Abstract
Sialic acids (Sias) are nonulosonic acid (NulO) sugars prominently displayed on vertebrate cells and occasionally mimicked by bacterial pathogens using homologous biosynthetic pathways. It has been suggested that Sias were an animal innovation and later emerged in pathogens by convergent evolution or horizontal gene transfer. To better illuminate the evolutionary processes underlying the phenomenon of Sia molecular mimicry, we performed phylogenomic analyses of biosynthetic pathways for Sias and related higher sugars derived from 5,7-diamino-3,5,7,9-tetradeoxynon-2-ulosonic acids. Examination of ≈1,000 sequenced microbial genomes indicated that such biosynthetic pathways are far more widely distributed than previously realized. Phylogenetic analysis, validated by targeted biochemistry, was used to predict NulO types (i.e., neuraminic, legionaminic, or pseudaminic acids) expressed by various organisms. This approach uncovered previously unreported occurrences of Sia pathways in pathogenic and symbiotic bacteria and identified at least one instance in which a human archaeal symbiont tentatively reported to express Sias in fact expressed the related pseudaminic acid structure. Evaluation of targeted phylogenies and protein domain organization revealed that the “unique” Sia biosynthetic pathway of animals was instead a much more ancient innovation. Pathway phylogenies suggest that bacterial pathogens may have acquired Sia expression via adaptation of pathways for legionaminic acid biosynthesis, one of at least 3 evolutionary paths for de novo Sia synthesis. Together, these data indicate that some of the long-standing paradigms in Sia biology should be reconsidered in a wider evolutionary context of the extended family of NulO sugars.
Keywords: legionaminic acid, phylogeny, pseudaminic acid, neuraminic acid, biosynthetic pathway
Sialic acids (Sias) are displayed in prominent positions on vertebrate cells and are critical for such physiological processes as cellular repulsion, renal filtration, and neuronal plasticity (1, 2). Many other Sia-dependent functions occur in conjunction with Sia-binding lectins, including down-modulation of complement activity and the regulation of leukocyte activation, migration, and apoptosis (1, 3). The divergence of the superphyla protostomes and deuterostomes† created a dichotomy in animal Sia expression, and heralded the emergence of widespread Sia-dependent biological functions in deuterostomes. Nearly 5 decades of research have confirmed that with few exceptions, these unique 9-carbon backbone sugars are conspicuously absent from many eukaryotic lineages, including most protostomes, plants, fungi, and protists (1, 4). Sia decoration by de novo biosynthesis or via metabolic scavenging pathways has been reported in more than a dozen pathogenic bacterial species (5, 6) and also was recently described in a human gut–associated methanogenic archaeon (7). During infection, microbes displaying Sia mimicry can exploit host factor H and/or Siglec-9 to down-regulate alternative complement deposition and neutrophil bactericidal activities (8–11).
Sias are 9-carbon backbone derivatives of neuraminic (Neu) and ketodeoxynonulosonic (Kdn) acids. They are actually part of a larger family of carbohydrate structures collectively called nonulosonic acids (NulOs)‡. A number of NulO sugars other than Sias have been found in microbes, all of which are derivatives of 4 isomeric 5,7-diamino-3,5,7,9-tetradeoxynon-2-ulosonic acids (12). At least 2 of these, the d-glycero-d-galacto isomer [legionaminic acid (Leg)] (13, 14) and l-glycero-l-manno isomer [pseudaminic acid (Pse)] (15, 16), have striking structural and biosynthetic similarities to Sias (Fig. 1). These commonalities among NulO pathways reflect the structural similarity of all of the NulO sugars, as well as their uniqueness compared with other monosaccharides. Similar steps in each NulO biosynthetic (NAB)§ pathway are catalyzed by homologous enzymes, including the condensation of a 6-carbon sugar intermediate with 3-carbon phosphoenolpyruvate (3C) to generate the 9-carbon backbone NulO sugar, followed by the activation of free NulO residues using cytidine triphosphate to form cytidine monophosphate (CMP)-NulO intermediates (Fig. 1). In Campylobacter species and other ε-proteobacteria, Pse modifications play critical roles in flagellar assembly and, consequently, motility (16, 17), an important physiological function in aquatic environments and for association with animals. Leg modifications also have been identified on flagellar subunits, but have less well-defined functions (14, 18, 19). Both Leg and Pse also have been identified as part of lipopolysaccharide (LPS) O antigens in some Gram-negative bacteria (12), where they conceivably could contribute to biofilm formation, resistance to phage predation, or animal associations. Despite the similarities of Leg and Pse to Sias, the potential roles of these sugars in host–pathogen interactions remain poorly defined, and their distribution among microbes has not yet been systematically investigated.
