Abstract
Mammalian Sonic hedgehog (Shh) signaling is essential for embryonic development and stem cell maintenance and has critical roles in tumorigenesis. Although core components of the Shh pathway are conserved in evolution, important aspects of mammalian Shh signaling are not shared with the Drosophila pathway. Perhaps the most dramatic difference between the Drosophila and mammalian pathways is that Shh signaling in the mouse requires a microtubule-based organelle, the primary cilium. Proteins that are required for the response to Shh are enriched in the cilium, but it is not clear why the cilium provides an appropriate venue for signal transduction. Here, we demonstrate that Kif7, a mammalian homologue of Drosophila Costal2 (Cos2), is a cilia-associated protein that regulates signaling from the membrane protein Smoothened (Smo) to Gli transcription factors. By using a Kif7 mutant allele identified in a reporter-based genetic screen, we show that, similar to Drosophila and zebrafish Cos2, mouse Kif7 acts downstream of Smo and upstream of Gli2 and has both negative and positive roles in Shh signal transduction. Mouse Kif7 activity depends on the presence of cilia and Kif7-eGFP localizes to base of the primary cilium in the absence of Shh. Activation of the Shh pathway promotes trafficking of Kif7-eGFP from the base to the tip of the cilium, and localization to the tip of the cilium is disrupted in a motor domain mutant. We conclude that Kif7 is a core regulator of Shh signaling that may also act as a ciliary motor.
Keywords: Gli2, intraflagellar transport, smoothened, neural tube, ENU
Although core components of the Hedgehog (Hh) pathway are conserved in evolution, important aspects of the pathway appear to diverge among animals. In particular, in mammals it is not clear how the membrane protein Smoothened (Smo) controls the activity of the Gli transcription factors that implement the pathway (1). In Drosophila, activation of the Hh pathway leads to formation of a protein complex that includes Costal2 (Cos2) on the cytoplasmic C-terminal tail of Smo, but vertebrate Smo lacks the major binding site for Cos2 that is crucial for complex formation (2). Kif7, a vertebrate homologue of Cos2, is important for Hh signaling in zebrafish (3), but cell-based assays have argued that Kif7 does not act in the mammalian pathway (2). Fused, a serine/threonine kinase that binds Drosophila Cos2, is important for Hh signaling in zebrafish (4), but the single mouse Fused homologue is not essential for Shh signaling and instead is important for the formation of motile cilia (5–7). Suppressor of fused (Sufu) plays a minor role in the Drosophila pathway that is only detected in the absence of Fused, but is an essential regulator of the pathway in zebrafish (4, 8), and is a very strong negative regulator of Shh signaling in the mouse (9, 10).
The most surprising difference between the Drosophila and mammalian pathways is that signaling from Smo to the Gli transcription factors in the mouse requires the primary cilium. Genetic studies have shown that a variety of proteins required for ciliogenesis, including the intraflagellar transport (IFT) machinery and basal body proteins that promote cilia formation and maintenance, are essential for all responses to mammalian Hedgehog (Hh) ligands in both early embryos and all other cell types that have been tested (11–14). In contrast, IFT proteins are not required for Hh signaling in Drosophila, and the role of cilia in the zebrafish pathway is controversial (15, 16). In the mouse, it appears that all of the core pathway proteins required for the response to Shh are associated with the cilium. Patched1 (Ptch1), the Shh receptor, is localized in cilia in the absence of ligand and moves out of cilia after exposure to ligand (17). In a similar time course, the membrane protein Smo moves into cilia in response to ligand, where it activates downstream signaling events (17, 18). Trafficking of Smo and Patched between vesicular and plasma membrane compartments in response to ligand also takes place in Drosophila, but not to any defined plasma membrane compartment (19), and Hedgehog-responding cells are not ciliated in Drosophila. In the mouse, the Gli transcription factors that implement Shh signals and the negative regulator Sufu are localized to cilia tips both in the presence and absence of ligand (20). Despite the clear connection between cilia and mammalian Hh signaling, it is not yet clear why the cilium is the site of Hh signal transduction.
One proposed explanation for these differences between Drosophila and mammals is that the IFT machinery that builds cilia substitutes for the function of Drosophila Cos2, a kinesin-related protein (1, 21, 22). Consistent with this hypothesis, experiments in mammalian cells indicated that the mouse proteins most similar to Cos2, Kif7 and Kif27, do not play a role in mammalian Hh signaling (2). Here, we show that, in contrast to this hypothesis, Kif7 is essential for mouse Shh signaling and that Kif7 protein is associated with cilia and may act as a ciliary motor. Thus, Kif7 and ciliary trafficking act in concert in mammalian Shh signaling; these findings provide perspectives on the connection between cilia and Hedgehog signaling.
Results
Mutation in Kif7 Expands Ventral Fates in the Mouse Neural Tube.
