Abstract
The cytosolic RNA-binding protein NAB1 represses translation of LHCII (light-harvesting complex of photosystem II) encoding mRNAs by sequestration into translationally silent mRNP complexes in the green alga Chlamydomonas reinhardtii. NAB1 contains 2 cysteine residues, Cys-181 and Cys-226, within its C-terminal RRM motif. Modification of these cysteines either by oxidation or by alkylation in vitro was accompanied by a decrease in RNA-binding affinity for the target mRNA sequence. To confirm the relevance of reversible NAB1 cysteine oxidation for the regulation of its activity in vivo, we replaced both cysteines with serines. All examined cysteine single and double mutants exhibited a reduced antenna at PSII caused by a perturbed NAB1 deactivation mechanism, with double mutations and Cys-226 single mutations causing a stronger and more distinctive phenotype compared with the Cys-181 mutation. Our data indicated that the responsible redox control mechanism is mediated by modification of single cysteines. Polysome analyses and RNA co-immunoprecipitation experiments demonstrated the interconnection of the NAB1 thiol state and its activity as a translation repressor in vivo. NAB1 is fully active in its dithiol state and is reversibly deactivated by modification of its cysteines. In summary, this work is an example that cytosolic translation of nucleus encoded photosynthetic genes is regulated via a reversible cysteine-based redox switch in a RNA-binding translation repressor protein.
Keywords: Chlamydomonas reinhardtii, light harvesting antenna, redox control, translation control
To compensate for changes in light intensity or spectral quality, plants have developed several short-term and long-term mechanisms to regulate the amount of light that is captured by each photosystem (1). One important long-term adaptation strategy of plant organisms involves the complex expression regulation of various nuclear-encoded light harvesting complex (Lhcb) genes (1). All levels of LHCII gene expression are targeted by regulation mechanisms (2–5) which rely on a complex retrograde and anterograde communication between plastid, nucleus, and cytosol (6). The cytosolic translation repressor NAB1, which was identified in a Chlamydomonas reinhardtii light acclimation mutant (4), is the center of interest within this work. NAB1 harbors 2 RNA-binding motifs and 1 of these motifs, located at the N terminus, belongs to the highly conserved family of CSD (cold shock domain) domains. Proteins containing a CSD motif are referred to as Y-box proteins and eukaryotic members of this large family generally contain a second auxiliary RNA-binding domain, which modulates the RNA affinity of the protein but can be dispensable for selective RNA recognition (7). In the case of NAB1, the CSD motif is combined with a C-terminal RRM (RNA recognition motif) domain, which was demonstrated not to be essential for selective RNA recognition (4). It was shown that NAB1 binds to the mRNA of LHCBM (major light-harvesting complex of photosystem II) genes, thereby preventing translation via sequestration of the message in translationally silent messenger ribonucleoprotein complexes (mRNPs). The LHCII complex of C. reinhardtii is constituted by 10 individual highly homologous LHCBM isoforms (8, 9), and NAB1 displays selectivity toward distinct isoforms with LHCBM6 mRNA being 1 of its main targets (4). It has been shown for numerous proteins that reversible modification of cysteine residues can act as an effective activity switch (10). In this work, we intended to investigate whether the composition of the light-harvesting antenna of PSII is controlled via the redox state of 2 cysteines, which are located in the C-terminal RRM domain of NAB1.
Results
Free Cysteines Are Required for Full RNA-Binding Activity of NAB1 in Vitro.
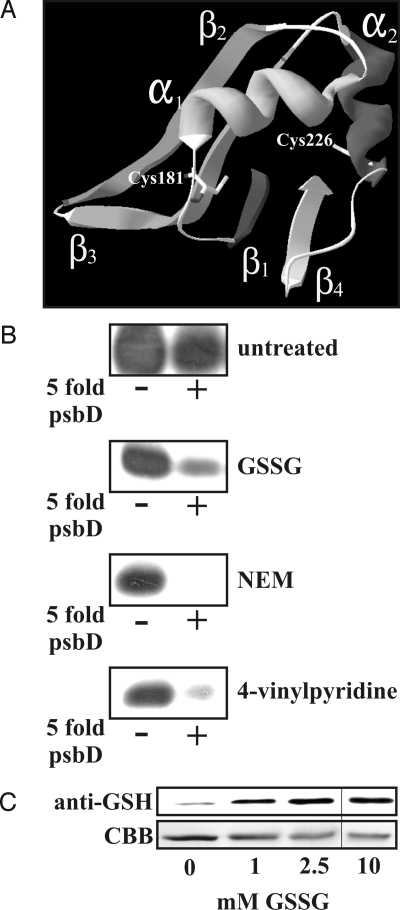