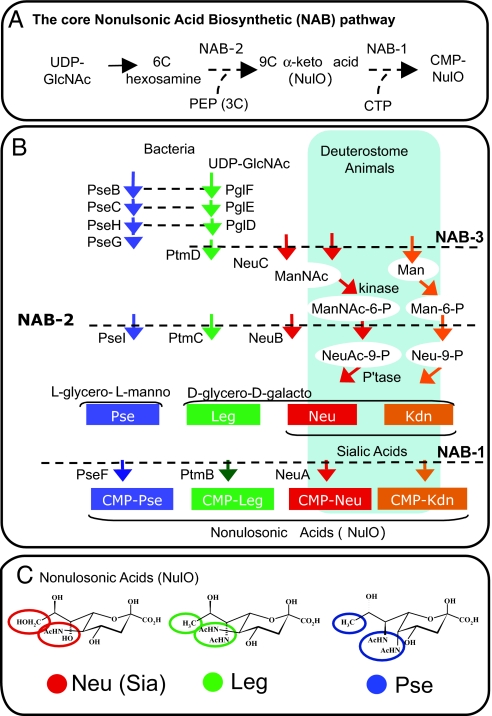
Fig. 1.
Related NAB pathways synthesize chemically related sugars. NulOs include all 9-carbon backbone α-keto acid sugars. NulO sugars described to date conform to one of several core backbones that can be further modified by epimerization or modification (1, 12). Major core backbones include the Neu, Kdn, Leg, and Pse acids. (A), The unique and shared “core” features of all NAB pathways include UDP-N-acetylglucosamine as a starting point, condensation of a 6-carbon intermediate with phosphoenolpyruvate to yield a 9-carbon α-keto acid (NAB-2), and formation of a CMP-activated NulO intermediate (NAB-1). (B), Architecture and nomenclature of NulO pathways. Horizontal dashed lines denote NAB enzymes in different pathways that share a common ancestor as deduced by amino acid sequence similarity. (C), Chemical structures of N-acetyl derivatives of Neu, Leg, and Pse.
In the present work, we probed the existing paradigms of Sia evolution using genomic, phylogenetic, and biochemical approaches to ask whether Sias were a unique innovation of the deuterostome lineage, whether bacterial mimicry of host Sias was the result of lateral gene transfer from an animal host or convergent evolution from microbial Sia-like biosynthetic pathways, and whether the chemical structure of Sias and related sugars can be predicted from genomic sequence information.
Results and Discussion
“Functional Clustering” Predicts a Remarkably Wide Distribution of NulO Sugar Expression Among Bacteria and Archaea.
To define the distribution of biosynthetic pathways for NulO sugars in members of bacteria and archaea, nearly 1,000 sequenced microbial genomes¶ were examined by BLAST for evidence of “functional clusters” (20) of NAB pathway genes. Unexpectedly, about 20% of all microbial phylogenetic types (phylotypes) sequenced to date were found to encode NAB pathway cassettes (Fig. 2).
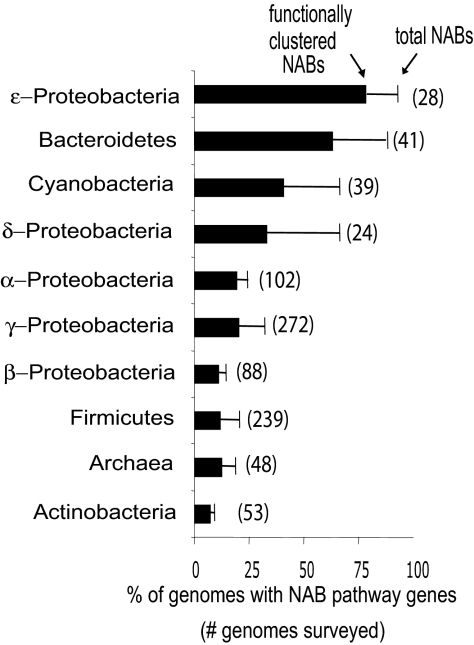
Fig. 2.
Distribution of predicted NAB pathways among bacteria and archaea. NAB enzymes NAB-1 and NAB-2 were identified by BLASTp in the genomes of various bacterial phyla and in sequenced archaeons. The number of genomes in each group is given in parentheses. Bars reflect the percentage of genomes in each group with one or more physically clustered NAB gene pairs, and thin line extensions reflect total NAB homolog pairs irrespective of functional clustering.
Many species/subsets, as well as entire phyla in which NulO sugars have never been documented, were found to have NAB enzymes in their genomes, including remarkably large proportions of available Bacteroidetes (36/41), Cyanobacteria (26/39), and δ-Proteobacteria (16/24), certain pathogenic members of the order Spirochaetales, and nearly 19% of sequenced Archaea (9/48) (Fig. 2). NAB pathways were identified in a larger fraction of the available genomes in bacterial phyla (divisions) previously known to include Sia-decorated pathogens (i.e., γ-Proteobacteria, β-Proteobacteria, and Firmicutes). Our analysis reveals a far wider distribution of NulOs and a deeper evolutionary history of this class of sugars than originally assumed.
Phylogenetic Prediction of NulO Types Reveals an Evolutionary Context for Sias and Sia-Like Sugars.