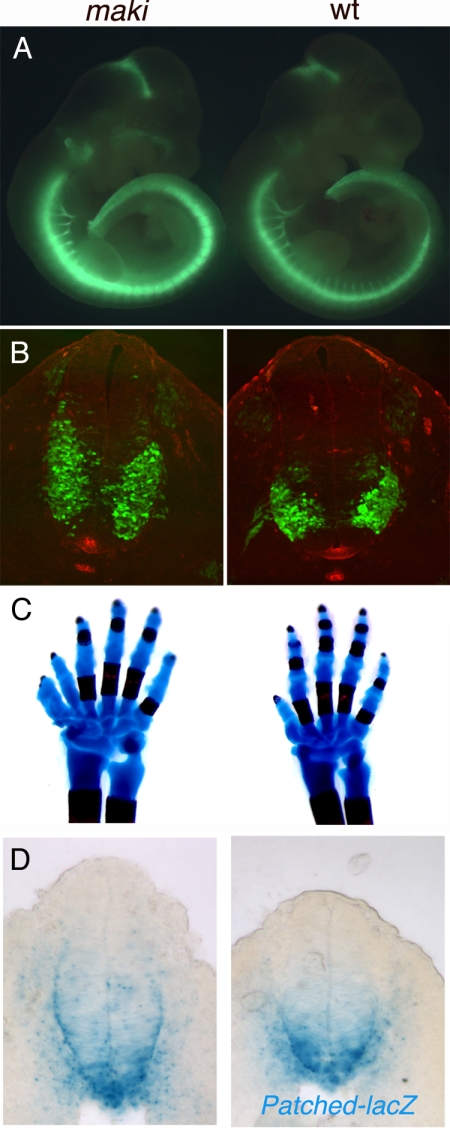
We carried out a genetic screen to identify new recessive N-ethyl-N-nitrosourea (ENU)-induced mutations that affect mammalian Shh signaling. Among its many functions in mammalian development, the role of Shh in the specification of ventral cell types in the developing neural tube is particularly well understood. For example, spinal motor neurons are induced in response to an intermediate level of Shh, and the number and position of motor neurons within the ventral neural tube is a sensitive indicator of Shh activity (23). We used an HB9-GFP transgenic reporter mouse that expresses GFP in motor neurons (24) (Fig. 1A and Fig. S1A) to identify mutations that altered Shh signaling. An interesting mutation, matariki (maki), was identified that caused an expanded motor neuron domain in the embryonic day 10.5 (E10.5) neural tube (Fig. 1A). Analysis of cross-sections confirmed that motor neurons were increased in number and expanded dorsally (Fig. 1B). The floor plate, which is specified by the highest level of Shh activity, appeared normal in the mutant embryos (Fig. 1B and Fig. S2). Nkx2.2 is expressed in the V3 interneuron progenitors that are specified by high levels of Shh signaling adjacent to the floor plate. The Nkx2.2 domain was expanded dorsally in maki embryos (Fig. 2 A), and markers of cell types that are specified by lower levels of Shh activity also shifted dorsally (Fig. S2). This neural patterning phenotype is consistent with a moderate elevation of Shh pathway activity. maki mutants died at the end of gestation, when other phenotypes characteristic of elevated Shh signaling were apparent, including preaxial polydactyly (Fig. 1C). To assess whether maki affected the Shh pathway directly, we analyzed the expression of a direct target gene of the Shh pathway, Ptch1. Ptch1-lacZ (25) was expressed in a modestly expanded domain in the maki neural tube, which confirmed that the mutation increased the activity of the Shh pathway (Fig. 1D).
Fig. 1.
The maki phenotype. (A) The expression of HB9-eGFP is stronger in maki than in wild-type E10.5 embryos. (B) Cross-sections through the brachial region of E10.5 embryos. HB9-eGFP is green; Shh protein expression is red. The domain of HB9-eGFP is expanded dorsally in maki embryos, whereas the domain of Shh expression in the floor plate is the same in maki and wild type. Dorsal, up. (C) Skeletal preparations of postnatal day 1 forelimbs show that maki embryos have preaxial polydactyly; this defect is completely penetrant. (D) Sections through the brachial neural tube of E9.5 embryos carrying a Ptch1-lacZ transgene, stained for β-galactosidase activity.
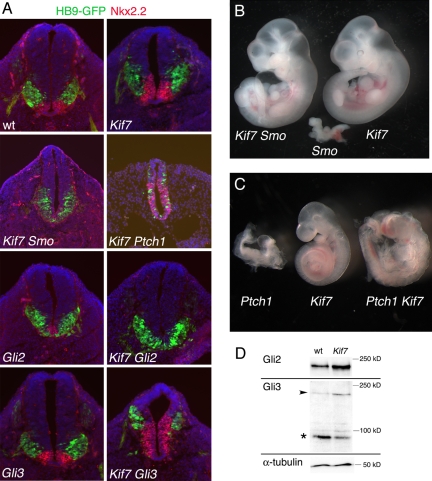
Fig. 2.
