localization of any FH1-FH2 domain mediates polarized growth, we performed domain swapping. We found that only the class II FH1-FH2, in combination with the PTEN-like domain, rescues polarized growth, because it cannot be replaced with a similar domain from a For1. We used in vitro polymerization assays to dissect the functional differences between these FH1-FH2 domains. We found that both the FH1 and the FH2 domains from For2 are required to mediate exceptionally rapid rates of actin filament elongation, much faster than any other known formin. Thus, our data demonstrate that rapid rates of actin elongation are critical for driving the formation of apical filamentous actin necessary for polarized growth.
Keywords: Physcomitrella patens, moss, profilin, tip growth, RNAi
Formin proteins are critical regulators of the actin cytoskeleton that drive cellular processes in all eukaryotes ranging from division and motility to cell polarity, including axonal morphogenesis (1–4). The defining features of formins are the formin homology domains (FH1 and FH2) (5). The FH1 domain is characterized by the presence of polyproline stretches known to interact with the small actin monomer binding protein, profilin (6). The FH2 domain promotes actin filament nucleation, and is located C-terminal to the FH1 domain (7, 8). Structural studies reveal that the FH2 domain forms a ring-like structure, which sits at the barbed end of an actin filament (9, 10). After nucleating a filament, the FH2 domain remains at the fast-growing filament end, and influences elongation rate as it moves processively with this end as additional monomers are incorporated. In vitro, the actin nucleating and elongating characteristics of individual formins can vary quite dramatically (3, 5). Because complex eukaryotes contain large formin gene families, the in vivo significance of these differences has been difficult to assess.
Plants have been particularly challenging, because most angiosperms contain many formin genes (11). For example, Arabidopsis thaliana has 21 formins that group into 2 distinct families based on the FH2 domain sequence (12). We have chosen to investigate formin function in an emerging model plant, the moss Physcomitrella patens. The Physcomitrella genome has recently been sequenced (13), revealing the presence of 9 formin genes. Eight of these formins group into the 2 angiosperm groups, with 6 in class I and 2 in class II. The final formin gene belongs to a newly identified plant group, class III, which so far has only been found in plants containing flagellate sperm (14). In this study, we have used a combination of loss-of-function and complementation analyses, together with biochemical characterization, to dissect the role and molecular functions of formins in plants.
Results
Silencing of All Class I Formins (For1) Affects Plant Size, but Not Polarized Growth.
Moss For1 form a monophyletic group that can be divided into 3 subgroups based on similarities within their FH2 domains and the presence or absence of certain sequence motifs (Fig. 1A) (14). We analyzed formin function using a rapid, transient RNAi assay, in which multiple family members are silenced simultaneously. This silencing is achieved by including sequences from each gene in the inverted repeat RNAi construct (15). The RNAi assay includes a robust internal control for monitoring the level of silencing (16).
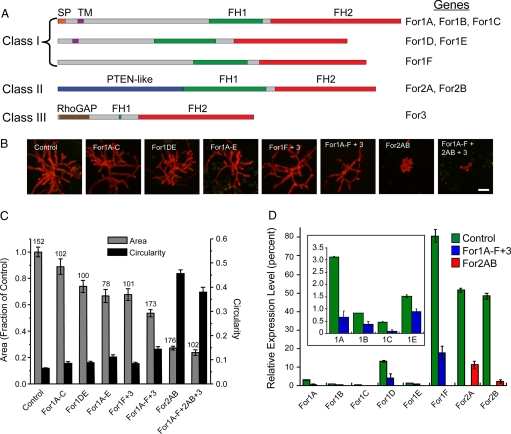
Fig. 1.
For2 are required for polarized growth. (A) Gene models for the 3 classes of formins present in Physcomitrella. SP, signal peptide (orange), TM, transmembrane domain (purple), FH1 (green), FH2 (red), PTEN (blue), RhoGAP-Rho GTPase activating domain (brown). (B) Representative micrographs of chlorophyll autofluorescence from 1-week-old moss plants, transformed with the indicated RNAi constructs. (Scale bar, 100 μm.) (C) Quantification of plant area (gray) and circularity (black). Area is determined from the area of chlorophyll autofluorescence, and is represented as a fraction of plants transformed with the control RNAi construct. Circularity is 4πArea/Perimeter2. Error bars represent SEM. Numbers above the bars represent the total number of plants measured for each condition. Statistical analyses of these data are presented in Table S1. (D) Real-time RT-PCR analysis of formin expression in 1-week-old moss plants. Expression levels were determined with respect to the Ubiquitin10 gene in control (green), For1A-F + 3 (blue) and For2AB (red) RNAi plants. Expression levels are represented as a percentage of total formin gene expression, with all For1 comprising 100% expression and all For2 comprising 100% expression. The For3 is not included, because it was not detected in plants of this age. (Inset) Magnification of the graph for For1A, B, C, and E.