To better illuminate the evolutionary history of these 9-carbon backbone NulO sugars and predict their distribution and structure, we performed phylogenetic analysis of the most highly conserved enzyme in the pathway (NAB-2) and overlaid this tree with published biochemical data [Fig. 3A; summarized with strain and sequence identifiers in supporting information (SI) Table S1]. NAB-2 condenses a 6-carbon intermediate with the 3-carbon molecular phosphoenolpyruvate to generate NulOs of different types (Fig. 1). Microbial NAB-2 homologs with published roles in Neu synthesis were identified in multiple clades that are phylogenetically distinct from the clade with animal NAB-2 homologs (Fig. 3A). Specifically, the analysis revealed 2 groups of bacterial Neu synthases, represented by clades “a” and “c” (Fig. 3A), which include sequences from well-known Sia-decorated pathogens (Table 1). These data point to the existence of at least 3 evolutionarily discrete types of Neu synthases that ultimately share a common ancestor (i.e., semiconvergence). Examination of other NAB pathway enzymes revealed similar phylogenetic signatures, indicating that most microbial NAB pathway cassettes persist as evolutionarily conserved units (Fig. S1). These data predict Neu expression in several pathogenic and commensal bacterial species in which NAB biosynthesis has not been characterized previously (Table 1). Moreover, the data strongly suggest that Neu expression is not limited to microbes that associate with animals, as has been commonly assumed (see the composition of clade “c” in Table 1).
Fig. 3.
NAB-2 phylogeny for predicting NulO structure. (A), A distance-based neighbor-joining tree was constructed as described in Materials and Methods, and published biochemical data were overlaid onto the tree (colored circles). A “cohesion group” approach (21) was used to infer enzyme specificities by extrapolation of the published biochemical data (13–15, 22–24) to phylogenetic clades supported by high bootstrap values (shown at relevant nodes). Color shading reflects the phylogenetic predictions of chemical structure for each clade. Clades are designated “a”–“h” for reference. Note that clade “h” is composed entirely of NAB-2 enzymes from animals. Organisms for which biochemical data are presented in later figures are indicated by name. (B), Percentages of NAB-2 clade affiliations were calculated and expressed as a function of the number of genomes surveyed in each NCBI-classified taxonomic group (see Materials and Methods). Note that “pruned” tree branches representing diverged NAB-2 enzymes were included in the tabulation of NAB-positive genomes as “unknown” NulO type (shown in black) if a nearby NAB-1 homolog was identified. Some individual genomes encode multiple NAB pathways leading to NAB-2 homolog/genome sampled values of >1. These data, with strain and sequence identifiers, are summarized in Table S1.
Table 1.
Phylogenetic prediction of sialic acid synthesis in bacteria
| Organism | NCBI class* | Host/environment | Disease | NulO known | |
|---|---|---|---|---|---|
| a | S.agalactiae | 4 | Human | MG, SP, PN | Neu |
| S. suis | 4 | Swine, human | MG, SP, endocarditis | Neu | |
| E. coli K1 | 3 | Human, avian | MG, SP, GE, cystitis | Neu | |
| R. gnavus | 4 | Human | NK, gut symbiont | NK | |
| F. nucleatum | 5 | Human, animal | Periodontal diseases | NK | |
| c | C. jejuni | 1 | Chicken, human | GE, autoimmunity | Neu |
| N. meningitidis | 2 | Human | MG, SP | Neu | |
| B. cereus G9241 | 4 | Soil, animal | Anthrax-like PN | NK | |
| H. acinonychis | 1 | Feline | Gastritis in cheetah | NK | |
| F. johnsoniae | 3 | Soil, aquatic | NK | NK | |
| F. psychrophilum | 3 | Aquatic, fish | Fry syndrome, fish | NK | |
| Algoriphagus sp. PR1 | 3 | Aquatic | NK | NK | |
| Flavobacterium sp. MED217 | 3 | Seawater | NK | NK | |
| I. ioihiensis | 4 | NK, hydrothermal vent | NK | NK |
MG; meningitis. SP, septicemia. PN, pneumonia. GE, gastroenteritis. NK; none known. Here ″a″ and ″c″ refer to phylogenetic clades shown in Fig. 3A.
*NCBI classifications are as follows: 1, ε-Proteobacteria; 2, β-Proteobacteria; 3, Bacteroidetes; 4, Firmicutes; 5, Fusobacteria.
One evolutionary explanation for the emergence of Sia mimicry is represented by highly supported nodes in the NAB-2 and NAB-3 trees showing that clade “a” (Neu-specific enzymes encoded exclusively in animal-associated bacteria) shares a common ancestor with clade “b” (Leg-specific enzymes encoded mostly by aquatic organisms) (Fig. 3A and Fig. S1). Interestingly, the phylogenetic relationship between NAB pathways represented by clades “a” and “b” do not reflect known evolutionary relationships between organisms represented in these clades. Consistent with the shared ancestry of clades “a” and “b,” organisms represented in these clades (but not those in clade “c”) encode acetyltransferases as part of their NAB gene cassettes. Previous studies have indicated that such acetyltransferases are required for overall NulO expression (16, 25, 26). For example, Campylobacter jejuni (clade “b”) requires PtmH for N-acetylation at the 7-carbon position of Pse residues (16), whereas Streptococcus agalactiae and Escherichia coli (clade “a”) use the homologous NeuD enzyme for O-acetylation at the same carbon position of Neu (26). These data indicate that Neu biosynthetic pathways in clade “a” were not acquired by lateral gene transfer from an animal host, but rather the Neu mimicry by organisms represented in clade “a” arose by recruitment and modification of an ancestral NulO pathway that requires an acetylation reaction at C7 (Fig. 1).