NAB1 harbors 2 cysteine residues, located at amino acid positions 181 and 226 within the C-terminal RRM domain. A structural model of the RRM domain of NAB1 was generated using the NMR structure of the highly homologous RRM motif of human RNA binding protein hnRNP M (Fig. 1A). Within this structure Cys-181 is part of a loop structure whereas Cys-226 is part of the α-helix α2 and both residues are separated by 14.97 Å. Exposition of these cysteines on the protein surface is a prerequisite for a potential reversible interaction with thiol modifying compounds in vivo. Modeling of the C terminus (Fig. S1) indicated that Cys-181 is buried in a groove-like structure together with 2 leucine residues and surrounded by uncharged amino acids creating an environment of low electrostatic potential. In contrast, Cys-226 could be more reactive because it is positioned in an exposed surface area at the interface of a negatively and positively charged patch. To analyze whether modification of cysteines within the RRM motif has an impact on the binding affinity toward its cellular mRNA target LHCBM6, RNA-binding studies with oxidized and reduced recombinant NAB1 were performed (Fig. 1B). For these experiments, a probe containing the CSDCS (cold shock domain consensus motif) motif of LHCBM6 was chosen, which was previously shown to bind NAB1 specifically (4). Because of the reducing conditions used for NAB1 purification, recombinant NAB1 proteins were maintained in a reduced state. Under this condition they efficiently bound a radioactive CSDCS probe derived from LHCBM6 (Fig. 1B, untreated). The presence of unlabeled competitor RNA (Fig. 1B, psbD +) had a negligible effect on binding efficiency, indicating sequence specificity of the protein-RNA interaction (4). In contrast, when shifted to an oxidized form by treatment with glutathione disulfide (GSSG), the binding signal of the NAB1-RNA complex was most strongly reduced after addition of competitor RNA (Fig. 1B, GSSG, psbD +), showing that oxidation mainly affects the specific binding activity of NAB1. To investigate whether oxidation of both cysteines via the formation of an intramolecular disulfide is crucial for the observed decrease in RNA affinity, thiol-alkylating compounds were used to modify NAB1 cysteines individually. The thiol-specific alkylator N-ethylmaleimide (NEM) and 4-vinylpyridine turned out to be potent inhibitors of the specific binding activity of NAB1, which was indicated by a complete absence of LHCBM6-NAB1 complexes after competitor addition (Fig. 1B). This result indicated that modification of RRM cysteines impairs the specific binding of NAB1 to LHCBM6 mRNA. An additional result was that intramolecular disulfide bridge formation is not a prerequisite for NAB1 deactivation in vitro. In line with these findings was that the treatment of recombinant NAB1 with GSSG resulted in the glutathionylation of single cysteines (Fig. 1C). To address the potential of intramolecular disulfide formation within NAB1 more precisely, a peptide mapping analysis with recombinant wild-type Wt-NAB1 and a recombinant Cys226Ser mutant was performed. Wt- and Cys-226-NAB1 protein was oxidized with diamide and subsequently digested by V8 protease treatment. Any intramolecular disulfide bridge formation in the Wt protein should result in an altered peptide fragment pattern compared with the corresponding Cys226Ser sample, for which intramolecular disulfide formation is not possible. However, nearly identical digestion patterns of Wt and Cys226Ser samples were obtained (Fig. S2). As an additional control, 1 part of each oxidized sample was rereduced with DTT before SDS/PAGE. After rereduction, the banding pattern remained unchanged, indicating again that no disulfide linked peptides were present in the oxidized digest samples. In summary, these results demonstrated that modification of individual cysteines impairs the activity of NAB1 in vitro and led us to investigate the in vivo relevance of thiol regulation as a mechanism to control the activity state of NAB1.
Fig. 1.
In silico and in vitro analyses to examine a potential cysteine regulation of NAB1. (A) Homology model of the C-terminal RRM domain of NAB1. Cys-181 and Cys-226 within the RRM domain are indicated. (B) In vitro RNA binding studies. Autoradiogram of UV-cross-linked and SDS/PAGE separated NAB1-LHCBM6-CSDCS complexes. Recombinant NAB1 was either left untreated (Top) or oxidized with 5 mM glutathione disulfide (GSSG, Middle) before addition of a radioactively labeled LHCBM6-CSDCS probe alone (−) or of a mixture containing labeled LHCBM6-CSDCS and a 5-fold molar excess of unlabeled psbD competitor RNA (+). N-ethylmaleimide (NEM, third panel from Top) or 4-vinylpyridine (4-vinylpyridine, Bottom) was applied for cysteine alkylation. (C) Treatment of recombinant NAB1 with different concentrations of glutathione disulphide (GSSG) and immunodetection of protein-glutathione adducts (Upper). Coomassie blue (CBB) stain of recombinant NAB1 after treatment with the indicated GSSG concentrations (Lower).
Replacement of RRM Domain Cysteines with Serine Creates a Small Antenna Phenotype.