Briefly, RNAi is performed in a transgenic moss line overexpressing a GFP-β-glucuronidase (GUS) fusion protein targeted to the nucleus. The RNAi constructs contain inverted repeats of the genes under investigation fused to 400 bp of the GUS coding sequence, resulting in simultaneous silencing of all sequences included in the construct. Within 24 h, high levels of expression of the RNAi construct are obtained, and by 48–72 h, one observes silencing of the nuclear GFP reporter. Selection for the RNAi construct is performed with antibiotics. Transformed 1-week-old plants lacking GFP fluorescence in the nucleus are undergoing active silencing, and are analyzed for a growth phenotype.
One-week old moss plants, regenerated from a single transformed protoplast, develop from polarized tip-growing protonemal cells. To quantify RNAi-induced phenotypes, plant area, as determined by the area of chlorophyll autofluorescence, is used as a direct measure of plant size and correlates with rates of growth. Because control plants are composed of protonemal tissue forming an extended and branched structure, polarized growth can be determined by quantifying plant morphology. Control plants have low circularity, which is a ratio of area to the square of the perimeter. In contrast, plants that lack polarized growth have lost polarized extensions, are more compact and, thus, have a large circularity value.
We generated RNAi constructs to silence all members within For1 subgroups, as well as constructs that silence multiple subgroups simultaneously. Interestingly, the growth phenotype varies in severity depending on the subgroup of For1 analyzed, and correlates with the relative expression levels of For1 determined by real-time RT-PCR (Fig. 1 A–D). For1A, B, and C, all have a predicted signal peptide and transmembrane domain, and silencing this subgroup has a weak effect on total plant area (Fig. 1 A–C). Silenced plants are ≈11% smaller than controls. Together, For1A, B, and C account for 4.4% of the expression of all For1 (Fig. 1D). For1D and E, which account for 14.8% of For1 levels, have a transmembrane domain and participate slightly more in growth, showing a 26% reduction in plant area (Fig. 1 A–D). Transformation with a construct that silences all 5 transmembrane domain containing For1 (15.2% of For1 levels) results in an additive growth defect, because 34% of plant area is reduced (Fig. 1 A–D). The remaining For1, For1F, has no obvious motifs at the N terminus, and is the most highly expressed For1, accounting for 80.7% of For1 expression (Fig. 1 A and D). For1F and For3 were included in the same RNAi construct before the discovery of For3 as an independent group (14), because its FH2 domain more closely resembles For1. However, the For3 transcript was undetectable in protonemal tissue (Fig. S1A). Nevertheless, For3 was included to ensure silencing of all class I-like formins. Expression of the For1F + 3 RNAi construct results in polarized plants 32% smaller than controls (Fig. 1 A–C). Because For3 is not expressed, it is likely that silencing of only For1F is responsible for the phenotype.
When all 6 For1 are silenced simultaneously, the plants are 46% smaller than controls, and contain less than half the number of cells (Fig. 1 B and C; Fig. S1B). Real-time RT-PCR analysis of class I silenced plants shows that all class I transcripts are reduced in expression (Fig. 1D). Overall, class I silenced plants have a 76% reduction in the total amount of class I transcripts. Although the plants are smaller, they retain highly polarized extensions reflected in their low circularity values (Fig. 1C). Retention of polarized growth indicates that even on silencing of all For1, tip growing cells maintain the necessary factors required to control cell polarity.
Because silencing of all For1 results in smaller plants with significantly fewer cells, we investigated whether loss of For1 may result in any defects in cell division. In moss, the apical cell grows by tip growth until it reaches a given size, at which point, the cell divides. Thus, smaller plants could result from slower tip growth and/or from a delay in cell division. To distinguish between these possibilities, we measured tip growth rates for individual cells from plants silencing all For1 or control plants. We determined that tip growth rates are indistinguishable between class I silenced and control plants (Fig. S1C). Therefore, For1 are most likely participating in cell division. The fact that we did not observe obvious defects, such as incomplete cell plates and/or binucleate cells, is not entirely surprising, because in plants, it has been shown that disruption of actin results in a delay in cytokinesis (17–19).
For2 Are Essential for Polarized Growth.
There are only 2 For2 genes, each containing a phosphatase tensin (PTEN)-like domain located at the N terminus, which has been hypothesized to mediate membrane targeting and subcellular localization (Fig. 1A) (20). Simultaneous silencing of both For2, with an 86% reduction of class II transcript levels, results in plants that contain clusters of small rounded cells lacking any polarized extensions (Fig. 1 B–D). Class II silenced plants also have fewer cells per plant (Fig. S1B), which may represent a delay in cell division. This delay is most likely because these cells no longer undergo tip growth, resulting in the inability to reach the appropriate size for cell division. Simultaneous knock down of all formin genes results in lethality, as evidenced by regions in the plants that exhibit a loss of chlorophyll fluorescence (Fig. S1D), an indication of senescence.