NulO Biosynthetic Pathways Originated and Diversified Early in the History of Cellular Life.
Based on the distribution of Sias among animals, it has been suggested that the biosynthetic pathway for this NulO sugar may have been an innovation of the deuterostome lineage (1). But the NAB-2-based phylogeny (Fig. 3) revealed novel phylogenetic clusters of microbial NAB homologs highly similar to components of animal Sia pathways (clades “f,” “g,” and possibly “e”), suggesting that Sia synthesis pathways of animals may have much deeper evolutionary roots. Notably, the branching pattern of the limited number of taxa within these clades is consistent with known evolutionary relationships among these organisms. Targeted phylogenetic trees and protein domain analyses of NAB-1 and NAB-2 pairs encoded in the animal and “animal-like” clades further support the deep but firm evolutionary relationship between these biosynthetic pathways (Fig. S2). A few of the organisms represented in these clades associate in various ways with animals, including the spirochetal zoonotic pathogen Leptospira interrogans and the Actinobacteria Brevibacterium linens and Thermobifida fusca, which that cause body odor and farmers' lung, respectively. All other microbes represented in these clades are environmentally associated, however.
A model of early cellular diversification of NAB pathways (Fig. S3), including those for Sias, is supported by multiple lines of evidence, including phylogenetic and protein domain comparisons (Fig. 3A and Fig. S2), the wide distribution of NAB pathways within members of Bacteria and the considerable diversity of their predicted sugar structures (Figs. 2 and 3B), and the diverse composition of taxa in the animal-like clades (Table S1). The presence of archaeal NAB sequences in several distinct clades of the phylogenetic tree (Fig. 3A) also supports the conclusion that paralogous gene duplications with divergence of enzymatic function occurred very early in cellular evolution. We conclude that NAB enzymes, which at first glance appear to be animal-like, should be more accurately considered present-day components of an ancient pathway that was universally adopted by deuterostome animals. The distant sequence relationship between animal and animal-like clades suggests that Sia biosynthesis did not arise by lateral transfer into the animal lineage, but rather that Sia synthesis was likely inherited in the traditional sense, accompanied by multiple gene losses along other eukaryotic lineages (27, 28).
Chemical Validation of the Phylogenomic/Phyloglycomic Approach.
To provide functional validation of our phylogenomic findings, we present 2 striking examples that illustrate the utility of a phylogenetic approach for predicting chemical structure and inferring the evolutionary history of this class of monosaccharides.
Photobacterium profundum Strains Encode Phylogenetically Distinct NAB Enzymes and Biochemically Distinct NulO Sugars.
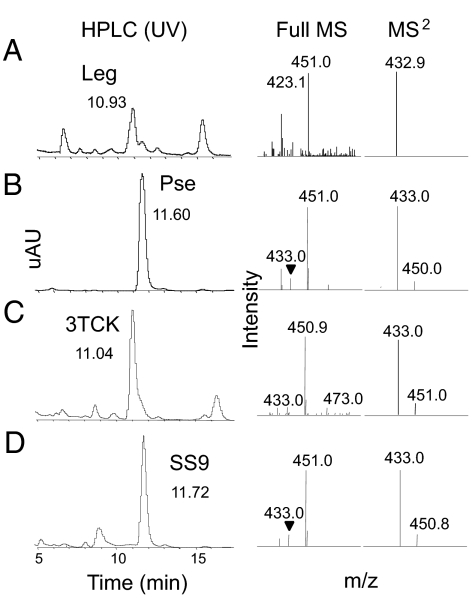
The phylogenetic data predict that 2 otherwise closely related strains of Photobacterium profundum (3TCK and SS9) encode biosynthetic pathways for distinct NulO sugar structures (Fig. 3). We tested this prediction using a well-validated approach for Sia structure identification. Leg and Pse acid standards were isolated from purified LPS preparations (29, 30) and, after fluorescent derivatization with 1,2-diamino-4,5-methylene dioxybenzene (DMB) (26), they eluted at distinct HPLC retention times (Fig. 4 A and B). Tandem mass spectrometry confirmed the expected masses of these eluted Leg and Pse standards, showing that the DMB-HPLC approach can be effectively applied to the broader class of NulO sugars. In another experiment, P. profundum SS9 and 3TCK strains were grown under optimized conditions, and mild acid hydrolysis was used to release NulO sugars. NulO sugars isolated from P. profundum genome strains 3TCK and SS9 exhibited the expected retention times and masses of Leg and Pse standards based on the phylogenetic prediction (Fig. 4 C and D).
Fig. 4.
Targeted chemical validation of Photobacterium profundum NAB pathways. LCMS analysis of NulO sugars isolated from purified LPS of Leg (A) or Pse (B) or from cultures of P. profundum genome strain (C) 3TCK or SS9 (D). Different HPLC retention times enable differentiation between Leg and Pse and reliable identification of NulO in 3TCK and SS9. MS and MS2 analyses provide additional confirmation of the expected mass for Leg and Pse for both standards and unknowns. In all cases, MS data are shown from 400–500 m/z.