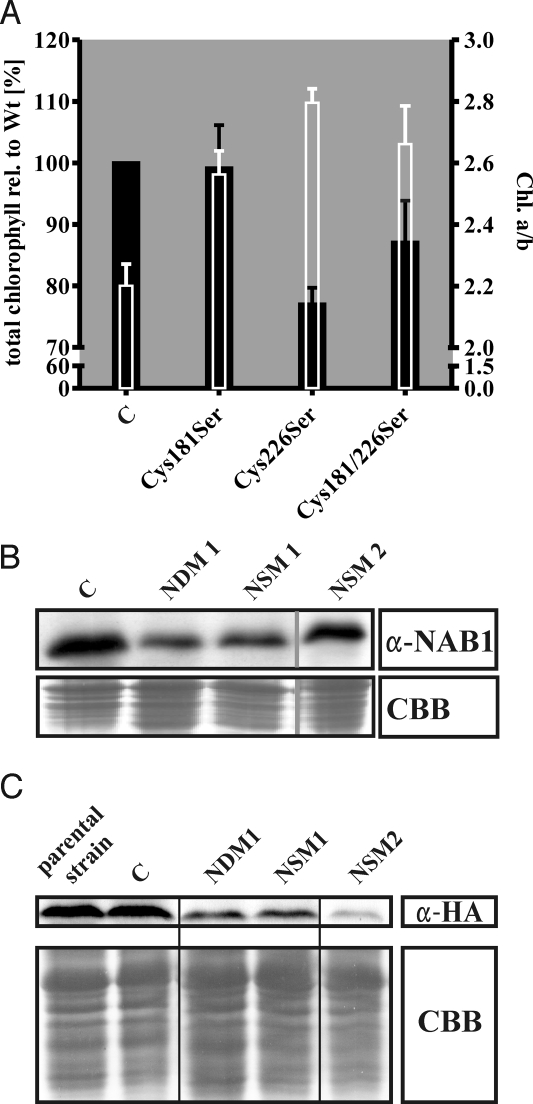
To determine whether the 2 cysteine residues have an essential regulative function in vivo, a mutagenesis strategy was applied. For this experiment, we selected a derivative strain of Stm3 that expresses an HA-epitope-tagged form of the LHCBM6 protein. The HA-tagged LHCBM6 isoform is expressed under the control of the PSAD promoter (4, 11), thus decoupling its transcription from LHC promoter activity. This NAB1-free strain was transformed with a mutated version of the NAB1 gene. This experimental setup allowed the direct correlation between NAB1 activity and post-transcriptional regulation of LHC isoform LHCBM6. Mutated constructs for each single cysteine (NSM) and for both cysteines (NDM) were obtained by site-directed mutagenesis which resulted in the exchange of cysteine with serine residues. After cotransformation of these constructs, 15 strains were successfully identified which stably expressed a mutated version of NAB1, carrying either the double mutation Cys-181/226Ser or the single mutations Cys181Ser and Cys226Ser. For precise chlorophyll analyses to investigate the relevance of both cysteines for LHC antenna size regulation (Fig. 2A), a control strain (C) was obtained by transformation of Stm3 with a Wt version of NAB1, which is expressed under control of the same promoter that drives expression of mutated NAB1 in the cysteine mutants. Based on this control strain, the Cys double mutants and the single Cys226Ser mutants showed a similar phenotype with an increase of Chl a/b ratios [from 2.2 in the control strain (C) to ≈2.6/2.8 in the mutants] accompanied by a 15–25% decrease of total chlorophyll content (Fig. 2A). Because Chl b is exclusively bound by light-harvesting proteins whereas Chl a is also present in the PS core complexes, the Chl. a/b ratio provides a parameter for the overall LHC antenna size compared with the entire PSI and PSII complexes. Consequently, the increased Chl a/b ratios are indicative for smaller light-harvesting antenna systems due to a regulatory dysfunction of NAB1. Of particular note was however, that the Cys-181 mutants exhibited a less distinctive phenotype with no differences in total chlorophyll content indicating functional differences between Cys-181 and Cys-226. To allow for more detailed analyses regarding the phenotypical characteristics caused by the mutations, we selected 1 representative strain for each single (NSM1: Cys181Ser; NSM2: Cys226Ser) and the double mutation (NDM1: Cys-181/226Ser). Immunoblot studies using a NAB1-specific antiserum demonstrated that the expression of NAB1 variants in these mutant strains was lower compared with the expression of Wt NAB1 in the control strain (Fig. 2B), excluding the risk that the significant decrease of antenna size was caused by increased levels of NAB1 protein.
Fig. 2.
Phenotypical analyses of NAB1-cysteine mutants. (A) Total chlorophyll content of NAB1-cysteine mutants relative to the control strain C expressing Wt-NAB1 (left y axis; black bars) and Chl. a/b ratios of cysteine mutants and control strain (right y axis; white bars). The data represent mean values of three independent chlorophyll measurements (using triplicates) performed with different strains for each cysteine mutation (10 strains expressing NAB1Cys181Ser; three strains expressing NAB1Cys226Ser; two strains expressing NAB1Cys-181/226Ser). Error bars indicate standard deviations (n = 30 for NAB1Cys181Ser; n = 9 for NAB1Cys226Ser; n = 6 for NAB1Cys-181/226Ser). (B) Anti-NAB1 immunoblot analyses to assess the NAB1 expression level in the Wt control strain and the cysteine mutant strains. (Upper) Representative immunoblot. (Lower) Coomassie blue stain (loading control). (C) Anti-HA-tag immunoblots to determine the expression of HA-epitope tagged LHCBM6 protein (Upper). (Lower) Coomassie blue-stained SDS protein gel (loading control).
Protein Expression of the Target mRNA LHCBM6 Is Down-Regulated by Cysteine Mutation.
Because all mutant strains expressed an HA-tagged version of the isoform LHCBM6, a comparative analysis of HA-LHCBM6 expression in the cysteine mutants, the parental strain, and in the control strain was a suitable method to analyze the effects of NAB1 cysteine mutation on its activity as a translation repressor in vivo. Transformation of the parental strain with mutagenized versions of NAB1, which lack 1 or both cysteines, yielded reduced HA-LHCBM6 amounts (Fig. 2C). Importantly HA-LHCBM6 expression of the mutant cell lines was also reduced compared with a cell line expressing wild-type NAB1 [Fig. 2C, lane 2 (C)].