Kif7 acts in the Shh pathway. (A) Expression of markers of motor neurons (expressing HB9-GFP; green) and V3 progenitors (marked by Nkx2.2; red) in single- and double-mutant neural tubes of E10.5 embryos. DAPI is blue. All sections are at the thoracic level; dorsal is up. Numbers of double-mutant embryos examined were as follows: 2 Ptch1 Kif7maki; 1 Smobnb Kif7maki; 4 Gli2 Kif7maki; 1 Gli3 Kif7maki. (B) The Kif7maki mutation rescues the early lethality of Smobnb. E10.5 embryos, from Left to Right, are as follows: Smobnb Kif7maki; Smobnb; Kif7maki. Smobnb Kif7maki double mutants resemble Kif7maki single mutants except that they show pericardial edema and an abnormal shape of the cranial neural tube. (C) The Kif7maki mutation rescues the early lethality and failure to turn of Ptch1. E9.5 embryos, from Left to Right, are as follows: Ptch1; Kif7maki; Ptch1 Kif7maki. Ptch1 Kif7maki double mutants complete embryonic turning and generate similar numbers of somites as the Kif7maki littermates, but have an open cranial neural tube. Heterozygosity for Kif7maki did not cause detectable dominant modification of any of these Shh pathway mutants. (D) Gli proteins in E10.5 whole-embryo extracts, analyzed by Western blot analysis. The arrowhead indicates full-length Gli3; the asterisk (*) indicates Gli3-repressor. In this experiment, the Kif7maki extract has 15% more Gli2 protein and a 3-fold lower ratio of Gli3-repressor/Gli3-full length than the wild-type control. The same trends were seen in other experiments.
We used meiotic recombination mapping to localize the gene responsible for the maki phenotype to a small interval on chromosome 7 and found that the maki phenotype was associated with a missense mutation (L130P) in a conserved region of the motor domain of Kif7 (Fig. S1 B and C) (see Experimental Procedures), which encodes a kinesin homologous to Drosophila Cos2. Because the motor domain of Drosophila Cos2 is required for its microtubule-dependent motility and point mutations in the Cos2 motor domain block its ability to repress Hh target gene expression (26, 27), the L130P mutation is likely to cause loss of Kif7 function. Both maki mutants and zebrafish Kif7 morphants show mild ectopic activation of Shh pathway targets (3), supporting the conclusion that the maki phenotype is caused by loss of Kif7 activity. Thus, contrary to a previous report (2), Kif7/Cos2 has a conserved role in mammalian Hh signaling.
Kif7 Acts as Both a Negative and Positive Regulator of Shh Signal Transduction.
Drosophila Cos2 is required to relay the signal from the transmembrane protein Smo to the Ci transcription factor. Cos2 acts as part of a protein complex that includes Fused and binds to the C-terminal tail of Smo (28). Because Shh signaling is not disrupted in mouse Fused mutants (5, 6) and the major Cos2 binding domain is not present in vertebrate Smo (2), it was important to define the step in the Shh pathway controlled by mouse Kif7.
Smo is required for embryos to survive past ≈E9.0 and for specification of all Shh-dependent ventral neural cell types (29). Smo Kif7maki double mutant embryos continued to develop until E10.5 (Fig. 2B) and generated ventral neural cell types, including motor neurons and some Nkx2.2+ cells (Fig. 2A). Thus, loss of Kif7 activated the pathway in the absence of Smo, indicating that Kif7 causes ligand-independent activation of the Shh pathway at a step downstream of Smo. Consistent with the hypothesis that Kif7 acts at a step downstream of Smo, we found that, as in wild type, Smo protein moves into cilia of Kif7maki mouse embryo fibroblasts in response to Shh.
The Gli2 and Gli3 transcription factors implement the responses to Shh in the neural tube (30–32). Gli2 is the principal transcriptional activator of Shh target genes. Gli2 mutants lack the most ventral cell type in the neural tube, the floor plate, and have a reduced number of Nkx2.2+ V3 progenitors (Fig. 2A) (33). In Gli2 Kif7maki double mutants, the motor neuron domain was not expanded dorsally and the floor plate and Nkx2.2+ domains were absent (Fig. 2A), indicating that the dorsal expansion of the motor neuron and Nkx2.2 domains seen in Kif7maki was due to increased activity of Gli2. The steady-state level of Gli2 protein was higher in E10.5 Kif7maki embryo extracts than in wild type (Fig. 2D), which may account, in part, for the elevated Gli2 activity in the mutants.
In wild-type embryos, Gli3 is proteolytically processed to a transcriptional repressor that keeps pathway targets off in the absence of ligand. Although Gli3 single mutants do not have defects in the pattern of motor neurons or V3 progenitors (34) (Fig. 2A), Gli3 Kif7maki double mutants showed a greater expansion of these ventral neural cell types than Kif7maki single mutants (Fig. 2A), indicating the Gli3 and Kif7 have independent roles in preventing ligand-independent activation of the pathway. We found, however, that the amount of processed Gli3 protein and the ratio of processed/full-length Gli3 were decreased in Kif7maki mutant extracts (Fig. 2D), suggesting that a decrease in the amount of transcriptional repressor could contribute to the expansion of ventral neural fates in Kif7maki embryos.