Methanobrevibacter smithii: A Case of Mistaken Identity.
The published genome sequence of the principal human gut–associated methanogen, Methanobrevibacter smithii, contains a gene cluster originally annotated to encode a Sia (Neu) biosynthesis pathway. This archaeon has a prominent capsule when grown in vitro, and previous biochemical assays plus lectin-binding immunohistochemical studies suggested that NulO sugars, presumably Neu5Ac, are expressed by the cultured-type strain (7). Further analyses using custom M. smithii GeneChips (see Materials and Methods) revealed that this gene cluster was present in 7/7 M. smithii isolates examined, and that expression of these genes is differentially regulated in vitro (Fig. S4).
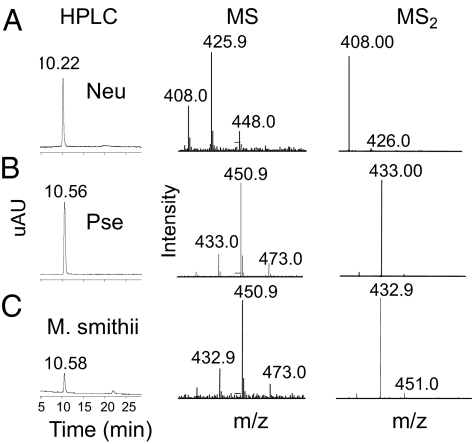
Here we show that the amino acid sequence of M. smithii NAB-2 clusters is within clade “d,” and thus is predicted to synthesize Pse rather than Neu acids (Fig. 3A). To test this hypothesis, we first compared the retention times and masses of DMB-derivatized α-keto acid standards Neu and Pse using reverse-phase HPLC with tandem mass spectrometry, and found that Neu and Pse have similar but distinct retention times and can be discriminated on the basis of mass-to-charge (m/z) ratio (Fig. 5 A and B). Parallel preparation and analysis of M. smithii NulO sugars in parallel with Neu and Pse standards clearly demonstrated a retention time and mass consistent with Pse expression (Fig. 5C). This finding again validates the phylogenetic approach to predicting NulO structure, and emphasizes that available methods for Sia detection can be successfully expanded to consider the larger family of NulO sugars.
Fig. 5.
The human gut methanogen M. smithii synthesizes pseudaminic, but not N-acetylneuraminic, acid. LCMS analysis of NulO sugars N-acetylneuraminic acid (Neu5Ac) (A), Pse from Pseudoalteromonas atlantica LPS (B), or isolated from cell pellets of M. smithii (C), as described in Materials and Methods. In all cases, MS data are shown from 400–500 m/z.
Prospectus.
In summary, our results provide insight into the evolutionary history of Sias by considering them in the larger phylogenetic context of related NulO sugars. We emphasize that the surprisingly wide distribution of NAB pathways among the 3 domains of life (Bacteria, Archaea, and Eukarya) is a reflection of many interwoven evolutionary processes, including gene duplications with functional divergence, gene loss, lateral gene transfer, and more specific adaptations of biosynthetic pathways. Clearly, much remains to be done to understand the biology and evolution of these remarkably common and diverse carbohydrate molecules found at the surfaces of contact between many bacteria and their external environments. These findings serve as a proof of principle for the utility of a phylogenomic/phyloglycomic approach to predicting NulO sugar types and strongly suggest that the expression of Sias and Sia-like sugars by bacteria may be advantageous in a wide range of animal body habitats. Determining the advantages of Sia mimicry in these different host contexts, as well as identifying those factors contributing to lateral dissemination of Sia gene cassettes (i.e., functional clusters) and their incorporation into various LPS or capsular polysaccharide biosynthetic pathways, are important areas for further investigation. These studies should provide a path for future investigation concerning the contributions of Sias and related sugars to the survival and persistence of microbes in both host and environmental reservoirs, as well as in disease pathogenesis.
Materials and Methods
Identification of Physically Clustered NAB Genes.
BLASTp, on the “BLAST with microbial genomes” webpage at the National Center for Biotechnology Institute (NCBI) website (www.ncbi.nlm.nih.gov/sutils/genom_table.cgi), was used to query 960 complete microbial genomes. Multiple NAB-1 and NAB-2 amino acid sequences were used for genomic query, with an emphasis on NAB enzymes of defined function or those encoded in organisms known to express specific NulO structures. Incomplete genomes also were queried using the nonredundant protein database and were included in the data set if deemed NAB-positive. Accession numbers for homologous NAB sequences were cataloged according to the NCBI taxonomic classification and examined for “functional clustering” (20) of NAB-1 and NAB-2 enzymes, as judged by proximal accession numbers. NAB-1 homologs were validated by phylogenetic analysis, and “contaminating” CMP-Kdo synthetases were removed; these distant NAB-1 homologs were not found in functional clusters with NAB-2 homologs. Note that Kdo is an 8-carbon α-keto acid that follows a similar biosynthetic pathway involving condensation of a 5-carbon sugar with phosphoenolpyruvate, followed by activation to CMP-Kdo (1). Results of the genomic profiling were compiled, and the proportion of genomes with one or more physical clusters of NAB-1 and NAB-2 genes was expressed as a function of total genomes surveyed in the different microbial taxa (Fig. 2).