Cysteine Mutants Are Unable to Enlarge Their LHCII Complexes Resulting in Impaired Growth Under Phototrophic Dim Light Conditions.
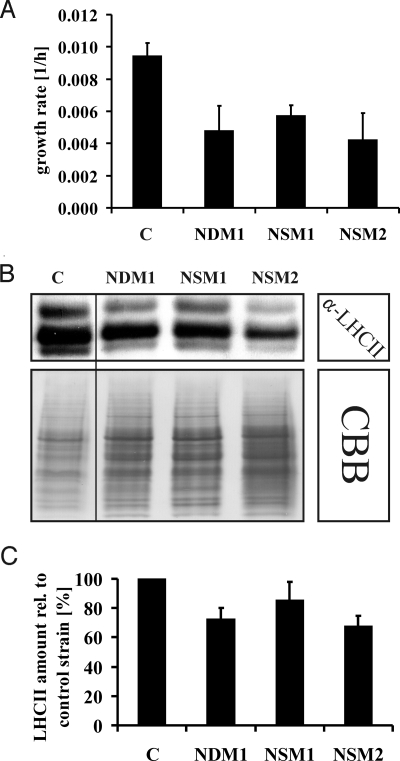
Chlorophyll measurements and LHCBM6 protein expression studies indicated that the presence of cysteine residues within its RRM domain is crucial for a deactivation of NAB1. Photoautotrophic growth experiments in minimal medium (HSM) under dim light (40 μmol m−2 s−1) were performed which force Wt Chlamydomonas cells to increase their light-harvesting antenna size to enhance the capture of photons for photosynthesis. All selected cysteine mutants showed a reduced growth rate under limiting light conditions when compared with the control strain (Fig. 3A). The exponential growth rates of the examined mutants varied between 0.0048Δ O.D.750 nm/h in the case of NDM1, 0.0058Δ O.D.750 nm/h for NSM1, and 0.0043Δ O.D.750 nm/h for NSM2 and were therefore significantly reduced in relation to the control strain (0.0095Δ O.D.750 nm/h). In good agreement with the results obtained under standard light conditions (see Fig. 2A), the Chl a/b ratio was highly increased in NDM1 and NSM2 only (2.24 ± 0.03 SD in control strain vs. 2.87 ± 0.03 SD in NDM1 and 3.09 ± 0.03 SD in NSM2), whereas the Cys-181 mutant NSM1 showed an increase to a much lesser extent (Chl a/b 2.32 ± 0.03 SD). In addition, total chlorophyll values in relation to the control strain (C) were only reduced in the double mutant NDM1 and strain NSM2 (76 ± 1% SD of control strain in case of NDM1 and 68 ± 2% SD for NSM2). Anti LHCII immunoblot studies (Fig. 3 B and C) confirmed that the amount of LHCII proteins was reduced in the cysteine mutants (72.8 ± 7.1% SE in the case of NDM1, 85.5 ± 12.3% SE for NSM1 and 67.7 ± 6.8% SE for NSM2 with the control strain being set to 100%) and demonstrated that the Cys-226 mutation again has a more severe effect on the LHC antenna size compared with the Cys-181 mutation (Fig. 3C). In conclusion, the observed phenotypes strongly indicate a direct correlation between the thiol state of the cysteines of NAB1 and the activity as a LHC translation repressor.
Fig. 3.
Growth and photosynthetic low light acclimation of control strain (C) and NAB1 cysteine mutants. (A) Growth rates within the exponential phase observed under phototrophic low light conditions (HSM medium; 40 μmol m−2 s−1). The growth rate was determined by measurements of the increase of the optical density per hour. Error bars indicate the standard deviation of three independent growth experiments. (B) Representative immunoblot using a LHCII-specific antiserum and Coomassie blue-stained SDS/PAGE gel. Protein samples were taken from cells grown under photoautotrophic dim light conditions (HSM medium; 40 μmol m−2 s−1). (C) Results from three independent LHCII immunoblot analyses after phototrophic growth (HSM medium; 40 μmol m−2 s−1) using samples of the control strain and the cysteine mutants. Signal intensities were quantified by densitometry and used to calculate the mean values represented by black bars. The amount of LHCII protein in the control strain was set to 100%. Standard errors are indicated by error bars.
Cys:Ser Replacement in NAB1 Prevents the Deactivation of LHCBM Translation Repression.