The analysis described thus far shows that mammalian Kif7, as in Drosophila and zebrafish (1, 3), defines a step in the pathway between Smo and the Gli proteins. The expansion of ventral neural cell types in Kif7maki mutants is much less pronounced than that seen in mutants that lack Patched1 or Sufu, core regulators of the pathway, or that lack IFT complex A proteins (8, 9, 25, 35, 36). The moderate activation of the Shh pathway could indicate that Kif7 has a relatively minor role in keeping the pathway off in the absence of ligand. Alternatively, like Drosophila Cos2, Kif7 could have both negative and positive roles in the pathway (37). Evidence that Kif7 is required for activation of the pathway came from the analysis of double mutants with Ptch1, the Shh receptor. In the absence of Ptch1, the pathway is constitutively activated. Ptch1 mutants arrest at ≈E9.0 and express the ventral markers with Nkx2.2 and FoxA2 throughout the neuroepithelium (9, 25). Ptch1 Kif7maki double mutants showed a milder phenotype and developed to E10 (Fig. 2C). In contrast to the complete ventralization of the Ptch1 neural plate, expression of Nkx2.2 and FoxA2 was restricted to the ventral three-fourths of the neural plate and Isl1+ motor neurons were scattered throughout the Ptch1 Kif7maki neuroepithelium (Fig. 2A). These findings suggest that, in addition to its role in negative regulation, Kif7 can promote activity of the Shh pathway.
Kif7 Activity Depends on IFT.
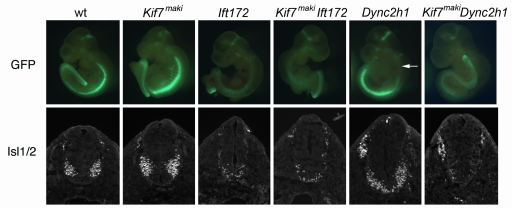
Genetic and cell biological experiments have shown that cilia are required for mouse Shh signaling and that proteins required for Shh signal transduction are associated with cilia (13, 17, 18, 20). Proteins required for ciliogenesis, including the Kinesin-II anterograde IFT motor and IFT complex B proteins, are required for Shh signal transduction at a step downstream of the membrane protein Smo and upstream of the Gli transcription factors (11, 13, 22), the same step of the pathway regulated by Kif7. Unlike mutants that lack Kinesin-II subunits (38–40), which have no cilia, Kif7maki cilia have wild-type morphology. To determine the relationship between Kif7 and cilia, we genetically ablated cilia in Kif7maki mutants. Loss of the IFT B complex protein IFT172 blocks cilia formation and prevents all responses to Shh (11). Ift172wim Kif7maki double mutants were indistinguishable from Ift172wim mutants, with few motor neurons, no Nkx2.2+ cells, and no floor plate (Fig. 3). Thus, the activity of Kif7 depends on the presence of cilia.
Fig. 3.
Kif7 activity depends on cilia. The distribution of motor neurons, marked by expression of HB9-GFP in whole E9.5 embryos and Isl1/2 in sections of the thoracic neural tube, in IFT single and double mutants. Ift172wim single mutants and Ift172wim Kif7maki double mutants both have a few scattered motor neurons in the ventral spinal cord. The Dync2h1mmi mutation was identified based on the loss of HB9-GFP expression in the cervical spinal cord (arrow). Dync2h1mmi Kif7maki double mutants have fewer motor neurons than Dync2h1mmi single mutants. Nkx2.2 was not expressed in Ift172wim, Kif7maki Ift172wim, or Kif7maki Dync2h1mmi double mutants. Numbers of double-mutant embryos examined were as follows: 2 Kif7maki Ift172wim; 2 Kif7maki Dync2h1mmi. No phenotypic variation was noted among embryos of the same genotype.
To probe the relationship between the Kif7 kinesin and trafficking within the cilium, we analyzed embryos that were homozygous for Kif7maki and also lacked normal retrograde trafficking within the cilium. In the HB9-GFP screen, we identified a mutant allele of the gene encoding the heavy chain of the dynein motor (Dync2h1) that powers retrograde transport within cilia (Fig. 3). The allele mei mei (mmi) carried a missense mutation (I1789N) in the motor domain. Like other Dync2h1 alleles (13, 41), Dync2h1mmi mutants lack motor neurons in the cervical spinal cord but have motor neurons that span the ventral midline in more caudal regions of neural tube. Like Ift172 Kif7maki embryos, Dync2h1mmi Kif7maki double mutants lacked ventral neural cell types, indicating that Dync2h1 is required for activation of the pathway in the Kif7maki embryos and suggesting that Kif7 affects a trafficking event that occurs before retrograde IFT. Remarkably, the Dync2h1mmi Kif7maki double mutants showed a stronger loss of Shh signaling than Dync2h1mmi single mutants: the double mutants had only a very small number of Isl1+ motor neurons at all rostrocaudal positions of the spinal cord (Fig. 3). The greater loss of ventral neural cell types in the absence of both Kif7 and Dync2h1 suggests that these 2 genes have additive, positive roles in Shh signaling, which is consistent with the positive role for Kif7 seen in the Ptch1 Kif7maki double mutants.