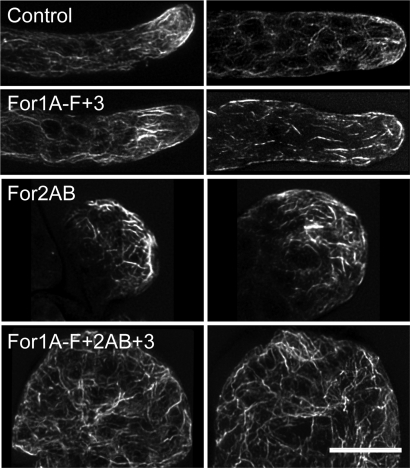
To determine the role of For1 versus For2 function in organizing actin filaments, we stained for filamentous (F-)actin in plants lacking formin function. Consistent with the observed phenotype that plants lacking For1 are highly polarized and maintain normal tip growth rates, F-actin localization is not dramatically affected in these plants (Fig. 2). In contrast, silencing of For2 results in significant disruption of F-actin organization (Fig. 2). Longitudinally oriented, bundles are replaced with an overall randomized orientation of filaments in the body of the cell. We quantified the reduction in longitudinal organization using a Fast Fourier Transform analysis. The elliptical shape of the resulting transform is a measure of orientational order (15, 21). Cells lacking class II function have a significant reduction in orientational order. However, F-actin does appear to accumulate at one end of the cell, suggesting that a polarizing signal is still present. Plants silencing all formins also have randomized actin organization (Fig. S2), with no accumulation of actin at one end of the cell, suggesting that these cells lack a polarity cue. Interestingly, these cells still contain F-actin, which could be attributed to the presence of other nucleators of actin polymerization.
Fig. 2.
For2 are required for proper actin organization. Representative images of fluorescent-phalloidin labeling of the F-actin cytoskeleton in cells from 1-week-old plants transformed with the indicated constructs. (Scale bar, 10 μm.)
Two For2 Isoforms in Moss Are Functionally Redundant.
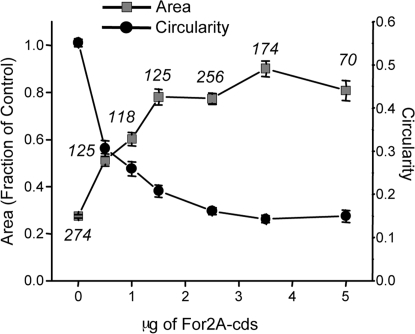
To determine whether the 2 For2 genes, which have similar levels of expression (For 2A, 52%; For 2B, 48%) (Fig. 1D), have nonoverlapping functions, we made specific RNAi constructs that target the 5′ UTR of each gene. Loss of either formin gene has no effect on plant size or polarity, demonstrating functional redundancy (Fig. S3 A and B). However, when 5′ UTRs from both genes were included in the RNAi construct, the For2 phenotype observed with the coding sequence RNAi construct was recapitulated (Fig. S3 A and B). Using UTRs to silence genes is a powerful tool, because it enables complementation by coexpression of a construct lacking the UTRs that is insensitive to the RNAi construct (15, 22). We cotransformed plants with the For2AB-5′ UTR construct and a cDNA construct containing only the coding sequence of For2A (For2Acds). Increasing amounts of For2Acds exhibited a dose-dependent rescue of plant size and polarity (Fig. 3; Fig. S3C). However, too much For2Acds was toxic, consistent with previous work showing that overexpression of formin causes growth defects (23, 24).
Fig. 3.
Complementation analysis of formin-RNAi plants. Complementation of formin RNAi phenotype by coexpression of For2A coding sequence construct (For2Acds). Quantification of plant area (gray circles) and circularity (black circles) as a function of the amount in micrograms of For2Acds. Error bars represent SEM. Numbers above the symbols in the graph represent the total number of plants measured for each condition.
PTEN-Like Domain Mediates Apical Localization of For2.