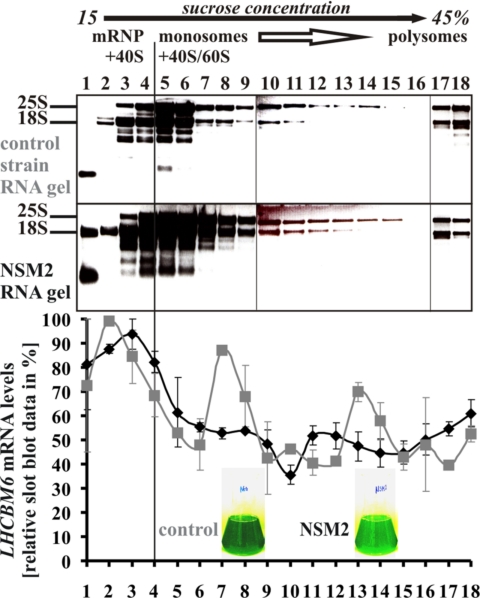
Polysome analyses were performed to investigate whether the observed reduced LHCII protein expression was caused by altered LHCBM6 mRNA translation efficiency (Fig. 4). Sucrose gradient fractionation of cytosolic extracts was performed to separate nontranslated subpolysomal mRNPs, monosomal and polysomal complexes (4). According to our previous findings regarding the prominent role of Cys-226 for NAB1 regulation we selected the Cys226Ser mutation strain NSM2 for this experiment. The results presented in Fig. 4 demonstrated that the distribution of LHCBM6 mRNA within the sucrose gradient of NSM2 was considerably different compared with the control strain. NSM2 displayed a high LHCBM6 content exclusively in subpolysomal nontranslated RNA fractions (Fig. 4, fractions 1–4) and only low amounts in monosomal/polysomal fractions (Fig. 4, fractions 5 and greater) whereas the control strain showed high amounts of LHCBM6 mRNA also in efficiently translated polysomal fractions. This result strongly indicated that the Cys-226 mutation causes an increased LHCBM6 mRNA sequestration, which is in good accordance to the observed reduced HA-LHCBM6 and LHCII expression levels in vivo (Figs. 2C and 3 B and C).
Fig. 4.
Polysome analysis of control strain and NSM2 cells grown phototrophically under dim light conditions. Cytosolic extracts were centrifuged through a 15–45% continuous sucrose gradient to separate subpolysomal mRNPs, monosomes and polysomes. RNA was extracted from 18 gradient fractions and analyzed by formaldehyde-agarose gel electrophoresis and ethidium bromide staining. The amount of LHCBM6 and β-ACTIN mRNA in each fraction was assessed by Northern slot-blot analysis. Slot-blot signals of LHCBM6 were quantified by densitometrical scanning and normalized to the corresponding ß-ACTIN signal. The strongest LHCBM6 blot signal obtained for each strain was set to 100%. Standard errors are based on three independent polysome fractionations.
Cys:Ser Mutation Results in a Higher Stability of NAB1-LHCBM6 Complexes Under Oxidative Stress Conditions in Vivo.
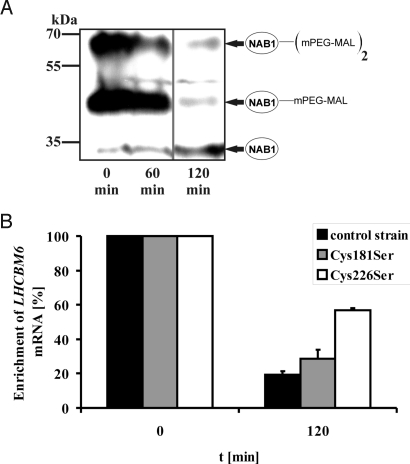
Characterization of the cysteine mutants strongly indicated that mutation of RRM cysteines perturbs the in vivo deactivation mechanism of NAB1. Consequently, we investigated whether oxidation of NAB1 changes its binding efficiency toward the target mRNA LHCBM6 in vivo by RNA coimmunoprecipitation (4). Control strain, NSM1 (Cys181Ser), and NSM2 (Cys226Ser) cultures were grown and cysteine oxidation of NAB1 was induced by the addition of diamide. To follow the in vivo thiol state of both NAB1 cysteines we applied the thiol alkylating compound mPEG-MAL. mPEG-MAL exclusively reacts with the SH-group of free cysteines and enables to determine the number of free cysteines present in a protein at a certain time. Because protein modification by mPEG-MAL results in a large shift in the electrophoretic mobility, the number of free cysteines is directly correlated to the apparent molecular weight and can be traced via immunodetection (12). Diamide addition resulted in a strong shift from the reduced to the oxidized NAB1 thiol state (Fig. 5A). After 120 min of incubation the bulk of NAB1 was shifted to a fully oxidized state, containing no reduced cysteines. This time point was chosen to analyze the RNA-binding activity of fully oxidized NAB1 compared with the reduced NAB1 thiol states found in the cell under normal, stress-free conditions. Cysteine oxidation resulted in a strong reduction of RNA-binding affinity in the control strain to 19 ± 2% and in the Cys-181 mutant NSM1 (28 ± 5%) compared with the reduced state (Fig. 5B). In the case of the Cys-226 mutant NSM2 however, more than half of the binding activity remained (57 ± 1%) demonstrating a higher resistance of NAB1Cys226Ser toward oxidative deactivation (Fig. 5B). Importantly, the amount of NAB1 was not significantly affected by the diamide treatment in all examined mutants and the control strain (Fig. S3). The results demonstrated that the oxidation of NAB1 cysteines in vivo is accompanied by a decreased binding toward the LHCBM6 target mRNA. The exchange of Cys-226 with serine strongly attenuates oxidative NAB1 deactivation, confirming that Cys-226 has a key function in the redox dependent activation of NAB1.
Fig. 5.