NAB-2 Phylogeny for Prediction of NulO Sugar Types.
Amino acid sequence comparisons indicated that NAB-2 sequences are better conserved than other NAB enzymes and form the most conclusive basis for prediction of specific NulO sugars in different organisms. NAB-2 amino acid sequences were collected using BLASTp at the NCBI nonredundant protein database and aligned using ClustalQt (Fig. S5) (31). Nexus files from the alignment were uploaded into PAUP* 4.0b10 (32) for exclusion of gaps and domains not found in all sequences, followed by construction of a neighbor-joining tree using the bootstrap/jackknife option with 1,000 replicates. Less-conserved sequences were apparent from visual inspection of the alignment and clustering of branches in a star shape at the base of the phylogenetic tree. Such branches were “pruned” from the tree in successive analyses to reveal significant monophyletic clades and improve aspects of the alignment that could better resolve sequence relationships between different clades. Analysis of the same alignment based on parsimony produced a nearly identical phylogenetic pattern as that from the distance-based approach. NulO types were predicted from extrapolations of published biochemical data (1, 12–16, 22–24, 33–35) to other members of clades supported by high bootstrap values (shown in Fig. 3A). To determine the distribution of NulO types among different microbial phylotypes, the percentages of NAB-2 clade affiliations (Fig. 3 A–G) were calculated and expressed as a function of the number of genomes surveyed in each NCBI-classified taxonomic group (Fig. 3B). “Pruned” tree branches representing diverged NAB-2 enzymes were included in the tabulation of NAB-positive genomes as “unknown” NulO type (shown in black in Fig. 3B) if a nearby NAB-1 homolog was identified. Note that many individual genomes encode multiple NAB pathways (e.g., ε-Proteobacteria, Bacteroidetes; see Table S1), sometimes leading to a ratio of NAB-2 homologs:genomes sampled of >1 in Fig. 3B.
Strains and Culture Conditions.
The 2 strains of Photobacterium profundum with complete genome sequences are close phylogenetic relatives but are adapted to different aquatic ecosystems (36). P. profundum SS9 is a piezophilic (pressure-loving) strain that grows optimally under low-temperature, high-pressure conditions. In contrast, P. profundum 3TCK is adapted to an aquatic niche much closer to the surface. High-pressure growth of SS9 was performed anaerobically at 16 °C in 2216 medium (Difco) supplemented with 20 mM glucose and 100 mM Hepes buffer (pH 7.5) (Sigma). Late-exponential phase cultures were diluted 500-fold into fresh medium and used to fill 4.5-mL polyethylene transfer pipettes (Samco). Transfer pipettes were heat-sealed with a hand-held heat-sealing clamp (Nalge) and incubated at 30 MPa in a stainless steel pressure vessel (37). 3TCK was cultivated in the same media but incubated at room temperature without shaking.
Three strains of M. smithii were obtained from the DSMZ culture collection (2374, 2375, and 11975), 4 strains were isolated from a single human fecal sample by selective culturing, and the sequenced-type strain (PS) was obtained from ATCC. M. smithii was grown in supplemented MBC medium under anaerobic conditions for 6 days at 37 °C as described previously (7).
GeneChip-Based Studies of M. smithii.
Genomic DNA was prepared and hybridized to a custom Affymetrix GeneChip containing probesets that recognize 99% of its 1,795 predicted protein coding genes. Similarly, RNA isolated under different in vitro growth conditions was hybridized to GeneChips as described previously (See SI Text) (7).
Chemical Analysis of NulO Acids.
M. smithii and P. profundum were harvested from cultures by centrifugation and washed twice with PBS. NulO residues were released from cells or purified LPS samples containing Leg (29) or Pse (30) acids by mild acid hydrolysis, and low molecular weight fractions were subjected to derivatization with DMB, followed by HPLC analysis and liquid chromatography–mass spectrometry (LCMS) as described previously (see SI Text) (38).
Supplementary Material
Acknowledgments.
We thank Russell F. Doolittle for many helpful discussions, Sandra Diaz for assisting with mass spectrometry, Doug Bartlett for providing P. profundum, Emiley Eloe for performing high-pressure cultivation of SS9, and Henning Seedorf for assisting with the GeneChip hybridizations. We appreciate the efforts of Olga Zagnitko in updating annotations of NAB pathways in The SEED, an annotation/analysis tool provided by the Fellowship for Interpretation of Genomes, as well as insightful comments from Aaron Carlin, Yung-Chi Chang, and Shannon Weiman. This work was funded by generous support from the University of California President's Postdoctoral Fellowship Program (to A.L.L.) and the Gianinni Family Foundation (to A.L.L.), along with National Institutes of Health grants HL057345 (to A.V.), DK30292 (to J.I.G.), and R01-HD051796 (to V.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Protostomes and deuterostomes are bilaterian animals with distinct developmental programs for gut tube formation in which the first opening of the embryo, the blastopore, becomes either the mouth (protostomes, “mouth first”) or the anus (deuterostomes, “mouth second”).