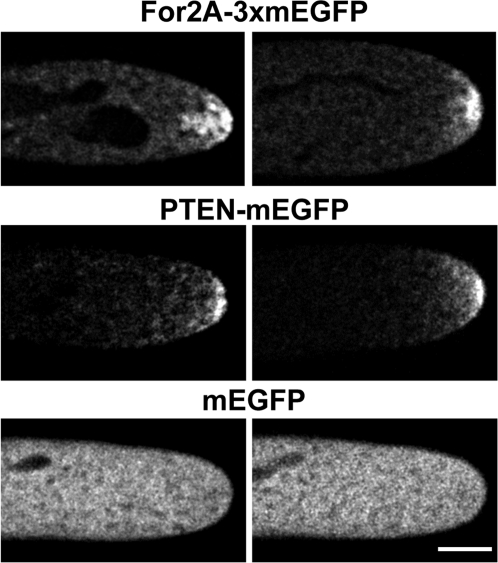
For2 are distinguished from For1 and For3 by the amino acid sequence in their FH2 domains and the PTEN-like domain at the N terminus. To determine the subcellular localization of For2A, we generated a stable moss line where the For2A locus was modified by homologous recombination. Sequences encoding for 3 tandem GFPs were integrated at the 3′ end of For2A. Fluorescence from For2A-3XmEGFP was found concentrated at the very apex of growing cells (Fig. 4). This localization reflects that of a functional protein, because For2A-3XmEGFP rescues the For2 RNAi phenotype (Fig. S4). To test whether the PTEN-like domain is sufficient for apical localization, we generated a moss line expressing PTEN-mEGFP. PTEN-mEGFP is also apically localized during growth (Fig. 4), suggesting that the PTEN-like domain confers apical membrane targeting. However, the PTEN-like or the FH1-FH2 domain is not sufficient for function, because neither domain alone rescues For2 RNAi (Fig. S4). Unlike recent studies in yeast and animals (25, 26), these data suggest that a properly localized actin elongating/nucleating activity is required for full For2 function.
Fig. 4.
For2A-3XmEGFP and PTEN-GFP are localized to the apical region of growing moss cells. Two representative fluorescence micrographs of transgenic moss plants containing the indicated constructs. (Scale bar, 10 μm.)
Class II FH1-FH2 Domain Is Critical for For2 Function in Vivo.
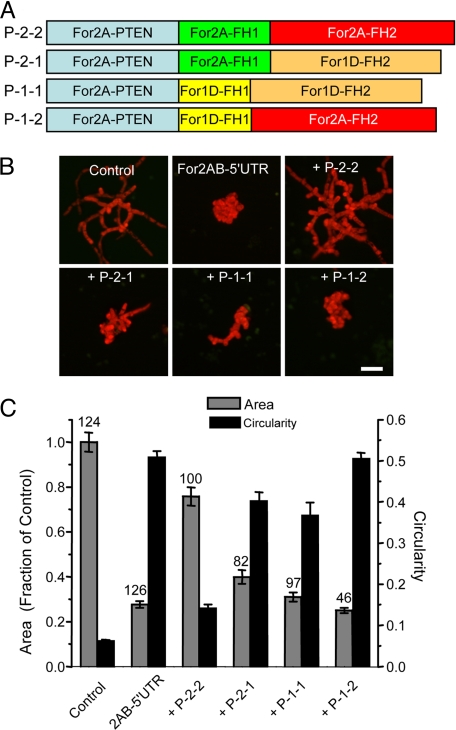
We hypothesized that the PTEN-like domain is critical for For2 function, because it determines proper localization to the site of polarized growth. This hypothesis raises the possibility that any formin FH1-FH2 domain may provide nucleating/elongating activity if properly targeted to the site of growth. To test this idea, we generated chimeras containing the N-terminal PTEN-like domain from For2A fused to different combinations of FH1 and FH2 domains stemming from either class I (For1D) or II (For2A) formins (Fig. 5A). To ensure that the linker regions between the domain junctions are identical, we constructed all of the chimeras and the For2A control (P-2-2) similarly (see Methods).
Fig. 5.
Class II FH1-FH2 domains are critical for polarized growth. (A) Schematic representation of constructs used for complementation studies. PTEN-like domain (blue); class II FH1 domain (green); class I FH1 domain (yellow); class II FH2 domain (red); class I FH2 domain (orange). (B) Fluorescence micrographs of 1-week-old moss plants transformed with the indicated constructs. Panels labeled with + “Construct” are all transformed with For2AB-5′ UTR in addition to the construct indicated. (Scale bar, 100 μm.) (C) Quantification of plant area (gray) and circularity (black). Error bars represent SEM. Numbers above the bars represent the total number of plants measured for each condition. Statistical analyses of these data are presented in Table S2.
Strikingly, none of the chimeras fully rescued the For2 RNAi phenotype (Fig. 5 A–C), suggesting that the combination of the FH1 and FH2 domains from For2A has a unique activity required for polarized growth. The chimeras containing a class I FH2 domain (constructs P-1-1, P-2-1) very weakly rescued the phenotype, regardless of what FH1 domain was present (Fig. 5 B and C). However, if a class I FH1 domain replaced the class II FH1 domain (construct P-1-2), absolutely no rescue was observed (Fig. 5B).