Effects of cysteine oxidation on the RNA-binding capacity of Wt-NAB1 (control strain), NAB1Cys181Ser, and NAB1Cys226Ser analyzed in vivo. (A) Examination of the Wt-NAB1 thiol state after 60 and 120 min after diamide addition (2 mM) to a liquid cell culture. The degree of cysteine modification was assessed by mPEG-MAL-labeling and subsequent anti-NAB1 immunoblot detection. (B) Coimmunoprecipitation of LHCBM6-mRNA using a NAB1-specific antiserum before and after diamide-induced oxidation of Wt -NAB1 (control strain), NSM1 (Cys181Ser) and NSM2 (Cys226Ser). The amount of coprecipitated LHCBM6-mRNA was quantified by RT-Q-PCR and the t0-value was set to 100% for each strain. Error bars indicate the standard error of four independent RT-Q-PCR measurements.
Discussion
The aim of the present study was to evaluate the possible relevance of 2 cysteine residues located in the RRM domain of NAB1 as central elements of an in vivo redox control mechanism, which determines its translation repressor activity. Preliminary studies carried out in vitro gave initial indications for the importance of the cysteines in controlling NAB1 activity, which were subsequently confirmed by intense in vivo studies. In vitro studies indicated that full RNA-binding activity of NAB1 requires cysteines in their SH-states (Fig. 1B) and that cysteine modification by either glutathionylation or alkylation inhibits the specific RNA-binding activity of NAB1 (Fig. 1 B and C). Peptide mapping analyses of oxidized and reduced protein samples of Wt-NAB1 and a Cys226Ser mutant did not reveal the existence of disulfide linked peptides, challenging the importance of intramolecular disulfide formation for NAB1 redox control (Fig. S2). Replacement of both cysteines with serine yielded in a distinct phenotype characterized by a perturbed expression of LHCII proteins thus proving that the cysteines are crucial for NAB1 regulation in vivo (Figs. 2 and 3). These findings strongly indicated that NAB1 was arrested in a permanently active repressor state after both cysteines were replaced by serine. Further in vivo analyses, however, demonstrated that the single cysteine mutants NAB1Cys181Ser and NAB1Cys226Ser displayed clear different phenotypical characteristics (Figs. 2, 3C, and 5). The phenotypes of NAB1Cys-181/226Ser double mutants and the NAB1Cys226Ser single mutants were very similar, which makes it feasible to suggest that modification of Cys-226, if compared with modification of Cys-181, has a larger impact on the activity state of NAB1 in vivo (Figs. 2, 3C, and 5). It should be noted, however, that oxidative treatment of a mutant recombinant NAB1Cys226Ser protein caused a significant decrease in its in vitro LHCBM6 RNA binding activity. This clearly demonstrates that Cys-181 is indeed involved in the deactivation of NAB1, although the phenotype of the corresponding Cys181Ser mutant cell lines is comparably milder than those of the Cys226Ser mutation. Finally polysome analyses together with mRNA-Coimmunoprecipitation studies (Figs. 4 and 5) fully demonstrated that in vivo deactivation of LHCBM6 mRNA sequestration and accordingly translation repression depends on cysteine modification of NAB1. As a final conclusion from the sum of our in vivo results the translation repressor activity of NAB1 is determined by the thiol state of 2 cysteines located in the RRM domain. Oxidized cysteines represent the off state of the repressor, whereas reduced cysteines represent the on state. It has already been shown that RRM containing proteins from plant organisms involved in translational regulation of photosynthetic genes can be activity-regulated via cysteine modification (13). However, these proteins were shown to be located in the plastid. NAB1 represents a eukaryotic example of a cytosolic RRM protein being subject to cysteine-based redox control. Apart from C. reinhardtii, NAB1 analogous proteins containing a combination of CSD and RRM domains were only identified in the genomes of closely related algal species C. incerta (14) and Volvox carteri (15). The position of both cysteines is conserved in all 3 genome sequences indicating that the mechanism of redox regulation is conserved at least within the Volvocales taxonomic group of green algae. The cysteine residues of cytosolic proteins are maintained in the reduced thiol state by action of thiol-based redox buffer systems (glutathione/glutaredoxins; thioredoxins/thioredoxin reductase). The total concentration of glutathione and the ratio of reduced to oxidized glutathione defines the cytosolic redox-state and undergoes considerable changes in response to a variety of environmental stresses (16). Disulfide bridge formation in proteins frequently tracks the oxidation state of the glutathione redox buffer (16). NAB1 forms mixed disulfides with glutathione under in vitro conditions, which in turn reduces its RNA-binding activity (Fig. 1 B and C). However, future experiments have to clarify whether glutahionylation of NAB1 occurs in vivo.
NAB1 fine-tunes the translation efficiency of plastid-targeted LHCII proteins and therefore the capacity of light-harvesting and rates of photosynthesis in the chloroplast of C. reinhardtii cells. Under conditions where the size and composition of the LHCII complex is not properly adjusted to the prevailing external situation, the increased/decreased need for LHCII protein synthesis has to be sensed by the translation repressor NAB1 through changes in the cytosolic redox-state.
