Abstract
The human parasite Plasmodium falciparum enzymatically digests hemoglobin during its intra-erythrocytic developmental stages in acidic food vacuole compartments. The released heme is rapidly detoxified by polymerization into the chemically inert pigment, hemozoin. Several heme-binding anti-malarial compounds, such as chloroquine, efficiently inhibit this process, and this is believed to be the predominant mechanism by which these drugs induce parasite toxicity. In an effort to expand the biochemical tools available for exploration of this pathogen's basic biology, we chose this heme-detoxification pathway as a model system for exploring the suitability of DNA aptamers for modulating this essential parasite biochemical pathway. In this report, we demonstrate that heme-binding DNA aptamers efficiently inhibit in vitro hemozoin formation catalyzed by either a model lipid system or parasite-derived extracts just as or more potently than chloroquine. Furthermore, when parasites are grown in red cells loaded with heme-binding aptamers, their growth is significantly inhibited relative to parasites exposed to non-heme-binding DNA oligonucleotides. Both the timing of parasite-induced toxicity and the concentration of heme-binding aptamer required for inducing toxicity correlate well with the uptake of red cell cytosolic components by the parasite, and the requirement for compounds with similar in vitro hemozoin inhibitory potency to preconcentrate within the parasite before observing toxicity. Thus, these heme-binding aptamers recapitulate the in vitro hemozoin inhibition activity and induce parasite toxicity in a manner consistent with inhibition of this pathway. Altogether, these data demonstrate that aptamers can be versatile tools with applicability in functionally dissecting important P. falciparum-specific pathways both in vitro and in vivo.
Each year, there are between 200–600 million cases of human malaria and close to 1 million resulting deaths (1, 2). The most severe form of the disease is caused by Plasmodium falciparum. Despite much effort, a vaccine has remained elusive, and disease treatment relies heavily on the use of anti-malarial drugs. However, resistance is becoming increasingly widespread (3, 4), making it critical to identify additional therapeutic targets. Unfortunately, only a very limited toolkit is available for elucidating biological function and validating potential drug targets in this organism.
Gene knockout strategies (5) have been used most commonly, but these are limited to studying non-essential genes. The applicability of RNA interference (RNAi), which has been extremely important in elucidating gene function in several eukaryotic organisms, is still being debated. Several groups have reported successfully using double-stranded RNA to specifically knock down target genes such as falcipains-1 and 2 (6, 7), dihydroorotate dehydrogenase (8), PfMyb1, a transcription factor (9), and several genes putatively involved in parasite infected RBC induction of endothelial cell apoptosis (10). However, whether this is mediated via RNAi per se or anti-sense mechanisms remains unclear, especially in light of bioinformatics data suggesting that key conserved components of the RNAi machinery are not encoded in the P. falciparum genome (11, 12). Inducible gene expression systems, unfortunately, are not routinely available for robustly controlling gene activity in functional studies with P. falciparum. However, there has been recent progress in this area with reports on a tetracycline-inducible transcription regulation system (13), a Shld1-regulatable FKBP destabilization domain system for regulating degradation of a target protein (14), and the use of a toyocamycin-regulated ribozyme for regulating a target mRNA (15). Presently, it is difficult to predict whether these strategies will facilitate full functional evaluation of the P. falciparum essential gene repertoire. Chemical biology approaches are increasingly being used to glean new insights into P. falciparum biology. Activity based probes have been used to identify and investigate the role of falcipain-1 in parasite invasion of RBCs (16) and that of the subtilisin-family serine protease, PfSUB1, and cysteine protease dipeptidyl peptidase in parasite release from infected RBCs (17). Similarly, selected enzyme targets are also being screened against large chemical libraries to identify specific inhibitors that can be used to modulate the activity of this target in situ and potentially serve as lead compounds in drug discovery efforts. A recent study used this approach to identify a family of 2,6,9-trisubstituted purines that implicated the calcium-dependent protein kinase 1 (PfCDPK1) in parasite motility and release from infected RBCs (18).
In an effort to extend the available chemical approaches for studying P. falciparum, we have initiated studies using nucleic acid aptamers, which are typically selected from extremely large libraries (≥ 1013 typically) using the easily accessible SELEX methodology (19–21). The potential for easy access to aptamers modulating the function of virtually any target of biological interest makes this approach particularly appealing, and indeed, successful regulation of many cellular targets (22) have previously been reported. Here, we demonstrate that aptamers can be successfully used to modulate an essential, P. falciparum-specific metabolic pathway.