Kif7 Moves from the Cilia Base to the Cilia Tip in Response to Activation of the Hh Pathway.
To define the subcellular localization of Kif7, we expressed an eGFP-tagged form of Kif7 in mammalian cell lines. Kif7 is a member of the Kinesin-4 family, with an N-terminal motor domain (42); this indicates that it should act like a conventional kinesin and carry cargo away from minus ends of microtubules, which are anchored at the base of the cilium. In MDCK cells, Kif7-eGFP was enriched at the base of the cilium (Fig. S3). In IMCD3 cells, a kidney epithelial line with long primary cilia, Kif7-eGFP was enriched at both the base and the tip of the primary cilia (Fig. S3). A Kif7-eGFP construct that carried the L130P mutation found in the maki allele encoded a protein that localized only to the base, but not the tip, of the primary cilia of IMCD3 cells (Fig. S3). Kif7 therefore associates with cilia and requires a normal motor domain to become enriched at the tips of cilia.
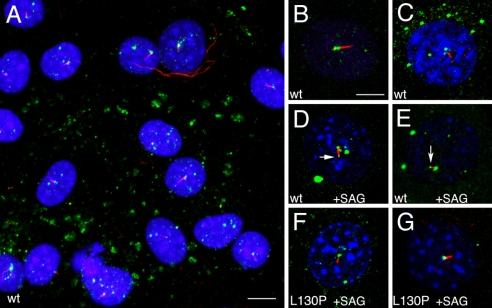
Kif7-eGFP was also enriched at the base of the cilium in NIH 3T3 cells, a Hedgehog-responsive fibroblast cell line (43) (Fig. 4 A–C). We activated the Shh pathway in these cells by treatment with Smoothened agonist (SAG) (44). When assayed 24 h after SAG addition, GFP was enriched at the cilia tips of 80% of the cells (Fig. 4 D and E). The time course of Kif7 movement to the cilia tip was comparable with that of Smo localization to cilia: Smo began to be detected in cilia at 1 h after SAG activation (17), and one-third of the Kif7-eGFP-expressing cells (4 of 12) had GFP at the tip 1 h after SAG addition. As in the IMCD3 cells, the Kif7L130P-eGFP mutant protein was detected at the base of the cilia of NIH 3T3 cells, but did not move to the cilia tips after addition of SAG (0 of 18 cells with tip GFP after SAG treatment; 16 of 18 had GFP at the cilia base). These findings indicate that activated Smo promotes the movement of Kif7 from the base of the cilia to the cilia tips and suggest that the relocalization of Kif7 to the cilia tip requires a functional motor domain.
Fig. 4.
Kif7 traffics within cilia. (A) A field of NIH 3T3 cells expressing Kif7-eGFP. In virtually all cells, Kif7-eGFP (green) is enriched at the base of the cilia, marked by expression of acetylated α-tubulin (red). Nuclei are visualized with DAPI (blue). (Scale bar: 10 μm.) (B and C) High-magnification view of Kif7-eGFP in NIH 3T3 cells not treated with SAG. Forty-nine of 50 cells examined by confocal microscopy had GFP at the base but not the tip of the cilium, and none had GFP at the cilia tip. (D and E) Kif7-eGFP at the tips of cilia (arrows) 24 h after treatment with 100 nM SAG. Base and tip GFP was seen in 20 of 25 cells analyzed, and 5 cells had GFP only at the base. (F and G) In cells expressing the mutant Kif7L130P-eGFP, GFP was detected at the base, but not the tip, of the cilium 24 h after addition of SAG. Zero of 18 cells had tip GFP after SAG treatment; 16 of 18 had GFP at the cilia base. The pattern of GFP detected was the same when detected as endogenous GFP or by using anti-GFP antibodies to detect the fusion protein. (Scale bar: B–G, 5 μm.)
Discussion
Our findings demonstrate that Kif7/Costal2 is a member of the core Shh signal transduction pathway that is conserved in Drosophila, zebrafish, and the mouse. Kif7 acts as a negative regulator of the pathway: loss of Kif7 activity causes an expansion of ventral neural cell types in the neural tube because of an expanded domain of expression of Shh target genes. The double-mutant analysis shows that Kif7 negatively regulates the pathway by preventing inappropriate activation of the Gli2 transcription factor in the absence of ligand. We also observe a decrease in the amount of Gli3 repressor in Kif7maki mutant embryos, which may contribute to activation of the pathway in the mutants. However, Gli3 repressor is present in Kif7 mutants, and the genetic analysis shows that Kif7 and Gli3 cooperate in the restriction of Shh activity and ventral fates in the neural tube.