Currently the knowledge of the interplay between the plastidic redox-state, which is to a large extent determined by photosynthetic electron transport activity, and the cytosolic redox-state is limited (17). Accordingly we currently cannot depict the complete retrograde signaling pathway of NAB1 redox-regulation. However, the finding that NAB1 is regulated via reversible thiol modification, and thus the cytosolic redox environment provides important insights into the mechanisms of redox-controlled translation regulation in the cytosol of photosynthetic organisms. Redox regulation of photosynthetic gene translation in the cytosol of plant cells was reported before (18, 19), but the molecular basis and the involvement of transcript-specific RNA-binding proteins remained to be elucidated. Because the active form of NAB1 contains cysteines in the reduced thiol state, NAB1 activation is linked to reducing conditions in the cytosol, whereas its deactivation is accompanied by shifts toward the more oxidized state. Under normal, stress-free conditions the cytosol of eukaryotic cells is in a highly reduced redox state (16). A key factor, required to maintain this reduced environment, is NADPH. Major sources of NADPH supply in the cytosol of plant organisms are the glucose consuming oxidative pentose phosphate cycle (17) and NAD(P)H exporting shuttle systems in the chloroplast envelope membrane (17, 20, 21), which are reliant on photosynthetic activity in the plastid. In our current working model, physiological conditions characterized by a sufficient provision of these photosynthates are connected to an active state of NAB1 and hence effective translation repression of LHCII transcripts. Within this model, a reduced photosynthetic performance caused by limited light supply in combination with a small antenna system oxidizes the cytosolic redox system. This in turn deactivates NAB1, thereby stimulating LHCII protein synthesis and facilitating an increase of the photosynthetic performance. For green alga, nuclear transcription activity of LHCII genes was shown to be regulated by a retrograde redox-signaling pathway emanating from the plastidic plastoquinon pool (3). With the identification of NAB1 (4) and with our recent findings, an additional mode of redox-regulated LHCII gene expression control was discovered that involves translation repression in the cytosol. Future studies targeted on the correlation between photosynthetic activity and the redox state of NAB1 could make important contributions to the understanding of retrograde signaling pathways in the context of photoacclimation processes.
Methods
Strains and Culture Conditions.
Liquid cultures of C. reinhardtii were either cultivated mixotrophically in TAP or phototrophically in HSM medium using low-light conditions of 40 μmol m−2 s−1 continuous white-light. Cultures growing in HSM medium were bubbled with 2% CO2. For details, see SI Materials and Methods.
Site-Specific Mutagenesis of NAB1 and Transformation.
Plasmid pGDNG1 was constructed by inserting the Wt NAB1 gene into the NdeI and EcoRI cloning sites of plasmid pGenD (13). The plasmids pGDNG1/Cys(181/226Ser), pGDNG1/Cys(181Ser), and pGDNG1/Cys(226Ser) were generated by site-specific replacement of thymine by adenine at positions 541 and 676 of the NAB1 gene in plasmid pGDNG1 (QuikChange Site-Directed Mutagenesis Kit; Stratagene. For primer details, see Table S1). These vectors were used to cotransform the NAB1-deficient cell line Stm3-HA-LHCBM6 (see SI Materials and Methods).
Coimmunoprecipitation (Co-IP) of NAB1 Targets.
Co-IPs were performed with liquid TAP cultures of control strain, strain NSM1, and strain NSM2 before and after a 2-h treatment with 2 mM diamide. RT-Q-PCR was applied to quantify the amounts of coprecipitated LHCBM6 and ß-ACTIN mRNA. ß-ACTIN served as a reference gene. For a detailed description of the procedure, see SI Materials and Methods.
Overexpression of Recombinant NAB1.
Recombinant NAB1 was purified under native conditions according to the QIAexpressionist manual (Qiagen). Reducing conditions during the purification process were maintained by addition of 5 mM β-mercaptoethanol to binding and wash buffer. Purified protein samples were supplemented with 10 mM DTT directly after elution.
NAB1-RNA-Binding Studies.
Recombinant NAB1 was subjected to oxidative treatment with 5 mM GSSG, alkylated with a 50-fold excess of NEM (N-ethylmaleimide) or a 5-fold excess of 4-vinylpyridine in respect to the sulfhydryls to blocked. Protein samples were then subjected to RNA-binding studies applying RNA probes derived from the C. reinhardtii LHCBM6 and psbD genes. The probe derived from the gene LHCBM6 was radioactively labeled, whereas the psbD probe was unlabeled and served as a competitor. For details, see SI Materials and Methods.
Subpolysome and Polysome Complex Fractionation.
Polysomes were fractionated as described before (5) and RNA was extracted from all 18 sucrose gradient fractions and analyzed in an agarose-formaldehyde denaturing gel. The RNA was slot-blotted on a positively charged nylon membrane (Hybond N+, Amersham) and hybridized with a digoxigenin-labeled LHCBM6- or ß-ACTIN-specific DNA probe. Signal intensity was quantified by densitometry and the LHCBM6 signal of each fraction was normalized to the corresponding ß-ACTIN signal. For experimental details, see SI Materials and Methods.
Gel Electrophoresis and Immunoblotting.
Proteins were separated by Tris-tricine or Tris-glycine-SDS/PAGE and detected by immunoblotting using enhanced chemiluminescence (ECL, Amersham). The NAB1-specific antiserum was obtained as already described (5) and anti-LHCII was provided by S. Jansson (Umeå, Sweden). HA-tagged proteins were detected with a HA-specific antibody (Roche Applied Science). NAB1-glutathione adducts were detected with a mouse monoclonal antibody directed against glutathione (101-A-250, Virogen). For a description of the procedure used for the detection of glutathionylated cysteines in recombinant NAB1 and the mPEG-MAL labeling procedure, see SI Materials and Methods.