Results and Discussion
Heme-Binding Aptamers Inhibit Hemozoin Formation.
During the intra-erythrocytic development of P. falciparum, Hb is taken up into vacuolar structures that are acidified to a pH between 4.5–5.5 (23–25), where it is sequentially digested by several proteases (26), and the released heme polymerized to hemzoin. Therefore, the initial objective was to examine whether heme-binding aptamers could first, bind heme at this pH, and second, inhibit hemozoin formation. For this proof-of-concept study, we elected to use several previously reported heme-binding DNA aptamers (27, 28) (Table 1), which were all discovered using SELEX carried out at pH 7–8.
Table 1.
The oligonucleotide sequences used in this study are summarized, along with apparent heme binding dissociation constants (KD) and IC50 values for hemozoin inhibition
| Name | Sequence | KD, μM | IC50, μM |
|---|---|---|---|
| PS2R | GCC CCC CCT CGT CGT CGA CGG TA | ND | ND |
| PS2M_Mod | GTG AAT AGA CGC GCT TGG AC | ND | ND |
| PS2M (29) | GTG GGT AGG GCG GGT TGG TA | 1.4 | 35 |
| PS21 (29) | ACG GGT AGG GTT TGG GCA GGA T | 1.2 | 43 |
| PS26 (29) | GAC GGG GTT GGG TCC GGG CGG TAG G | 2.6 | 28 |
| OKA_26–5 (30) | ATG GGG TCG GGC GGG CCG GGT GTC AT | 1.2 | 21 |
| OKA_26–3 (30) | ATG GTG GAC GGA GAT GGG ACG TAG | 0.5 | 47 |
| CQ | Chloroquine | 5 (31) | 13 |
Sequences shown in bold italic do not bind heme, whereas heme-binding sequences are shown in bold. ND, Not Detectable.
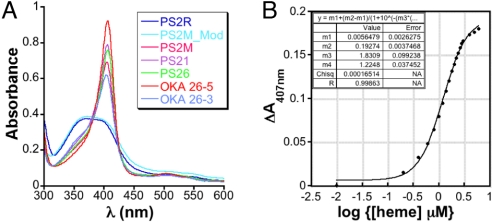
The heme-binding properties of these aptamers were studied at pH 5.2 by electron absorption (UV-visible) spectroscopy. Qualitatively, the aptamers induced a sharp, hyperchromic heme Soret with a bathochromic shift to 408 nm indicative of a single spectroscopic species and a specific mode of heme binding to the aptamers, while the heme Soret with control DNA sequences PS2R (random DNA sequence) and PS2M_Mod (permuted PS2M sequence) remained broad with maxima at 396 nm, which is typical for unbound heme in solution (Fig. 1A). These spectral properties are identical to those observed at pH 7, suggesting that pH does not significantly impact the nature of the interaction between the aptamer and heme.
Fig. 1.
(A) Absorption spectra of aptamers and control oligonucleotides (10 μM) incubated with heme (10 μM) at pH 5. The heme binding aptamers in complex with heme exhibit a Soret at approximately 408 nm, while the non-heme binding oligonucloetides have a broad Soret centered around 396 nm. (B) Spectral titration of OKA_26–5 with heme.
To obtain a more quantitative measure of the affinity of the DNA aptamers for heme, attempts were made to carry out UV-visible monitored heme titrations at pH 5.2. Unfortunately, the poor solubility of heme at this pH prevented these studies. However, heme is sufficiently soluble at pH 7 and since the aptamer-heme complex had virtually identical spectra at pH 5 and 7, titrations were carried out at pH 7 instead. A typical titration curve obtained in these experiments and fit to Eq. 1 (Materials and Methods) is shown in Fig. 1B. From these data, dissociation constants ranging between 0.5–3 μM were obtained (Table 1). Not surprisingly, these dissociation constants are tightly clustered, most likely a reflection of the similarity in the selection pressure applied during SELEX. Importantly, these dissociation constants are very similar to the 5 μM previously reported for chloroquine (29), suggesting these aptamers might inhibit hemozoin formation with similar efficacy. Altogether, these data indicate that the selected heme-binding aptamers continue to bind heme at pH 5, and at pH 7 have dissociation constants similar to the heme-chloroquine complex.
