Abstract
Inhibition of bacterial gene expression by RNase P-directed cleavage is a promising strategy for the development of antibiotics and pharmacological agents that prevent expression of antibiotic resistance. The rise in multiresistant bacteria harboring AAC(6′)-Ib has seriously limited the effectiveness of amikacin and other aminoglycosides. We have recently shown that recombinant plasmids coding for external guide sequences (EGS), short antisense oligoribonucleotides (ORN) that elicit RNase P-mediated cleavage of a target mRNA, induce inhibition of expression of aac(6′)-Ib and concomitantly induce a significant decrease in the levels of resistance to amikacin. However, since ORN are rapidly degraded by nucleases, development of a viable RNase P-based antisense technology requires the design of nuclease-resistant RNA analog EGSs. We have assayed a variety of ORN analogs of which selected LNA/DNA co-oligomers elicited RNase P-mediated cleavage of mRNA in vitro. Although we found an ideal configuration of LNA/DNA residues, there seems not to be a correlation between number of LNA substitutions and level of activity. Exogenous administration of as low as 50 nM of an LNA/DNA co-oligomer to the hyperpermeable E. coli AS19 harboring the aac(6′)-Ib inhibited growth in the presence of amikacin. Our experiments strongly suggest an RNase P-mediated mechanism in the observed antisense effect.
Keywords: aminoglycoside, antibiotic resistance, antisense, nucleic acids analogs, RNase P
Emergence and spread of antibiotic resistance genes among bacterial pathogens is becoming a serious problem worldwide. The semisynthetic aminoglycoside amikacin (Ak) is very useful in the treatment of multiresistant infections because only a limited number of modifying enzymes, such as AAC(6′)-I-type acetyltransferases, are able to inactivate it (1). Unfortunately, the rise in multiresistant strains harboring AAC(6′)-I-type enzymes, specially AAC(6′)-Ib, has seriously limited the successful use of aminoglycosides including Ak (1, 2). Compounding the problem, while new antibiotics to treat multiresistant gram positives are in the pipeline, the number of potential new antimicrobial candidates against problematic Gram-negative bacterial pathogens, the usual hosts of AAC(6′)-Ib, is very short (3, 4). As a consequence, there is an urgent need to develop new antibiotics and strategies to preserve the activity of existing ones. A variety of antisense strategies to silence resistance genes and achieve phenotypic conversion to susceptibility have been explored (5, 6).
A promising antisense strategy takes advantage of the characteristics of RNase P, a ribozyme responsible for generating the mature 5′ end of tRNA that includes an RNA component (M1) that is the catalytic subunit and a cofactor protein (C5) (7–9). Short oligoribonucleotides, known as external guide sequences (EGSs), can elicit RNase P-mediated cleavage of a complementary RNA molecule (10). However, in most cases where RNase P-mediated inhibition of expression of genes has been achieved, the EGSs were transcribed from plasmids inside the cytoplasm (8, 11, 12). Therefore, this general strategy is not viable as the final stage in development of an RNase P-based antisense technology. The EGSs must be added from outside and penetrate the cells to exert their action. Since oligoribonucleotides are rapidly degraded by nucleases, successful development of this technology depends on finding nuclease-resistant analogs that induce RNase P-mediated degradation of the target mRNA.
We have recently designed an EGS, EGSC3, that when transcribed from a recombinant clone reduces the host cell's levels of resistance to Ak (13). In this work, we tested isosequential nuclease-resistant oligonucleotide analogs to determine their ability to elicit RNase P-mediated cleavage of aac(6′)-Ib mRNA at levels comparable to those elicited by the oligoribonucleotide EGSC3. Our results showed that locked nucleic acids (LNA)/DNA co-oligomers act as effective EGSs.
Results
Nuclease-Resistant Oligonucleotide Analogs as External Guide Sequences.