Supplementary Material
Acknowledgments.
We are grateful to J. Beckmann (University of Bielefeld) and C. Claus [Department of Clinical Research (DCR), University of Bern), Bern, Switzerland] for help with transformation and cloning. This work was supported by Deutsche Forschungsgemeinschaft Grant FOR387 and KR1585–5/1 (to O.K.) and by NI390–4/1 (to J.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900670106/DCSupplemental.
References
- 1.Walters RG. Towards an understanding of photosynthetic acclimation. J Exp Bot. 2005;56:435–447. doi: 10.1093/jxb/eri060. [DOI] [PubMed] [Google Scholar]
- 2.Pursiheimo S, Mulo P, Rintamaki E, Aro EM. Coregulation of light-harvesting complex II phosphorylation and lhcb mRNA accumulation in winter rye. Plant J. 2001;26:317–327. doi: 10.1046/j.1365-313x.2001.01033.x. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y-B, Durnford DG, Koblizek M, Falkowski PG. Plastid regulation of Lhcb1 transcription in the Chlorophyte Alga Dunaliella tertiolecta. Plant Physiol. 2004;136:3737–3750. doi: 10.1104/pp.104.038919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mussgnug JH, et al. Nab1, an RNA-binding protein involved in the light-regulated differential expression of the light-harvesting antenna of Chlamydomonas reinhardtii. Plant Cell. 2005;17:3409–3421. doi: 10.1105/tpc.105.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKim SM, Durnford DG. Translational regulation of light-harvesting complex expression during photoacclimation to high-light in. Chlamydomonas reinhardtii. Plant Physiol Biochem. 2006;44:857–865. doi: 10.1016/j.plaphy.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 6.Koussevitzky S, et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- 7.Bouvet P, Matsumoto K, Wolffe AP. Sequence-specific RNA recognition by the Xenopus Y-box proteins. J Biol Chem. 1995;270:28297–28303. doi: 10.1074/jbc.270.47.28297. [DOI] [PubMed] [Google Scholar]
- 8.Stauber EJ, et al. Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryotic Cell. 2003;2:978–994. doi: 10.1128/EC.2.5.978-994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turkina MV, et al. Environmentally modulated phosphoproteome of photosysynthetic membranes in the green alga Chlamydomonas reinhardtii. Mol Cell Proteomics. 2006;5:1412–1425. doi: 10.1074/mcp.M600066-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Buchanan BB, Balmer Y. Redox regulation: A broadening horizon. Annu Rev Plant Biol. 2005;56:187–220. doi: 10.1146/annurev.arplant.56.032604.144246. [DOI] [PubMed] [Google Scholar]
- 11.Fischer N, Rochaix J-D. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol Genet Genomics. 2001;265:888–894. doi: 10.1007/s004380100485. [DOI] [PubMed] [Google Scholar]
- 12.Katzen F, Beckwith J. Role and location of the unusual redox-active cysteines in the hydrophobic domain of the transmembrane electron transporter DsbD. Proc Natl Acad Sci USA. 2003;100:10471–10476. doi: 10.1073/pnas.1334136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alergand T, Peled-Zehavi H, Katz Y, Danon A. The chloroplast protein disulfide isomerase RB60 reacts with a regulatory disulfide of the RNA-binding protein RB47. Plant Cell Physiol. 2006;47:540–548. doi: 10.1093/pcp/pcj023. [DOI] [PubMed] [Google Scholar]
- 14.Popescu CE, Borza T, Bielawski JP, Lee RW. Evolutionary rates and expression level in Chlamydomonas. Genetics. 2006;172:1567–1576. doi: 10.1534/genetics.105.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nematollahi G, Kianianmomeni A, Hallmann A. A Quantitative analysis of cell-type specific gene expression in the green alga. Volvox carteri BMC Genomics. 2006;7:321. doi: 10.1186/1471-2164-7-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. J Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 17.Foyer CH, Noctor G. Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid Redox Signal. 2009;11:1–45. doi: 10.1089/ars.2008.2177. [DOI] [PubMed] [Google Scholar]
- 18.Petracek ME, Dickey LF, Huber SC, Thompson WF. Light-regulated changes in abundance and polyribosome association of ferredoxin mRNA are dependent on photosynthesis. Plant Cell. 1997;9:2291–2300. doi: 10.1105/tpc.9.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherameti I, et al. Polyribosome loading of spinach mRNAs for photosystem I subunits is controlled by photosynthetic electron transport. Plant J. 2002;32:631–639. doi: 10.1046/j.1365-313x.2002.01452.x. [DOI] [PubMed] [Google Scholar]
- 20.Kelly GJ, Gibbs M. Non-reversible D-glyceraldehyde 3-phosphate dehydrogenase of plant tissues. Plant Physiol. 1973;52:111–118. doi: 10.1104/pp.52.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner WT, Waller JC, Snedden WA. Identification, molecular cloning and functional characterization of a novel NADH kinase from Arabidopsis thaliana (thale cress) Biochem J. 2005;385:217–223. doi: 10.1042/BJ20040292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.