For2 FH1-FH2 Domain Mediates Exceptionally Rapid Rates of Actin Elongation.
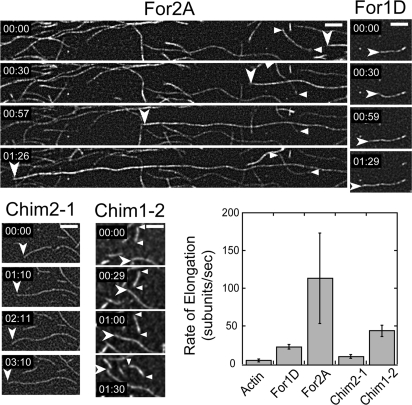
Our in vivo studies suggest that the class II FH1-FH2 domain combination is essential for formin-mediated polarized growth. This requirement raises the possibility that class I and II FH1-FH2 domains have different activities with respect to actin polymerization. To test this idea, we measured their activities in vitro in the presence of profilin, which interacts with the FH1 domain to drive actin-filament elongation (27, 28). Moss has 3 profilin isoforms that are 73–86% identical and functionally redundant (15). We chose profilin A (PRFA) for these studies because it is the most highly expressed isoform in protonemal tissue (15). Using evanescent wave (total internal reflection; TIRF) microscopy, we determined the rate of elongation of actin filament barbed ends bound to the FH1-FH2 domains arising from the following: For2 (For2A), For1 (For1D), chimeric protein containing For2A FH1 fused to For1D FH2 (Chim2-1), and chimeric protein containing For1D FH1 fused to For2A FH2 (Chim1-2) (Fig. 6). Rates of actin elongation for formin-bound and free barbed ends were measured for every sample. The rate of For2A-mediated barbed-end elongation (at least 115 subunits per second) is faster than any of the other FH1-FH2 combinations tested. On average it is 2.7 times faster than Chim1-2 (43 subunits per second), 5 times faster than For1D (24 subunits per second), and 19 times faster than Chim2-1 or free actin barbed ends (6 subunits per second) (Fig. 6).
Fig. 6.
The FH1-FH2 domains from For2A, For1D, Chim2-1, and Chim1-2 have distinct actin elongating activity. Direct visualization by TIRF microscopy of the effect of FH1-FH2 domains of For2A, For1D, Chim2-1, or Chim1-2 on actin filament elongation by profilin/actin complex. The following concentrations were used: 1.2 μM actin-alexa-532, 3.6 μM moss profilin, 2 nM For2A, or 100 nM For1D; 0.8 μM actin-Alexa-568, 2.4 μM profilin, 10 nM Chim2-1, or 5 nM Chim1-2. Frames were taken at the indicated time during polymerization. Arrowheads mark the end of actin filaments growing at the rate consistent with the presence of a formin. Triangles mark actin filaments growing at the rate of a free barbed end (see yellow arrows in Movies S1–S8). Rates of elongation for control and formin associated filaments come from a measure of example filaments such as the ones marked with triangles and arrowheads, respectively, for each formin construct tested. Quantification of the data is presented in the graph. Error bars represent SD. (Scale bar, 5 μm.)
Formins enhance both elongation and nucleation of actin filaments (7, 8, 27, 28). The TIRF assay allows us to measure only the rate of actin filament elongation in the presence of moss formins, not to determine their actin nucleation efficiency. To characterize the actin nucleation activities of the FH1-FH2 domains tested in the TIRF assay, we performed a bulk pyrene fluorescence assay. We found that in the presence of moss profilin, the For2A and Chim1-2 are significantly more active than the For1D and Chim2-1 (compare Fig. S5 A and C with Fig. S5 B and D). The nucleation efficiency, which is indicated by the concentration of filament ends in the bulk experiment (29), was calculated using the elongation rates measured in the TIRF assays (see Methods; Fig. 6). These results indicate that For2A, For1D, and Chim2-1 have similar nucleation efficiencies (Fig. S5E). Chim1-2 is a better nucleator (Fig. S5E); however, its rate of actin elongation is slower than that of For2A (Fig. 6). Together, these data suggest that the actin elongation activity, not the nucleation activity of the FH1FH2 domains from For2, is more critical for tip growth.
Interestingly, the actin polymerization activity of both For2A and For1D is lower in the presence of human profilin than moss profilin (compare orange and gray curves in Fig. S5 A and B). This difference suggests that the sequence context of the polyproline tracks is important for proper interaction with profilin. Also, the source of actin may affect to some extent the rates of formin-mediated actin elongation.