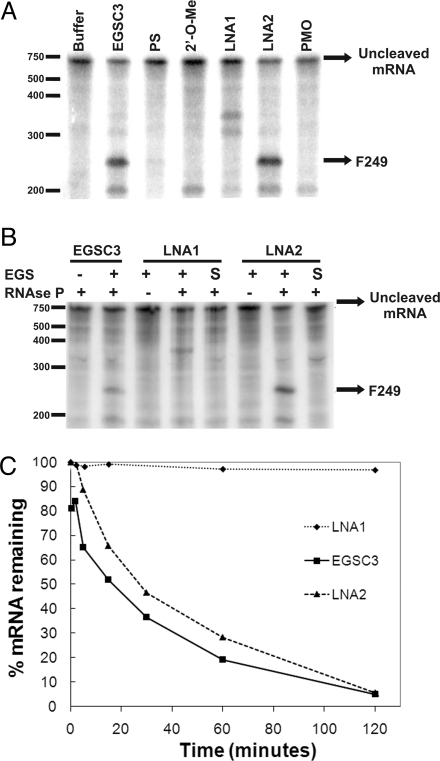
Several isosequencial oligomers were designed using phosphorothioate oligodeoxynucleotides (PS), 2′-O-methyl oligoribonucleotides (2′-O-Me), phosphorodiamidate morpholino oligomers (PMO), or LNA. EGSs were designed to include 2 regions: a 13-nucleotide segment antisense to the target mRNA (AS region) followed at the 3′ end by the ACCA sequence (ACCA region) that interacts with the UGG sequence within the P15-loop of the M1 RNA component of the E. coli RNase P (7, 9, 12). While the PS and PMO were substituted with analogs in all positions, the 2′-O-Me and LNA oligomers were designed as 2′-O-Me/RNA or LNA/DNA chimeric co-oligomers. Their structural details and their names are shown in Table 1. There are advantages in using co-oligomers instead of oligomers containing all units as LNA or 2′-O-Me. The right mix of nucleotides and analog monomers results in co-oligomers that usually possess enhanced resistance to nucleases present in serum, have lower toxicity, and show a proper balance between efficiency and specificity (14, 15). This is especially important in the case of the LNA-bearing compounds because every substituted nucleotide monomer dramatically increases the thermal stability of duplexes with complementary RNA or DNA (16). Furthermore, these compounds exhibit reduced toxicity when compared to other analogs (17). LNA1 contains 6 substitutions, all of them within the AS region leaving all 4 residues in the ACCA region unsubstituted. LNA1, as well as the PS, 2′-O-methyl and PMO compounds failed to induce significant cleavage of aac(6′)-Ib mRNA (Fig. 1A). Interestingly, the recent results of Shen et al. (18) showed that a PMO when conjugated to a peptide induced cleavage of a target mRNA. Failure of LNA1 to elicit cleavage is in keeping with previous results by Perreault and Altman (19) who observed that the 2′-OH at the C residue located toward the 5′ end of the ACCA region is essential for full EGS activity. In this work they showed that replacing this C by a dC residue in an otherwise all RNA EGS results in a 10-fold reduction in activity when the incubation is carried out in the presence of M1 RNA. LNA2, which includes 3 substitutions at the 5′ end of the AS region (CAA) and 3 substitutions at the 3′end (CCA) (Table 1), induced cleavage at a level similar to that of the control EGSC3 generating the expected 249-nucleotide fragment (F249) (Fig. 1A). No products were detected when the enzyme was omitted from the reaction mixture indicating that cleavage was RNase P-mediated (Fig. 1B). The replacement of DNA by LNA residues at the ACCA region seems to have an effect similar to that observed by Perreault and Altman (19) when DNA were replaced by RNA residues in the ACCA region. In all incubations, even in the absence of RNase P, we observed an approximate 190-nucleotide band of unknown origin (13). Sequence specificity was tested using as EGSs the sense versions of LNA1 and LNA2, as expected no mRNA cleavage was detected (Fig. 1B). The rates of mRNA cleavage using LNA2 or EGSC3 at 0.05 μM were nearly identical and more than 95% of the aac(6′)-Ib mRNA was digested after 2 h (Fig. 1C). Interestingly, although LNA1 did not induce detectable cleavage at this concentration (Fig. 1C), it showed higher binding affinity to mRNA than LNA2 (SI Text, Fig. S1) indicating that high binding affinity between the EGS and its target is not enough to elicit RNase P-mediated degradation. Binding must result in the appropriate interactions of EGS with the RNase P components to elicit cleavage.
Table 1.
Oligomers used in this study
| Oligomer | Sequence and chemistry |
|---|---|
| EGSC3 | caaguacuguuccacca |
| EGSC3DNA | CAAGTACTGTTCCACCA |
| Phosphorothioate oligodeoxynucleotide (PS) | caaguacuguuccacca |
| 2′-O-methyl oligoribonucleotide (2′-O-Me) | CAAguacuguuccaCCA |
| Phosphorodiamidate morpholino oligomers (PMO) | CAAGTACTGTTCCACCA |
| LNA1 | CAAGTACTGTTCCACCA |
| LNA2 | CAAGTACTGTTCCACCA |
| LNA3 | CAAGTACTGTTCCACCA |
| LNA4 | CAAGTACTGTTCCACCA |
| LNA5 | CAAGTACTGTTCCACCA |
| LNA6 | CAAGTACTGTTCCACCA |
| LNA7 | CAAGTACTGTTCCACCA |
| LNA8 | CAAGTACTGTTCCACCA |
| LNA9 | CAAGTACTGTTCCACCA |
| LNA10 | CAAGTACTGTTCCACCA |
| LNA11 | CAAGTACTGTTCCACCA |
| LNA1Sense | GGAACAGTACTTGACCA |
| LNA2Sense | GGAACAGTACTTGACCA |
| LNA9Sense | GGAACAGTACTTGACCA |
| LNA10Sense | GGAACAGTACTTGACCA |
| LNA9alk-phos | AGGCATCTATACCACCA |
| LNA10alk-phos | AGGCATCTATACCACCA |
Plain font, DNA; lower case, RNA; italics, PS; bold, 2′-O-Me; bold italics, PMO; underlined, LNA.
Fig. 1.