There currently is no established nomenclature defining the 9 carbon α-keto acids as a group. Following discussions with Hans Kamerling, Roland Schauer, and Nathan Sharon, here we use the abbreviation “NulO” for non-2-ulosonic acids, which assumes Nul for non-2-uloses and maintains the discrimination between aldonic acids and uronic acids, such as glucuronic acid. We suggest that the use of the term “sialic acid” (Sia) continue to be limited to its original use in describing Neu, Kdn, and their derivatives in deuterostomes and their pathogens, and that NulO be used to encompass the entire group of 9 carbon α-keto acids.
NAB pathways have been defined in various bacterial pathogens. Enzymes in each of these pathways have been given different designations, beginning with “Neu” for neuraminic, “Pse” for pseudaminic, and “Ptm” for posttranslational modification (see Fig. 1 for published designations). To avoid confusion, here we use “NAB” to refer to homologous steps in these related biosynthetic pathways, numbering the enzymatic steps as shown in Fig. 1.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902431106/DCSupplemental.
These sequences had been previously deposited in the Genbank database. For a list of accession numbers, see Table S1. Annotations have been updated in The SEED, an annotation/analysis tool provided by the Fellowship for Interpretation of Genomes.
References
- 1.Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: An evolutionary perspective. Chem Rev. 2002;102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 2.Eylar EH, Doolittle RF, Madoff MA. Sialic acid from blood cells of the lamprey eel. Nature. 1962;193:1183–1184. doi: 10.1038/1931183a0. [DOI] [PubMed] [Google Scholar]
- 3.Varki A. Glycan-based interactions involving vertebrate sialic acid–recognizing proteins. Nature. 2007;446:1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- 4.Warren L. The distribution of sialic acids in nature. Comp Biochem Physiol. 1963;10:153–171. doi: 10.1016/0010-406x(63)90238-x. [DOI] [PubMed] [Google Scholar]
- 5.Vimr E, Lichtensteiger C. To sialylate, or not to sialylate: That is the question. Trends Microbiol. 2002;10:254–257. doi: 10.1016/s0966-842x(02)02361-2. [DOI] [PubMed] [Google Scholar]
- 6.Kelm S, Schauer R. Sialic acids in molecular and cellular interactions. Int Rev Cytol. 1997;175:137–240. doi: 10.1016/S0074-7696(08)62127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samuel BS, et al. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci USA. 2007;104:10643–10648. doi: 10.1073/pnas.0704189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damian RT. Molecular mimicry in biological adaptation. Science. 1965;147:824. doi: 10.1126/science.147.3660.824-b. [DOI] [PubMed] [Google Scholar]
- 9.Carlin AF, Lewis AL, Varki A, Nizet V. Group B streptococcal capsular sialic acids interact with siglecs (immunoglobulin-like lectins) on human leukocytes. J Bacteriol. 2007;189:1231–1237. doi: 10.1128/JB.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ram S, et al. A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J Exp Med. 1998;187:743–752. doi: 10.1084/jem.187.5.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlin AF, et al. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood. 2009;113:3333–3336. doi: 10.1182/blood-2008-11-187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knirel YA, Shashkov AS, Tsvetkov YE, Jansson PE, Zähringer U. 5,7-Diamino-3,5,7,9-tetradeoxynon-2-ulosonic acids in bacterial glycopolymers: Chemistry and biochemistry. Adv Carbohydr Chem Biochem. 2003;58:371–417. doi: 10.1016/s0065-2318(03)58007-6. [DOI] [PubMed] [Google Scholar]
- 13.Glaze PA, Watson DC, Young NM, Tanner ME. Biosynthesis of CMP-N,N′-diacetyllegionaminic acid from UDP-N,N′-diacetylbacillosamine in Legionella pneumophila. Biochemistry. 2008;47:3272–3282. doi: 10.1021/bi702364s. [DOI] [PubMed] [Google Scholar]
- 14.McNally DJ, et al. Targeted metabolomics analysis of Campylobacter coli VC167 reveals legionaminic acid derivatives as novel flagellar glycans. J Biol Chem. 2007;282:14463–14475. doi: 10.1074/jbc.M611027200. [DOI] [PubMed] [Google Scholar]
- 15.Schoenhofen IC, McNally DJ, Brisson JR, Logan SM. Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: Synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology. 2006;16:8C–14C. doi: 10.1093/glycob/cwl010. [DOI] [PubMed] [Google Scholar]
- 16.McNally DJ, et al. Functional characterization of the flagellar glycosylation locus in Campylobacter jejuni 81–176 using a focused metabolomics approach. J Biol Chem. 2006;281:18489–18498. doi: 10.1074/jbc.M603777200. [DOI] [PubMed] [Google Scholar]
- 17.Schirm M, et al. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol Microbiol. 2003;48:1579–1592. doi: 10.1046/j.1365-2958.2003.03527.x. [DOI] [PubMed] [Google Scholar]