Discussion
By systematically silencing all formin family members, we demonstrate that For2, not For1, nor For3 are critical for polarized plant cell growth. It was previously suggested that For1 participate in polarized growth of both pollen tubes and root hairs (23, 24, 30). In contrast, our results do not show a dramatic effect on polarization of cells lacking For1. Because the pollen tube and root hair studies were based on overexpression (23, 24, 30), it is difficult to compare them with our results. Disruption of additional class I isoforms in seed plants should help clarify this point. The only For1 loss of function phenotype known in Arabidopsis is for AtFH5, which indicates that this protein is important for cytokinesis (31). Our results in moss are consistent with this observation. We show that plants lacking For1 perform tip growth at the same rate as control plants. Because growth in moss is tightly coupled to cell division, our results demonstrate that silencing of For1 results in cell division delays. However, the precise mechanism of class I function in moss remains to be determined.
Plants lacking For2 are unable to perform polarized growth and have highly disorganized F-actin. This phenotype strongly resembles plants lacking profilin function (15). Also, profilin containing a lesion in the polyproline binding site is unable to rescue polarized growth or actin localization (15), indicating that formins must work together with profilin in vivo to generate actin structures at the apex of the cell that promote polarized growth. Interestingly, both For2 and profilin (15) deficient plants appear to retain some form of polarization of F-actin, suggested by the accumulation of a dense network of cortical actin at one end of the cell. This result suggests that a polarizing signal is still transduced to the actin cytoskeleton, either via the Arp2/3 complex, which is known to contribute to polarized growth (32, 33), or by the For1. Our data supports the latter, because lack of cortical actin polarization in cells lacking all formins suggests that For1 contribute to the polarization signal. However, this contribution is not sufficient to promote actin polymerization required for polarized growth.
Our data demonstrates that rapid actin elongation is required at the cell apex to generate apical actin structures that promote polarized growth. We show that the PTEN-like domain of For2 is tip-localized similar to the localization of full-length For2. Thus, the PTEN-like domain presumably responds to a polarizing signal at the cell tip; the nature of this interaction remains to be determined.
We show that the For2A FH1-FH2 domain allows an exceptionally rapid rate of actin elongation at the barbed ends, at least 2 times faster than mouse Dia1 and Caenorhabditis elegans CYK1, the fastest formins tested so far (34, 35). Interestingly, this rapid rate of elongation requires both the FH1 and FH2 domains from For2A. Replacement of either domain with the equivalent domains from the slower formin, For1D, reduces the elongation rate. Chim1-2 is 2–3 times slower than For2A, and Chim2-1 no longer facilitates actin elongation, only actin nucleation. This specificity suggests that there is a synergistic actin elongation activity between the FH1 and FH2 domains from a formin of a given class, which is consistent with recent studies demonstrating that profilin isoform specificity is not entirely dependent on the FH1 domain alone (36).
It is striking that for Chim2-1 the very large FH1 domain from For2A, containing 18 stretches of profilin-binding polyproline tracks, does not appear to modulate the rate of actin elongation. This observation suggests that there is an incompatibility between the FH1 and FH2 domains from For2A and For1D to cooperate to increase the rate of actin elongation, without affecting actin nucleation activity. Chim1-2 has the For1D FH1 domain containing 6 polyproline tracks, 3 times less than the For2A FH1 domain. Interestingly, the elongation rate is 2–3 times slower than For2A, suggesting that for Chim1-2 the FH1 domain from For1D may be contributing to actin elongation and is not entirely incompatible with the For2A FH2 domain.
In vivo, P-2-1 and For1D exhibit similarly weak rescue of the For2 RNAi phenotype. Interestingly P-1-2 exhibits no rescue; its FH1FH2 domains mediate faster actin elongation rates than the corresponding FH1FH2 domains of P-2-1 and For1D. However, in vitro, we observed that the FH1FH2 domain of P-1-2 weakly bundles actin filaments (Fig. S6), a property not observed with any of the other constructs tested. This observation suggests that the lack of complementation could result from increased bundling in addition to slower rates of actin elongation. Together the in vivo complementation studies with the in vitro elongation analyses, we show that only the class II FH1-FH2 combination is capable of fully supporting polarized growth, suggesting that very rapid rates of actin elongation are required to build the apical actin network underlying polarized growth.
This study provides mechanistic insights into the molecular basis of plant cell polarized growth. Using systematic silencing of a large gene family, coupled with in vivo and in vitro functional studies, we demonstrate that plant cells require For2, not For1, to drive tip growth. We also show that For2 are tip-localized and exceptionally rapid actin-elongating factors. This fast polymerization engine must work in concert with profilin (15) to properly organize a dynamic network of elongating actin filaments.
Methods
Tissue Culture and Protoplast Transformation.
All moss tissue culture and transformation procedures were as in Vidali et al. (15).
DNA Constructs, Image Acquisition, and Real-Time RT-PCR.