Ability of LNA/DNA chimeric co-oligomers to elicit RNase P-mediated cleavage of aac(6′)-Ib mRNA. (A) Induction of cleavage of 32P-labeled aac(6′)-Ib mRNA by EGSC3 and LNA/DNA co-oligomers at 0.5 μM. Cleavage reactions were carried out as described in the Methods for 2 h in the presence of RNase P and the indicated co-oligomers (see Table 1). The products were analyzed by PAGE and phosphorimaging. Location of RNA molecular size standards (nucleotides) are shown to the left and the position of the reaction product, F249 is shown to the right. (B) RNase P and sequence-dependence of 32P-labeled aac(6′)-Ib mRNA cleavage. Reactions were carried out as in A in the presence (+) or absence (-) of RNase P or the compounds indicated on top of the gel at 0.5 μM. S indicates that the co-oligomer has the sense sequence. (C) Kinetics of RNase P cleavage. Reactions were performed as in A for 0, 2, 5, 15, 30, 60, and 120 min using 0.05 μM of the indicated compound. The bands were quantified by densitometry.
Effect of LNA Substitutions at Different Positions of the Oligomer.
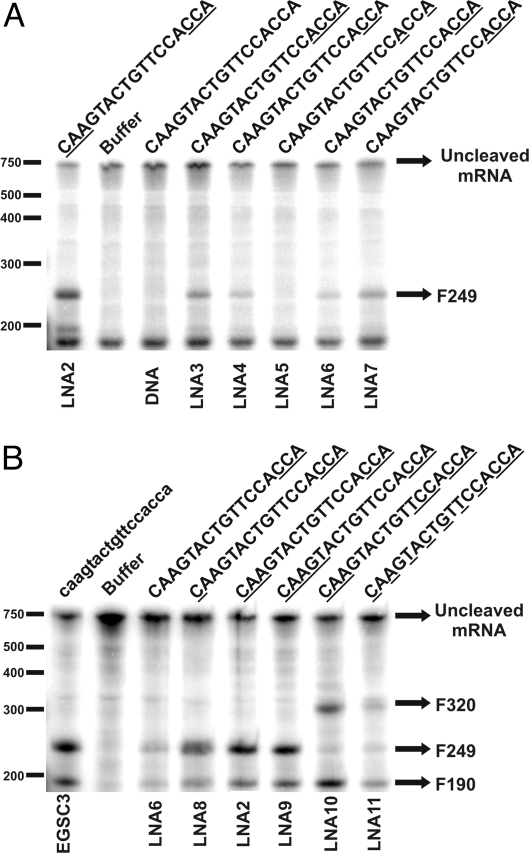
The results described so far suggested that besides replacements in the AS region, at least some of the nucleotides in the ACCA region must also be substituted to induce efficient RNase P cleavage of the target mRNA (Fig. 1 C and Table 1). Therefore, we analyzed the effect of LNA replacements in the ACCA region of the EGS. Several LNA/DNA co-oligomers with deoxynucleotides in the AS region (no substitutions) and a different number and configuration of LNA substitutions in the ACCA region were designed and tested (Table 1, EGSC3DNA, LNA3 to LNA7). Fig. 2A shows that while in our conditions DNA did not induce cleavage activity, the substitution by LNAs of the 2 central Cs of the ACCA region (LNA4) was sufficient to elicit some cleavage of mRNA. Replacement of the terminal 3 positions (LNA6) showed no additional activity respect to LNA4. The activity of the compounds was maximal within this group of EGSs when the 3 inner (LNA7) or all 4 (LNA3) residues were substituted by LNAs. On the other hand, replacement of only the adenine residues of the ACCA region (LNA5) resulted in an oligomer that was not able to induce the cleavage reaction.
Fig. 2.
Activities of LNA/DNA co-oligomer EGSs. (A) Induction of cleavage of 32P-labeled aac(6′)-Ib mRNA by LNA/DNA co-oligomers with different substitution configurations in the ACCA region and LNA2. (B) Induction of cleavage of 32P-labeled aac(6′)-Ib mRNA by EGSC3 (low case) and LNA/DNA co-oligomers with different substitution configurations in the AS region while leaving ACCA region constant.
An increase in the thermal stability of duplexes between the ACCA region and the UGG sequence within the M1 RNA P15 loop (9, 20, 21) and/or the presence of an O at the 2′ position of residues in the ACCA region [as described by Perreault et al. (19)] might explain the enhancement in the ability of the EGS to elicit RNase P-mediated cleavage described in the previous paragraph. While the 2 central C residues within the ACCA region are essential in the interaction EGS/M1 RNA, the A residue located at the 3′ end seems to play a secondary role.