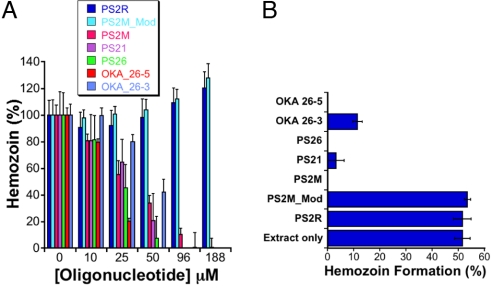
Next, we examined whether aptamers interfere with hemozoin formation in vitro using 1-mono-oleoylglycerol (MOG) (30) as an initiator. In these assays, MOG and heme were co-incubated in pH 5.2 buffer and the nucleic acid concentration titrated to attain the desired final concentration. After incubating overnight at ambient temperatures, the amount of heme remaining in the supernatants of each reaction was quantified spectrophotometrically. Hemozoin formation resulted in a decrease in the amount of free heme present in the supernatant, and this was quantified relative to a control reaction from which the lipid catalyst had been omitted. As shown in Fig. 2A, hemozoin formation is significantly and specifically inhibited by the heme-binding aptamers and not the control DNA oligonucleotides. Similar to CQ, these compounds lead to a concentration-dependent reduction in hemozoin formation. In contrast, the non-heme binding sequences PS2R and PS2M_Mod did not significantly impact hemozoin formation. The IC50 for aptamer inhibition of hemozoin formation ranged from 21–47 μM (Table 1), and is comparable to that for CQ (13 μM). In addition to efficaciously inhibiting hemozoin formation in the MOG-initiated assay, we also showed that these heme-binding aptamers similarly interfere with hemozoin formation catalyzed by late ring-early trophozoite stage-derived parasite extracts (Fig. 2B). These results demonstrate that aptamers interfere with both model assay and, more importantly, parasite-based hemozoin formation mechanisms, indicating they can potentially inhibit this process in vivo.
Fig. 2.
(A) Aptamer-mediated inhibition of hemozoin formation using MOG as an initiator. (B) Hemozoin formation catalyzed by parasite lysates is specifically inhibited by heme-binding aptamers, but not by control oligonucleotides that do not bind heme.
Nuclease-Resistant Aptamers Retain Their Heme-Binding Affinity.
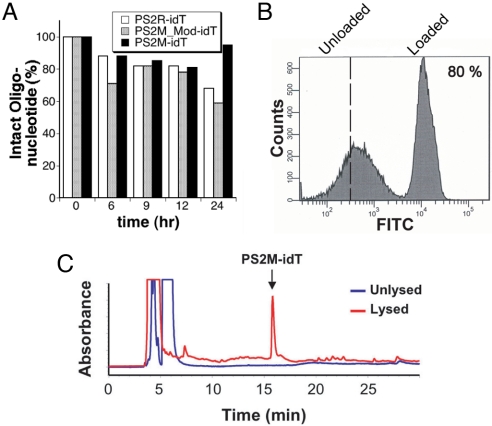
In preparation for evaluating the impact of these heme-binding aptamers on cultured P. falciparum, both aptamers and selected oligonucleotides were modified with 3′-inverted deoxythymidine (idT) to protect them against nuclease degradation. This single modification, which is readily incorporated during solid phase DNA synthesis, has previously been used to confer significant nuclease resistance to several target DNA oligonucleotides (31–33). The stabilities of the control oligonucleotides PS2R-idT and PS2M_Mod-idT, and the heme-binding aptamer PS2M-idT were evaluated in buffered, nuclease-rich 10% FBS. Samples were incubated for 0, 6, 9, 12, and 24 h at 37 °C, and aliquots were removed and analyzed by C18 reverse-phase chromatography. Areas under the peaks corresponding to the sample DNA were determined, and peak areas were normalized to time = 0. These results, shown in Fig. 3A, demonstrate that after 24 h, only 20–40% degradation had occurred, confirming that this modification leads to significant protection of our test samples. Importantly, this level of protection is well suited to the timescale of the parasite growth experiments (vide infra). Having verified the stability of the 3′-idT aptamer and control oligonucleotides, we confirmed that this modification had not significantly impacted the native heme-binding properties of the aptamer by doing UV-visible spectral heme titrations with PS2M-idT, as before. From these experiments, a KD = 1.9 ± 0.6 μM was determined for the binding interaction of PS2M-idT with heme, in very good agreement with the KD = 1.4 μM determined for PS2M. Thus, this modification represents a facile strategy for stabilizing heme-binding aptamers without perturbing their binding properties.