All constructs were generating using conventional and Gateway (Invitrogen) recombinant DNA techniques. For detailed information, see the SI Methods. Image Acquisition and real-time RT-PCR detailed methods are described in SI Methods.
F-Actin Staining.
To visualize F-actin, an adaptation of the method developed by Tewinkel et al. (37) was used, together with the addition of chemical cross-linkers (38), the protocol is described in detail in Vidali et al. (15). Quantification of F-actin images was performed as described in Vidali et al. (15).
High-Resolution Imaging of Growing Moss Cells.
Stable lines containing fluorescently tagged constructs were verified by PCR, and were imaged using a Nikon C1 laser scanning confocal (Nikon Instruments). We developed moss culturing and imaging conditions to enable consistent observation of growing cells in the microscope (see SI Methods).
TIRF Fluorescence Microscopy.
Time course of alexa-532 (or 568)-actin polymerization was observed on an Olympus IX-71 inverted microscope equipped with a 60×, 1.45 NA Planapo objective (Olympus), and modify according to Amann and Pollard (39). The time course of actin polymerization was acquired at 10-s time intervals with a Hamamatsu ORCA-ER camera (Hamamatsu Photonics) with metamorph version 6.2r6 (Universal Imaging Media).
Actin Nucleation Assay.
Actin nucleation was performed essentially as described by Higgs et al. (29). Actin-profilin complexes (10% pyrene labeled) were polymerized at room temperature in the presence or absence of formins by the addition of 1/10 volume of 10× KMEI. The polymerization was followed using a Xenius SAFAS (Sfas).
Actin Filament Elongation/Nucleation Data Analysis.
Elongation rates were determined by measuring filament lengths with metamorph during actin filament elongation in the TIRF assay. Rates were converted from micrometers per second to subunits per second by using 370 actin monomers per micrometer. We calculated the concentration of barbed ends according to Higgs et al. (29), using the rate of subunit addition at the barbed ends determined by TIRF microscopy.
Supplementary Material
Acknowledgments.
We thank Peter Hepler, Caleb Rounds, Pat Wadsworth, and Wei-Lih Lee for careful review of the manuscript; Sam Hazen, Shuang Wu, and Liang Song for assistance with real-time PCR; and Scotty Fay, Kelli Pattavina, and Cécile Prunier for technical support. R.C.A. was supported by the Plant Biology Graduate Program R. J. Davis Botany Fund and the National Science Foundation Integrative Graduate Education and Research Traineeship to the Institute for Cellular Engineering. This work was supported by the National Science Foundation Grants MCB-0516702, MCB-0640530, and MCB0747231 (M.B.); the David and Lucille Packard Foundation (M.B.); and the Agence Nationale de la Recherche Grant ANR-06-PCVI-0022-02 (to L.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. FJ997271 (For2A) and FJ997272 (For2B)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0901170106/DCSupplemental.
References
- 1.Evangelista M, et al. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 2.Feierbach B, Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr Biol. 2001;11:1656–1665. doi: 10.1016/s0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- 3.Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 4.Matusek T, et al. Formin proteins of the DAAM subfamily play a role during axon growth. J Neurosci. 2008;28:13310–13319. doi: 10.1523/JNEUROSCI.2727-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higgs HN. Formin proteins: A domain-based approach. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- 8.Pruyne D, et al. Role of formins in actin assembly: Nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, et al. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- 10.Otomo T, et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 11.Blanchoin L, Staiger CJ. Plant formins: Diverse isoforms and unique molecular mechanism. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbamcr.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Deeks MJ, Hussey PJ, Davies B. Formins: Intermediates in signal-transduction cascades that affect cytoskeletal reorganization. Trends Plant Sci. 2002;7:492–498. doi: 10.1016/s1360-1385(02)02341-5. [DOI] [PubMed] [Google Scholar]
- 13.Rensing SA, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 14.Grunt M, Zarsky V, Cvrckova F. Roots of angiosperm formins: The evolutionary history of plant FH2 domain-containing proteins. BMC Evol Biol. 2008;8:115. doi: 10.1186/1471-2148-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vidali L, Augustine RC, Kleinman KP, Bezanilla M. Profilin is essential for tip growth in the moss Physcomitrella patens. Plant Cell. 2007;19:3705–3722. doi: 10.1105/tpc.107.053413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bezanilla M, Perroud PF, Pan A, Klueh P, Quatrano RS. An RNAi system in Physcomitrella patens with an internal marker for silencing allows for rapid identification of loss of function phenotypes. Plant Biol. 2005;7:251–257. doi: 10.1055/s-2005-837597. [DOI] [PubMed] [Google Scholar]