LNA2, which has 3 replacements in both the AS and ACCA region, showed significantly higher activity than the co-oligomers that include replacements only at the ACCA region (Fig. 2A), indicating that further substitutions on the AS region improves the oligomer's activity. Hence, we tested co-oligomers with various replacement configurations within the AS region while leaving the ACCA region constant (listed in Table 1, LNAs 2, 6, and 8–11). We decided to replace only the 3 terminal nucleotides within the ACCA region to minimize the number of replacements and maximize nuclease resistance. Co-oligomers LNA6, LNA8, LNA2, and LNA9 have 0, 1, 3, and 5 LNA replacements at their 5′ ends, respectively (Table 1 and Fig. 2B). LNA10 and LNA11 have been designed as a gapmer and a mixmer, respectively (Table 1 and Fig. 2B). We observed that increasing the number of replacements in the 5′ end resulted in higher RNase P-mediated cleavage, probably by increasing affinity of the EGS for the mRNA (Fig. 2B and SI Text, Fig. S2). Among these EGSs, LNA2 and LNA9 exhibited the highest mRNA cleavage-eliciting efficiency. LNA10 (gapmer configuration) also showed strong eliciting activity. Conversely, LNA11 (mixmer configuration) showed poor activity (Fig. 2B), probably by changing structural details of the helix in the stem region. It has been shown before that these changes have an additive effect on the interaction with M1 RNA resulting in a decrease of cleavage activity (19). To ensure that there were no smaller cleavage products, a gel was run for a shorter time (SI Text, Fig. S3).
Our results indicate that rather than a direct correlation between number of substitutions and level of activity there is an ideal number and configuration of substitutions. It is of interest that LNA10 showed a high level of activity but only a small proportion of cleaved product was F249. The main product of cleavage was an about 320-nucleotide fragment (F320) indicating that the point of cleavage in the presence of LNA10 did not occur at the predicted location (Fig. 2B). Binding kinetics showed that LNA10 has the highest affinity, followed by LNA9 and EGSC3, which have similar binding affinities, and LNA2 had the lowest affinity (SI Text, Fig. S2). F320 was also the main cleavage product detected by incubation in the presence of LNA11 (Fig. 2B). It is of interest that Shen et al. (18) have recently found a peptide-PMO oligomer that elicit RNase P-mediated cleavage of mRNA at different locations.
Stability of LNAs When Exposed to Cell Suspensions and Cell Extracts.
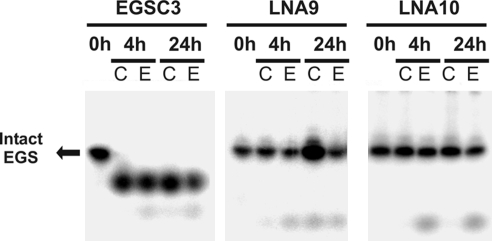
Although LNA/DNA co-oligomers are known to be resistant to serum nucleases (15, 22, 23), their resistance to bacterial nucleases is not well-established. Therefore, radioactively labeled EGSC3, LNA9, and LNA10 were incubated for several hours with either E. coli cultures or E. coli sonicates. About 50% and 100% of the unsubstituted oligoribonucleotide was degraded after incubation for 30 min and 24 h, respectively (Fig. S4). Conversely, degradation of the LNA/DNA co-oligomers was virtually undetected after 24 h of exposure to cell extracts or cell suspensions (Fig. 3).
Fig. 3.
Nuclease resistance of EGSC3 and LNA/DNA co-oligomers. 32P-labeled EGSC3 (RNA), LNA9, and LNA10 were incubated in the presence of cells in culture (C) or extracts obtained by soft sonication (E) for 0, 4, and 24 h and analyzed by PAGE.
Cellular Uptake of LNA EGSs.
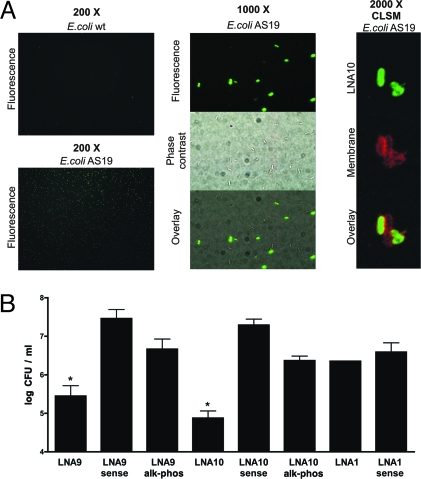
Uptake of LNA EGS by bacterial cells was assessed by exposing the hyperpermeable E. coli AS19 to Alexa Fluor 488-conjugated LNA10. Examination by epifluorescence microscopy showed that nearly 50% of the cells were associated with fluorescence. Conversely, fluorescence was associated to a very low proportion of the wild-type E. coli DH5α (Fig. 4A). A set of equivalent experiments using radioactively labeled LNA10 showed similar results (Fig. S5) indicating that cell penetration of the LNA EGS was not affected by conjugation to Alexa Fluor 488. Further analysis of 0.7-μm section images obtained by confocal laser scanning microscopy (CLSM) of E. coli AS19 stained with the membrane specific dye FM5–95 and exposed to Alexa Fluor 488-conjugated LNA10 confirmed that the co-oligomer reached the cytoplasm (Fig. 4A).
Fig. 4.