- 18.Logan SM, et al. Identification of novel carbohydrate modifications on Campylobacter jejuni 11168 flagellin using metabolomics-based approaches. FEBS J. 2009;276:1014–1023. doi: 10.1111/j.1742-4658.2008.06840.x. [DOI] [PubMed] [Google Scholar]
- 19.Twine SM, et al. Flagellar glycosylation in Clostridium botulinum. FEBS J. 2008;275:4428–4444. doi: 10.1111/j.1742-4658.2008.06589.x. [DOI] [PubMed] [Google Scholar]
- 20.Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonner CA, et al. Cohesion group approach for evolutionary analysis of TyrA, a protein family with wide-ranging substrate specificities. Microbiol Mol Biol Rev. 2008;72:13–53. doi: 10.1128/MMBR.00026-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou WK, Dick S, Wakarchuk WW, Tanner ME. Identification and characterization of NeuB3 from Campylobacter jejuni as a pseudaminic acid synthase. J Biol Chem. 2005;280:35922–35928. doi: 10.1074/jbc.M507483200. [DOI] [PubMed] [Google Scholar]
- 23.Hao J, Balagurumoorthy P, Sarilla S, Sundaramoorthy M. Cloning, expression, and characterization of sialic acid synthases. Biochem Biophys Res Commun. 2005;338:1507–1514. doi: 10.1016/j.bbrc.2005.10.113. [DOI] [PubMed] [Google Scholar]
- 24.Suryanti V, Nelson A, Berry A. Cloning, over-expression, purification, and characterisation of N-acetylneuraminate synthase from Streptococcus agalactiae. Protein Expr Purif. 2003;27:346–356. doi: 10.1016/s1046-5928(02)00633-2. [DOI] [PubMed] [Google Scholar]
- 25.Daines DA, Wright LF, Chaffin DO, Rubens CE, Silver RP. NeuD plays a role in the synthesis of sialic acid in Escherichia coli K1. FEMS Microbiol Lett. 2000;189(2):281–284. doi: 10.1111/j.1574-6968.2000.tb09244.x. [DOI] [PubMed] [Google Scholar]
- 26.Lewis AL, Hensler ME, Varki A, Nizet V. The group B streptococcal sialic acid O-acetyltransferase is encoded by neuD, a conserved component of bacterial sialic acid biosynthetic gene clusters. J Biol Chem. 2006;281:11186–11192. doi: 10.1074/jbc.M513772200. [DOI] [PubMed] [Google Scholar]
- 27.Salzberg SL, White O, Peterson J, Eisen JA. Microbial genes in the human genome: Lateral transfer or gene loss? Science. 2001;292:1903–1906. doi: 10.1126/science.1061036. [DOI] [PubMed] [Google Scholar]
- 28.Doolittle RF. Gene transfers between distantly related organisms. In: Syvanen M, Kado C, editors. Horizontal Gene Transfer. 2nd Ed. New York: Academic; 2002. pp. 269–275. [Google Scholar]
- 29.Knirel YA, Rietschel ET, Marre R, Zähringer U. The structure of the O-specific chain of Legionella pneumophila serogroup 1 lipopolysaccharide. Eur J Biochem. 1994;221:239–245. doi: 10.1111/j.1432-1033.1994.tb18734.x. [DOI] [PubMed] [Google Scholar]
- 30.Perepelov AV, et al. Structure of an acidic polysaccharide from the agar-decomposing marine bacterium Pseudoalteromonas atlantica strain IAM 14165 containing 5,7-diacetamido-3,5,7,9-tetradeoxy-L-glycero-L-manno-non-2-ulosonic acid. Carbohydr Res. 2005;340:69–74. doi: 10.1016/j.carres.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinform. 2002 doi: 10.1002/0471250953.bi0203s00. Unit 2.3. [DOI] [PubMed] [Google Scholar]
- 32.Swofford DL. Sunderland, MA: Sinauer Associates; 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods) Version 4. [Google Scholar]
- 33.Sundaram AK, et al. Characterization of N-acetylneuraminic acid synthase isoenzyme 1 from Campylobacter jejuni. Biochem J. 2004;383(Pt 1):83–89. doi: 10.1042/BJ20040218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gunawan J, et al. Structural and mechanistic analysis of sialic acid synthase NeuB from Neisseria meningitidis in complex with Mn2+, phosphoenolpyruvate, and N-acetylmannosaminitol. J Biol Chem. 2005;280:3555–3563. doi: 10.1074/jbc.M411942200. [DOI] [PubMed] [Google Scholar]
- 35.Shashkov AS, et al. Structure of the O-antigen of Providencia stuartii O20, a new polysaccharide containing 5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-d-galacto-non-2-ulosonic acid. Carbohydr Res. 2007;342:653–658. doi: 10.1016/j.carres.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Campanaro S, et al. Laterally transferred elements and high pressure adaptation in Photobacterium profundum strains. BMC Genomics. 2005;6:122. doi: 10.1186/1471-2164-6-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eloe EA, Lauro FM, Vogel RF, Bartlett DH. The deep-sea bacterium Photobacterium profundum SS9 utilizes separate flagellar systems for swimming and swarming under high-pressure conditions. Appl Environ Microbiol. 2008;74:6298–6305. doi: 10.1128/AEM.01316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis AL, Nizet V, Varki A. Discovery and characterization of sialic acid O-acetylation in group B Streptococcus. Proc Natl Acad Sci USA. 2004;101:11123–11128. doi: 10.1073/pnas.0403010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.