In addition to its role as a negative regulator, genetic experiments revealed that Kif7 also promotes Shh signaling. The positive role of Kif7 was seen in 2 double mutants: the neural tube of Kif7maki Ptch1 double mutants is less strongly ventralized than that of Ptch1 single mutants, and Kif7maki Dync2h1 double mutants lack motor neurons, a stronger dorsalization than seen in Dync2h1 single mutants. The dual negative and positive roles suggest that Kif7-containing protein complexes may be reorganized in response to ligand, as is the case for Cos2 (28). For example, Kif7 may act in one complex to negatively regulate the pathway in the absence of ligand and become incorporated into a distinct, positively acting complex in response to Shh.
Our results contrast with experiments in which Kif7 knockdown or overexpression had no detectable effect on Hh signaling in NIH 3T3 cells (2). In that study, Sufu knockdown caused a clear elevation of Gli-luciferase reporter assays, but no elevation of reporter activity was observed when Kif7 activity was reduced. Because Kif7 has both negative and positive roles in the pathway, its removal in the embryo causes a net activation of the pathway that is much less than that seen in Sufu mutant embryos. It is therefore possible that the cell-based reporter assays were not sensitive enough to detect changes in pathway activity that are clear in the embryo.
Like Cos2 (27), Kif7 changes its subcellular localization in response to pathway activation: Kif7-eGFP is enriched at the base of the cilium in the absence of ligand and moves to the cilia tip when the pathway is activated. We therefore propose that the negative activity of Kif7 takes place at the base and its positive role takes place at the tip. Because zebrafish Kif7 binds Gli proteins directly (3), we suggest that, in the absence of pathway activation, Kif7 at the base of the cilium negatively regulates the pathway by targeting Gli2 away from the cilium or promoting Gli2 turnover at the basal body, where proteasomes are enriched (45). In response to activation of Smo, Kif7 is loaded onto axonemal microtubules and moves to the cilia tip. At the cilia tip, Kif7 may interact with the Gli proteins and Sufu that are enriched in that compartment (20). Cos2 is also required for responses to high levels of Drosophila Hh, and this has been attributed to antagonism of Sufu (37). Similarly, Kif7 at the cilia tip may antagonize Sufu to promote activation of Gli proteins.
Although Cos2 has been shown to have motor activity, the Cos2 motor lacks conserved motifs important for binding of ATP and microtubules and moves 4- to 10-fold more slowly than a conventional kinesin (27). It has therefore been proposed that Cos2 acts primarily as a scaffold protein for cytoplasmic signaling complexes (46, 47). In contrast, the Kif7 motor domain includes all of the sequence motifs necessary for ATP and microtubule binding, and the maki mutation demonstrates that the motor domain is essential for its function in the Shh pathway and its ability to move to the cilia tip. These findings raise the possibility that the ancestral Kif7/Costal2 required efficient motor activity and that the lineage that led to Drosophila might have lost both full motor activity of Cos2/Kif7 and the association of Hedgehog signaling with the primary cilium in parallel.
The data suggest that, once activated, Smo promotes translocation of Kif7 into the ciliary axoneme, and Kif7 may act as an anterograde IFT motor to transport itself and perhaps other cargo to the cilia tip. These results suggest that Kif7 is an essential component of the Shh pathway and may also act as an IFT motor. Such a dual function would be sufficient to explain the coupling of mammalian Shh signaling to the primary cilium.
Experimental Procedures
Mouse Strains and Phenotypic Analysis.
ENU mutagenesis and the genetic screen were carried out as described in ref. 48, except that mutagenized C57BL6/J males and F1 males were mated with females homozygous for an HB9-eGFP reporter in the FVB genetic background, made by using a previously described construct (24) (Fig. S1A). Crosses and analysis of neural patterning were performed as described in ref. 49. Mutant strains were as follows: Ptch1tm1Mps (25), Smobnb (50), Ift172wim (11), Gli2 (33), and Gli3Xt (51); genotyping was performed as described. Alcian blue and Alizarin red were used to stain cartilage and bone, as described in ref. 52. For Western blot analysis, whole E10.5 embryos were lysed in RIPA buffer (0.15 mM NaCl/0.05 mM Tris·HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) and processed using standard methods. Gli3 antibodies were the kind gift of B. Wang (Weill Cornell Medical College, New York), and Gli2 antibodies were the kind gift of J. T. Eggenschwiler (Princeton University, Princeton, NJ).
Identification of the maki Mutation.