Internalization and in vivo effect of DNA/LNA co-oligomers. (A) Epiflourescence microscopy images of E. coli AS19 or E. coli wild-type and CLSM images of E. coli AS19 preincubated with Alexa488-labeled LNA10 for 30 min are shown at the indicated magnification. (B) Effect of Ak on the survival of E. coli AS19 (pFC9) cells preincubated with LNA9 (50 nM), LNA10 (5 μM), or LNA1 (5 μM). Results are expressed as log of mean± SD of colony forming units per milliliter (log CFU/mL). Experiments were performed by triplicate. Similar results were observed in 3 independent assays. *, P < 0.05.
In Vivo Effect of LNA EGSs on Ak Resistance.
To determine whether LNA9 or LNA10 interfere with expression of antibiotic resistance in vivo E. coli AS19 harboring pFC9, a plasmid that includes aac(6′)-Ib, was cultured in the presence of each 1 of these compounds before being exposed to 50 μg/mL Ak. The same strain was cultured in the presence of sense versions of these LNAs (LNA9Sense and LNA10Sense, Table 1) or EGSs with the same LNA/DNA configuration but targeting the alkaline phosphatase gene (LNA9alk-phos and LNA10alk-phos, Table 1) as controls. Fig. 4B shows that growth of cultures that had been exposed to 5 μM LNA10 or 50 nM LNA9 is significantly inhibited in the presence of antibiotic, indicating that the appropriate antisense LNAs interfere with the expression of Ak resistance. Experiments using different concentrations of each antisense co-oligomer show dose dependence on the biological effect (Fig. S6). Fig. 4B also shows that LNA1, which did not elicit RNase P cleavage in vitro, did not interfere with Ak resistance in vivo, strongly suggesting that the effect of LNA9 and LNA10 occurs through activation of the E. coli AS19 RNase P. This result supports the observation that LNA substitutions in the ACCA region are essential to obtain active LNA/DNA EGSs that can elicit RNase P-mediated degradation of the target mRNA.
Discussion
AAC(6′)-Ib, an enzyme responsible for resistance to several aminoglycosides including Ak, is present in over 70% of AAC(6′)-I-producing Gram-negative clinical isolates (1, 2). We have recently shown that the external guide sequence EGSC3, when expressed from a recombinant plasmid present in cells harboring aac(6′)-Ib, elicits cleavage of the mRNA by RNase P leading to a significant decrease in the level of resistance (13). However, for this general strategy to be viable as the final stage in development of RNase P-based antisense technologies the EGSs must be added from outside of the cells. Since oligoribonucleotides are rapidly degraded by nucleases, 1 of the problems to solve for the successful development of this strategy is to find nuclease-resistant analogs that are active as EGSs. Comparison of several analogs showed that LNA/DNA co-oligomers were the most efficient analogs to induce RNase P-mediated degradation of the aac(6′)-Ib mRNA. On the basis of these results, we carried out a comprehensive analysis of LNA substituted oligomers. However, the possibility that the analogs that did not elicit aac(6′)-Ib mRNA cleavage in this study could induce RNase P activity in replacement configurations different from those tested here or when targeting other RNA molecules should not be ruled out.
Analyses of EGSC3 isosequential LNA/DNA co-oligomers with different numbers and locations of LNA substitutions suggest that different configurations must be tested to identify the oligomer with the desired characteristics, that is, it promotes high levels of RNase P cleavage, it acts specifically on the mRNA target, and it is resistant to the action of nucleases. While the DNA EGSC3 with no LNA substitutions failed to promote any detectable cleavage, substitution of the 2 Cs within the ACCA region was sufficient to induce detectable levels of RNase P cleavage of the target aac(6′)-Ib mRNA. Interestingly, we noticed that there was higher RNase P activity in presence of LNA7 rather than LNA6 suggesting that besides the 2 central Cs the A located at the 3′ end also interacts with P15 loop of M1 RNA. These results are in keeping with previous work by Kirsebom and colleagues (7, 9, 12). Further studies will be necessary to fully understand the molecular basis of the increase in the ability to elicit RNase P-mediated digestion of the target mRNA showed by LNA-containing oligomers as opposed to full DNA EGSs, and to determine if the increase in thermal stability of duplexes with complementary DNA or RNA is at all related to this effect (15). The fraction of mRNA molecules cleaved increased when selected residues within the AS region were also replaced by LNA residues. However, replacement of too many residues in the oligomer can increase unspecific interaction with RNA molecules other than the target reducing specificity or increasing the binding affinity but generating a complex that is not recognized by RNase P. The most appropriate configuration LNA/DNA was obtained empirically analyzing several co-oligomers. These assays were carried out using co-oligomers that have the 3 terminal CCA nucleotides replaced rather than all 4 residues in the ACCA region to minimize unspecific binding to aac(6′)-Ib and other mRNAs. Although one could expect that replacing the ACC nucleotides leaving the terminal A unreplaced could have higher activity, we decided against this possibility because these co-oligomers would be more susceptible to degradation. Co-oligomers with replacements at the 5′ and 3′ ends (LNA2 and LNA9) were efficient and induced digestion at the expected location of aac(6′)-Ib mRNA. The gapmer LNA10, which includes a total of 9 LNA replacements, also showed significant levels of activity but the product of digestion induced was not that one expected by interaction between the oligomer and the target mRNA. This could be the result of non-specific binding due to the enhanced affinity conferred by too many LNA monomers. Productive LNA10 binding to nucleotides 310 to 326 on the aac(6′)-Ib mRNA, where the 6 LNA replacements in AS region could form Watson-Crick bonds would explain the generation of F320. An alternative explanation is that the 3-dimensional structure of the mRNA and/or the EGS may be such that there is a site that, although not perfectly complementary, is more readily available for interaction than the target region. In this case, the unusually high thermostability of the complex between the mRNA and the LNA10 would permit a productive interaction. Further research is needed to determine the molecular basis of the anomalous cleavage location. LNA11, a mixmer that includes 10 replacements, shows lower efficiency, and the product of digestion is also the anomalous one. The recent work by Shen et al. (18) also describes unexpected cleavage digestion when using a peptide-PMO-EGS analog. In this case the authors postulate that the positively charged peptide could interact with the target mRNA or the M1 RNA changing their structure and leading to formation of a non-obvious available site.