- 17.Higaki T, Kutsuna N, Sano T, Hasezawa S. Quantitative analysis of changes in actin microfilament contribution to cell plate development in plant cytokinesis. BMC Plant Biol. 2008;8:80. doi: 10.1186/1471-2229-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valster AH, Pierson ES, Valenta R, Hepler PK, Emons A. Probing the plant actin cytoskeleton during cytokinesis and interphase by profilin microinjection. Plant Cell. 1997;9:1815–1824. doi: 10.1105/tpc.9.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoneda A, Akatsuka M, Hoshino H, Kumagai F, Hasezawa S. Decision of spindle poles and division plane by double preprophase bands in a BY-2 cell line expressing GFP-tubulin. Plant Cell Physiol. 2005;46:531–538. doi: 10.1093/pcp/pci055. [DOI] [PubMed] [Google Scholar]
- 20.Cvrckova F, Novotny M, Pickova D, Zarsky V. Formin homology 2 domains occur in multiple contexts in angiosperms. BMC Genomics. 2004;5:44. doi: 10.1186/1471-2164-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marga F, Grandbois M, Cosgrove DJ, Baskin TI. Cell wall extension results in the coordinate separation of parallel microfibrils: Evidence from scanning electron microscopy and atomic force microscopy. Plant J. 2005;43:181–190. doi: 10.1111/j.1365-313X.2005.02447.x. [DOI] [PubMed] [Google Scholar]
- 22.Augustine RC, Vidali L, Kleinman KP, Bezanilla M. Actin depolymerizing factor is essential for viability in plants, and its phosphoregulation is important for tip growth. Plant J. 2008;54:863–875. doi: 10.1111/j.1365-313X.2008.03451.x. [DOI] [PubMed] [Google Scholar]
- 23.Cheung AY, Wu HM. Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell. 2004;16:257–269. doi: 10.1105/tpc.016550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deeks MJ, et al. Arabidopsis group Ie formins localize to specific cell membrane domains, interact with actin-binding proteins and cause defects in cell expansion upon aberrant expression. New Phytol. 2005;168:529–540. doi: 10.1111/j.1469-8137.2005.01582.x. [DOI] [PubMed] [Google Scholar]
- 25.Gao L, Bretscher A. Polarized growth in budding yeast in the absence of a localized formin. Mol Biol Cell. 2009 doi: 10.1091/mbc.E09-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Guo F, Rubinstein B, Li R. Actin-driven chromosomal motility leads to symmetry breaking in mammalian meiotic oocytes. Nat Cell Biol. 2008;10:1301–1308. doi: 10.1038/ncb1788. [DOI] [PubMed] [Google Scholar]
- 27.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romero S, et al. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 29.Higgs HN, Blanchoin L, Pollard TD. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- 30.Yi K, et al. Cloning and functional characterization of a formin-like protein (AtFH8) from Arabidopsis. Plant Physiol. 2005;138:1071–1082. doi: 10.1104/pp.104.055665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingouff M, et al. Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat Cell Biol. 2005;7:374–380. doi: 10.1038/ncb1238. [DOI] [PubMed] [Google Scholar]
- 32.Harries PA, Pan A, Quatrano RS. Actin-related protein2/3 complex component ARPC1 is required for proper cell morphogenesis and polarized cell growth in Physcomitrella patens. Plant Cell. 2005;17:2327–2339. doi: 10.1105/tpc.105.033266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perroud PF, Quatrano RS. The role of ARPC4 in tip growth and alignment of the polar axis in filaments of Physcomitrella patens. Cell Motil Cytoskel. 2006;63:162–171. doi: 10.1002/cm.20114. [DOI] [PubMed] [Google Scholar]
- 34.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 35.Neidt EM, Skau CT, Kovar DR. The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J Biol Chem. 2008;283:23872–23883. doi: 10.1074/jbc.M803734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neidt EM, Scott BJ, Kovar DR. Formin differentially utilizes profilin isoforms to rapidly assemble actin filaments. J Biol Chem. 2009;284:673–684. doi: 10.1074/jbc.M804201200. [DOI] [PubMed] [Google Scholar]
- 37.Tewinkel M, Kruse S, Quader H, Volkmann D, Sievers A. Visualization of actin filament pattern in plant-cells without pre-fixation - a comparison of differently modified phallotoxins. Protoplasma. 1989;149:178–182. [Google Scholar]
- 38.Lovy-Wheeler A, Wilsen KL, Baskin TI, Hepler PK. Enhanced fixation reveals the apical cortical fringe of actin filaments as a consistent feature of the pollen tube. Planta. 2005;221:95–104. doi: 10.1007/s00425-004-1423-2. [DOI] [PubMed] [Google Scholar]
- 39.Amann KJ, Pollard TD. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using TIRF fluorescence microscopy. Proc Natl Acad Sci USA. 2001;98:15009–15013. doi: 10.1073/pnas.211556398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.