The maki mutation was induced in a C57BL6/J chromosome and all outcrosses were to the FVB strain (Fig. S1A). We were therefore able to map the maki mutation to a segment of chromosome 7 based on the identification of regions homozygous for C57BL6/J polymorphisms in DNA from mutant embryos on genome-wide SNP panel (53). Further genetic mapping localized the gene between 2 SNPs, rs37700257 (85.4 Mb) and rs32302579 (86.95 Mb) (Ensembl build M37; www.ensembl.org). Contrary to a previous report (2), we found that Kif7 is widely expressed in the midgestation embryo, so we were able to sequence RT-PCR products amplified from E10.5 mutant and wild-type cDNAs. The Kif7maki mutation is a T-to-C missense substitution that disrupts an AluI restriction site present in the wild-type sequence, creating a restriction fragment length polymorphism (RFLP) that was used for genotyping. PCR amplification from genomic DNA (primers, 5′-GGTGGTGGTGGTGGATACTT and 5′-GCAGATCTCGGAACTCTTCCT) generated a 214-bp PCR product containing the RFLP. AluI digestion does not cleave the mutant sequence but cleaves wild type to give 132- and 82-bp fragments.
Kif7-eGFP Expression and Localization.
C-terminally tagged Kif7wt-eGFP and Kif7maki-eGFP constructs were generated by subcloning full-length Kif7 and Kif7maki cDNAs into the pEGFPN3 mammalian expression vector (Clontech). To generate NIH 3T3 cells stably expressing Kif7-eGFP or Kif7maki-eGFP, cells were transfected with Lipofectamine2000 (Invitrogen) and split 1:10 the following day. After 48 h of transfection, cells were selected in 500 μg/mL G418. Primary antibodies to mouse acetylated α-tubulin (1:1000; Sigma) and secondary antibodies tagged with Alexa Fluor 568 (Invitrogen; 1:1000) were used to detect cilia. NIH 3T3 cell culture and immunofluorescent staining were performed as described in ref. 17. To assay Kif7-eGFP localization in response to Shh pathway activation, NIH 3T3 cells were grown to confluence and serum starved with the addition of 100 nM SAG for 24 h before fixation and staining; the eGFP-tagged fusion protein was detected either by fluorescence or with mouse anti-GFP antibodies (1:1250; Invitrogen). Confocal microscopy was performed by using an upright Leica TCS SP2 AOBS laser-scanning microscope. Images were taken with a 63× objective and 2× zoom. Confocal datasets were processed by using the Volocity software package (Improvision).
Note Added in Proof.
While this article was in press, two articles (54, 55) appeared that described similar phenotypes of targeted null mutations in Kif7.
Supplementary Material
Acknowledgments.
We thank Ivo Lieberam and T. M. Jessell (Columbia University, New York), Jonathan Eggenschwiler, Baolin Wang, Alexandra Joyner (Sloan-kettering Institute, New York), and Matthew Scott (Stanford University, Stanford CA) for constructs, mouse strains, and reagents; the Memorial Sloan-Kettering Molecular Cytology and Mouse Genetics Core facilities for technical assistance; Tamara Caspary and Christine Larkins for technical advice; and Sarah Goetz for comments on the manuscript. The work was supported by grants from Project ALS and National Institutes of Health Grant NS044385. Monoclonal antibodies to neural patterning markers were from the Developmental Studies Hybridoma Bank.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906944106/DCSupplemental.
References
- 1.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varjosalo M, Li SP, Taipale J. Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev Cell. 2006;10:177–186. doi: 10.1016/j.devcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 3.Tay SY, Ingham PW, Roy S. A homologue of the Drosophila kinesin-like protein Costal2 regulates Hedgehog signal transduction in the vertebrate embryo. Development. 2005;132:625–634. doi: 10.1242/dev.01606. [DOI] [PubMed] [Google Scholar]
- 4.Wolff C, Roy S, Ingham PW. Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr Biol. 2003;13:1169–1181. doi: 10.1016/s0960-9822(03)00461-5. [DOI] [PubMed] [Google Scholar]
- 5.Chen MH, Gao N, Kawakami T, Chuang PT. Mice deficient in the fused homolog do not exhibit phenotypes indicative of perturbed hedgehog signaling during embryonic development. Mol Cell Biol. 2005;25:7042–7053. doi: 10.1128/MCB.25.16.7042-7053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merchant M, et al. Loss of the serine/threonine kinase fused results in postnatal growth defects and lethality due to progressive hydrocephalus. Mol Cell Biol. 2005;25:7054–7068. doi: 10.1128/MCB.25.16.7054-7068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson CW, et al. Fused has evolved divergent roles in vertebrate Hedgehog signalling and motile ciliogenesis. Nature. 2009;459:98–102. doi: 10.1038/nature07883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koudijs MJ, et al. The zebrafish mutants dre, uki, and lep encode negative regulators of the hedgehog signaling pathway. PLoS Genet. 2005;1:e19. doi: 10.1371/journal.pgen.0010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper AF, et al. Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development. 2005;132:4407–4417. doi: 10.1242/dev.02021. [DOI] [PubMed] [Google Scholar]