The ability of EGSs to interfere with resistance to Ak was studied in E. coli AS19 cells harboring aac(6′)-Ib. LNA9 was the most efficient compound, which caused significant growth inhibition at 50 nM. Prior work showed a correlation between binding affinity of the EGS to mRNA in vitro and inhibition of Ak resistance when the EGSs were transcribed from a resident plasmid (13). We speculated that the inhibitory effect of a given EGS could be predictive on the basis of its binding affinity. However, although this may be correct when comparing RNA EGSs with different sequences, the results shown here indicate that isosequential co-oligomers with different substitution configurations may have comparable binding affinities and significantly differ in their inhibitory ability. LNA9 and LNA10 showed high binding affinities but LNA9 was active at 100-fold lower concentrations than LNA10. However, it was of interest that in the dose–response experiment the reduction of colony forming units/mL induced by LNA10 was higher than that shown by LNA9. We do not know yet why despite being more efficient, LNA9 does not reach the LNA10 level of reduction.
The internalization experiments showed that the co-oligomers are taken up by E. coli AS19 but not a wild-type strain. These results, taken together with those showing interference with expression of amikacin resistance, indicate that the LNA/DNA co-oligomers are active once inside the bacterial cell, but are not readily taken up by wild-type E. coli. Therefore, a method to induce internalization must be developed before these compounds can be developed to induce phenotypic conversion to susceptibility. Several groups have experimented with a variety of strategies to induce uptake of oligonucleotides and analogs into the cytosol. These strategies included liposome encapsulation, modification of the oligonucleotide by introduction of structures like hairpins, or attachment of cell-permeabilizing peptides (18, 24–30). We are presently carrying out assays using these approaches to induce penetration of LNA/DNA co-oligomers into E. coli and other bacteria. Development of internalization techniques will permit us to carry out assays using infection model systems.
In conclusion, the development of nuclease resistant analogs that can elicit RNase P-mediated degradation of bacterial mRNA and inhibit gene expression when added to the culture medium represents a step in the right direction toward utilization of nuclease resistant analog EGSs as therapeutic agents to inhibit expression of antibiotic resistance genes or inhibit growth of pathogens by targeting essential genes.
Materials and Methods
Bacterial Strains, Plasmids, and Oligonucleotide Analogs.
E. coli DH5α (31) was used as host for all plasmids. The permeable E. coli AS19 (32) was used for in vivo inhibition assays. This strain was periodically isolated and tested for lysozyme sensitivity to avoid accumulation of revertants as described before (32). Bacterial cultures were carried out in Lennox Luria (L) broth. The HPLC-purified oligonucleotides were purchased from IDT Technologies. The sequences of the oligonucleotides used in this study are shown in Table 1.
General Procedures.
Plasmid DNA was extracted using the QIAspin miniprep kit (Qiagen). M1 RNA, aac(6′)-Ib mRNA, and the C5 protein were prepared as described before (13). The aac(6′)-Ib mRNA and the oligoribonucleotides and analogs were 5′-end-labeled as described before (33). Radioactivity was visualized using a STORM 840 PhosphorImager and quantified using ImageQuant (Molecular Dynamics). Nucleic acids were quantified using Beckman Coulter DU 530 spectrophotometer (Beckman Coulter). Gel electrophoresis was performed in Miniprotean Tetra cell system (BioRad) using GTG buffer (USB).
In Vitro RNase P Assays.
EGS-mediated digestion of 5′-end-labeled aac(6′)-Ib mRNA was assayed basically as described by Li et al. (34). Radiolabeled aac(6′)-Ib mRNA (2.5 pmol) was preincubated with the appropriate EGS (5 pmol) at 25 °C for 2 h in a volume of 3 μL. The components of RNase P were mixed (2.5 pmol M1 RNA and 70 pmol C5 protein) in buffer containing 20 mM Hepes-KOH (pH 8.0), 400 mM ammonium acetate, 10 mM magnesium acetate, and 5% glycerol and preincubated at 37 °C for 15 min in a final volume of 7 μL. After preincubation, both solutions were combined and incubated at 37 °C for 2 h. The reaction was stopped by heating and phenol/cholorform extraction. Then sample was heat-denatured 2 min at 85 °C after the addition of 1 volume of gel loading buffer (95% formamide, 1 mM EDTA, pH 8, Bromophenol Blue 0.01 w/m) and analyzed by 5% denaturing GTG-PAGE.
Nuclease Resistance Assays.
Overnight cultures of E. coli DH5α were diluted 1/100 in LB and grown until OD600≈1. One milliliter of culture was sonicated for 5 s 6 times on ice. Then 20 pmol radioactively labeled EGS were mixed with 6 μL of the cell cultures or 6 μL of the cell lysates and incubated for the indicated times. Then, 1 volume of gel loading buffer was added and sample was heated for 2 min at 85 °C. Sample was analyzed by 15% denaturing GTG/PAGE.
Cell Incorporation of EGS.
Cells (106) of E. coli DH5α or E. coli AS19 cells were incubated in saline buffer with 5 μM Alexa Fluor 488-conjugated LNA10 for 30 min at 37 °C in absence of light. Then the cells were washed twice by centrifugation, resuspended in 10 μL saline buffer. Then FM5–95 was added to a final concentration of 10 μM and cells were on ice for 1 to 2 min. Bacteria were then poured on 2% agarose saline buffer and covered. Images were taken by epifluorescence microscopy in a BX60 Olympus microscope (Olympus) or by laser scanning confocal microscopy (LSCM) in a Zeiss Pascal confocal microscope (Zeiss).
In Vivo Activities of EGS.
A single colony of E. coli AS19 (pFC9) cells was cultured ON in LB medium supplemented with ampicillin (100 μg/mL). Then 105 cells were loaded by triplicate for each treatment on a 96-multiwell plate at the indicated concentrations of LNA in a final volume of 150 μL. After 2 h of culture at 37 °C with 150 rpm agitation Ak was added to a final concentration of 40 μg/mL. Cells were cultured in the same conditions for 1 additional hour, serially diluted, and spread on LB plates. After overnight incubation at 37 °C, colonies were counted for analysis. In all cases, sense and missense LNA (alk-phos) were used as controls. The experiments were repeated 3 times and the results are expressed as mean ± SD of a representative assay done by triplicate. Statistical significance was analyzed by 1-way ANOVA 2-tailed test, and Tukey's test for comparison was used to determined significant differences. Values of P < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments.
We thank S. Altman for encouragement, useful discussion and suggestions, and for making his manuscript available before its publication. This study was supported by National Institutes of Health Public Health Service Grant 2R15AI047115 (to M.E.T.) and X-240 Universidad de Buenos Aires (to A.Z.). A.Z. is a career member of Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET). A.J.C.S.B. was supported by CONICET and American Society for Microbiology International Fellowship for Latin America. J.C.J. was supported by LA Basin Minority Health and Health Disparities International Research Training Program (MHIRT) 5T37MD001368-09 (National Center on Minority Health and Health Disparities).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906529106/DCSupplemental.
References
- 1.Tolmasky ME. Aminoglycoside-modifying enzymes: Characteristics, localization, and dissemination. In: Bonomo R, Tolmasky ME, editors. Enzyme-Mediated Resistance to Antibiotics: Mechanisms, Dissemination, and Prospects for Inhibition. Washington, DC: ASM Press; 2007. pp. 35–52. [Google Scholar]
- 2.Vakulenko SB, Mobashery S. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev. 2003;16:430–450. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Neill AJ. New antibacterial agents for treating infections caused by multi-drug resistant Gram-negative bacteria. Expert Opin Investig Drugs. 2008;17:297–302. doi: 10.1517/13543784.17.3.297. [DOI] [PubMed] [Google Scholar]
- 4.Taubes G. The bacteria fight back. Science. 2008;321:356–361. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen LC, Sperling-Petersen HU, Mortensen KK. Hitting bacteria at the heart of the central dogma: Sequence-specific inhibition. Microb Cell Fact. 2007;6:24. doi: 10.1186/1475-2859-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodford N, Wareham DW. Tackling antibiotic resistance: A dose of common antisense? J Antimicrob Chemother. 2008 doi: 10.1093/jac/dkn467. [DOI] [PubMed] [Google Scholar]
- 7.Altman S. A view of RNase P. Mol Biosyst. 2007;3:604–607. doi: 10.1039/b707850c. [DOI] [PubMed] [Google Scholar]
- 8.Guerrier-Takada C, Salavati R, Altman S. Phenotypic conversion of drug-resistant bacteria to drug sensitivity. Proc Natl Acad Sci USA. 1997;94:8468–8472. doi: 10.1073/pnas.94.16.8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirsebom LA. RNase P RNA mediated cleavage: Substrate recognition and catalysis. Biochimie. 2007;89:1183–1194. doi: 10.1016/j.biochi.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Forster AC, Altman S. External guide sequences for an RNA enzyme. Science. 