- 10.Svärd J, et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 11.Huangfu D, et al. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 12.Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong SY, Reiter JF. The primary cilium at the crossroads of mammalian hedgehog signaling. Curr Top Dev Biol. 2008;85:225–260. doi: 10.1016/S0070-2153(08)00809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lunt SC, Haynes T, Perkins BD. Zebrafish ift57, ift88, and ift172 intraflagellar transport mutants disrupt cilia but do not affect hedgehog signaling. Dev Dyn. 2009;238:1744–1759. doi: 10.1002/dvdy.21999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aanstad P, et al. The extracellular domain of Smoothened regulates ciliary localization and is required for high-level Hh signaling. Curr Biol. 2009;19:1034–1039. doi: 10.1016/j.cub.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 18.Corbit KC, et al. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 19.Denef N, Neubüser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell. 2000;102:521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- 20.Haycraft CJ, et al . Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: Conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- 22.Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- 23.Briscoe J, Novitch BG. Regulatory pathways linking progenitor patterning, cell fates and neurogenesis in the ventral neural tube. Philos Trans R Soc Lond B Biol Sci. 2008;363:57–70. doi: 10.1098/rstb.2006.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 25.Goodrich LV, Milenković L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 26.Ho KS, Suyama K, Fish M, Scott MP. Differential regulation of Hedgehog target gene transcription by Costal2 and Suppressor of Fused. Development. 2005;132:1401–1412. doi: 10.1242/dev.01689. [DOI] [PubMed] [Google Scholar]
- 27.Farzan SF, et al. Costal2 functions as a kinesin-like protein in the hedgehog signal transduction pathway. Curr Biol. 2008;18:1215–1220. doi: 10.1016/j.cub.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, et al. Hedgehog-regulated Costal2-kinase complexes control phosphorylation and proteolytic processing of Cubitus interruptus. Dev Cell. 2005;8:267–278. doi: 10.1016/j.devcel.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Zhang XM, Ramalho-Santos M, McMahon AP. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R symmetry by the mouse node. Cell. 2001;106:781–792. [PubMed] [Google Scholar]
- 30.Motoyama J, et al. Differential requirement for Gli2 and Gli3 in ventral neural cell fate specification. Dev Biol. 2003;259:150–161. doi: 10.1016/s0012-1606(03)00159-3. [DOI] [PubMed] [Google Scholar]
- 31.Bai CB, Stephen D, Joyner AL. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev Cell. 2004;6:103–115. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 32.Lei Q, Zelman AK, Kuang E, Li S, Matise MP. Transduction of graded Hedgehog signaling by a combination of Gli2 and Gli3 activator functions in the developing spinal cord. Development. 2004;131:3593–3604. doi: 10.1242/dev.01230. [DOI] [PubMed] [Google Scholar]
- 33.Matise MP, Epstein DJ, Park HL, Platt KA, Joyner AL. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development. 1998;125:2759–2770. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- 34.Persson M, et al. Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes Dev. 2002;16:2865–2878. doi: 10.1101/gad.243402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran PV, et al. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat Genet. 2008;40:403–410. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cortellino S, et al. Defective ciliogenesis, embryonic lethality and severe impairment of the Sonic Hedgehog pathway caused by inactivation of the mouse complex A intraflagellar transport gene Ift122/Wdr10, partially overlapping with the DNA repair gene Med1/Mbd4. Dev Biol. 2009;325:225–237. doi: 10.1016/j.ydbio.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang G, Amanai K, Wang B, Jiang J. Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes Dev. 2000;14:2893–2905. doi: 10.1101/gad.843900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nonaka S, et al. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 39.Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc Natl Acad Sci USA. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeda S, et al. Left-right asymmetry and kinesin superfamily protein KIF3A: New insights in determination of laterality and mesoderm induction by kif3A−/− mice analysis. J Cell Biol. 1999;145:825–836. doi: 10.1083/jcb.145.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.May SR, et al. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 42.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: Insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 43.Taipale J, et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 44.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci USA. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15:303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Sisson JC, Ho KS, Suyama K, Scott MP. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90:235–245. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 47.Robbins DJ, et al. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- 48.García-García MJ, et al. Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc Natl Acad Sci USA. 2005;102:5913–5919. doi: 10.1073/pnas.0501071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 50.Caspary T, et al. Mouse Dispatched homolog1 is required for long-range, but not juxtacrine, Hh signaling. Curr Biol. 2002;12:1628–1632. doi: 10.1016/s0960-9822(02)01147-8. [DOI] [PubMed] [Google Scholar]
- 51.Hui CC, Joyner AL. A mouse model of Greig cephalopolysyndactyly syndrome: The Extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat Genet. 1993;3:241–246. doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]
- 52.Shen J, et al. Skeletal and CNS defects in Presenilin-1-deficient mice. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 53.Moran JL, et al. Utilization of a whole genome SNP panel for efficient genetic mapping in the mouse. Genome Res. 2006;16:436–440. doi: 10.1101/gr.4563306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheung HO, et al. The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci Signal. 2009;2:ra29. doi: 10.1126/scisignal.2000405. [DOI] [PubMed] [Google Scholar]
- 55.Endoh-Yamagami S, et al. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr Biol. 2009 doi: 10.1016/j.cub.2009.06.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.