1990;249:783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- 11.Ko JH, Izadjoo M, Altman S. Inhibition of expression of virulence genes of Yersinia pestis in Escherichia coli by external guide sequences and RNase P. RNA. 2008;14:1656–1662. doi: 10.1261/rna.1120508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKinney JS, Zhang H, Kubori T, Galan JE, Altman S. Disruption of type III secretion in Salmonella enterica serovar Typhimurium by external guide sequences. Nucleic Acids Res. 2004;32:848–854. doi: 10.1093/nar/gkh219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soler Bistue AJ, et al. External guide sequences targeting the aac(6′)-Ib mRNA induce inhibition of amikacin resistance. Antimicrob Agents Chemother. 2007;51:1918–1925. doi: 10.1128/AAC.01500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur J Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 15.Wahlestedt C, et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci USA. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersen M, Wengel J. LNA: A versatile tool for therapeutics and genomics. Trends Biotechnol. 2003;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 17.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 18.Shen N, et al. Inactivation of expression of several genes in a variety of bacterial species by the EGS technology. Proc Natl Acad Sci USA. 2009;106:8163–8168. doi: 10.1073/pnas.0903491106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perreault JP, Altman S. Important 2′-hydroxyl groups in model substrates for M1 RNA, the catalytic RNA subunit of RNase P from Escherichia coli. J Mol Biol. 1992;226:399–409. doi: 10.1016/0022-2836(92)90955-j. [DOI] [PubMed] [Google Scholar]
- 20.Kirsebom LA, Svard SG. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 1994;13:4870–4876. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svard SG, Kagardt U, Kirsebom LA. Phylogenetic comparative mutational analysis of the base-pairing between RNase P RNA and its substrate. RNA. 1996;2:463–472. [PMC free article] [PubMed] [Google Scholar]
- 22.Kurreck J, Wyszko E, Gillen C, Erdmann VA. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30:1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt KS, et al. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004;32:5757–5765. doi: 10.1093/nar/gkh862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fillion P, Desjardins A, Sayasith K, Lagace J. Encapsulation of DNA in negatively charged liposomes and inhibition of bacterial gene expression with fluid liposome-encapsulated antisense oligonucleotides. Biochim Biophys Acta. 2001;1515:44–54. doi: 10.1016/s0005-2736(01)00392-3. [DOI] [PubMed] [Google Scholar]
- 25.Eriksson M, Nielsen PE, Good L. Cell permeabilization and uptake of antisense peptide-peptide nucleic acid (PNA) into Escherichia coli. J Biol Chem. 2002;277:7144–7147. doi: 10.1074/jbc.M106624200. [DOI] [PubMed] [Google Scholar]
- 26.Harth G, Zamecnik PC, Tang JY, Tabatadze D, Horwitz MA. Treatment of Mycobacterium tuberculosis with antisense oligonucleotides to glutamine synthetase mRNA inhibits glutamine synthetase activity, formation of the poly-L-glutamate/glutamine cell wall structure, and bacterial replication. Proc Natl Acad Sci USA. 2000;97:418–423. doi: 10.1073/pnas.97.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harth G, Zamecnik PC, Tabatadze D, Pierson K, Horwitz MA. Hairpin extensions enhance the efficacy of mycolyl transferase-specific antisense oligonucleotides targeting Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2007;104:7199–7204. doi: 10.1073/pnas.0701725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurupati P, Tan KS, Kumarasinghe G, Poh CL. Inhibition of gene expression and growth by antisense peptide nucleic acids in a multiresistant beta-lactamase-producing Klebsiella pneumoniae strain. Antimicrob Agents Chemother. 2007;51:805–811. doi: 10.1128/AAC.00709-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikravesh A, et al. Antisense PNA accumulates in Escherichia coli and mediates a long post-antibiotic effect. Mol Ther. 2007;15:1537–1542. doi: 10.1038/sj.mt.6300209. [DOI] [PubMed] [Google Scholar]
- 30.Tilley LD, Mellbye BL, Puckett SE, Iversen PL, Geller BL. Antisense peptide-phosphorodiamidate morpholino oligomer conjugate: dose-response in mice infected with Escherichia coli. J Antimicrob Chemother. 2007;59:66–73. doi: 10.1093/jac/dkl444. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 32.Sekiguchi M, Iida S. Mutants of Escherichia coli permeable to actinomycin. Proc Natl Acad Sci USA. 1967;58(6):2315–2320. doi: 10.1073/pnas.58.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarno R, Ha H, Weinsetel N, Tolmasky ME. Inhibition of aminoglycoside 6′-N-acetyltransferase type Ib-mediated amikacin resistance by antisense oligodeoxynucleotides. Antimicrob Agents Chemother. 2003;47:3296–3304. doi: 10.1128/AAC.47.10.3296-3304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Guerrier-Takada C, Altman S. Targeted cleavage of mRNA in vitro by RNase P from Escherichia coli. Proc Natl Acad Sci USA. 1992;89:3185–3189. doi: 10.1073/pnas.89.8.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.