Fig. 3.
(A) The 3′-idT derivatives of control oligonucleotides and heme-binding aptamers are resistant to nuclease degradation. (B) A typical histogram observed during erythrocyte loading with 5′-fluorescein labeled aptamers. (C) HPLC chromatograms of the supernatant from PS2M-loaded erythrocytes that are unlysed (blue) and lysed (red).
Aptamers Can Be Efficiently Loaded into RBCs.
Having prepared the nuclease-resistant heme-binding aptamers and control oligonucleotides, we sought to efficiently make them bioavailable to parasitized RBCs. We chose to encapsulate aptamers into RBCs using a hypotonic lysis and resealing (HL/R) method based on previous studies demonstrating that parasites infecting RBCs preloaded with non-toxic fluorescent or biotin-labeled dextrans accumulate these tracers in the acidic organelles in which Hb degradation and hemozoin formation occur (23, 34). The HL/R method consistently resulted in 65–85% of RBCs per sample being loaded with oligonucleotide as determined by FACS using 5′-fluorescein-labeled PS2M-idT (Fig. 3B). Using analytical HPLC in combination with the FACS data on loading efficiency, we confirmed that the average encapsulated nucleic acid concentration is similar to that present in the bulk solution. Additionally, specific encapsulation of loaded nucleic acids within erythrocytes was confirmed qualitatively by fluorescence microscopy and quantitatively by HPLC analysis (Fig. 3C) of the supernatant derived from preloaded cells deliberately lysed by hypotonic shock. As expected, only after lysis could aptamer be detected in the supernatant.
Parasite Growth in Heme-Binding Aptamer Loaded RBCs Is Inhibited.
Before examining the impact of aptamers on parasite growth, we confirmed that the HL/R method did not interfere with the ability of erythrocytes to support parasite growth. To evaluate this, sham-loaded erythrocytes (lysed and resealed in the absence of any nucleic acid) and similarly washed but unlysed erythrocytes were infected in parallel with late stage parasites. Parasite growth was determined at the end of 72 h by FACS. These data show that both sets of erythrocytes supported parasite growth equally well at 3 initial parasitemia levels tested (Fig. S1A), and hence this method for preloading erythrocytes is compatible with our experimental objectives.
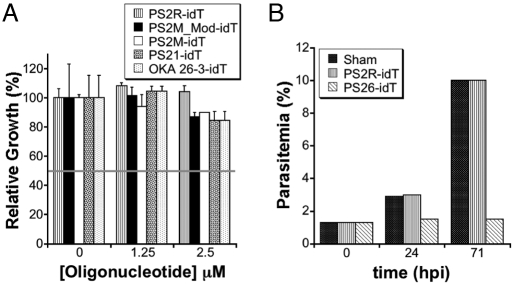
Parasite growth in erythrocytes preloaded with 2 control oligonucleotides, PS2R-idT and PS2M_Mod-idT, and 2 aptamers, PS2M-idT and PS26-idT were evaluated. The control oligonucleotides induced a modest but constant toxicity (≈20%) at the concentrations tested relative to sham-loaded cells (no oligonucleotide encapsulated during the HL/R procedure). On the other hand, a dose-dependent increase in aptamer-induced toxicity is observed relative to PS2R-idT-loaded cells (Fig. 4 and Fig. S2). This differential toxicity is unlikely to have arisen from residual aptamer bound to the external surface of the loaded RBCs. Indeed, oligonucleotides and other polyanionic macromolecules have been demonstrated to interfere with merozoite invasion into RBCs, with DNA oligonucleotides (10 μM) significantly inhibiting parasite growth (35, 36). Therefore, to conclusively eliminate this inhibitory mechanism, intact RBCs in isotonic buffer (no RBC lysis) were exposed to the same final concentrations of PS2R-idT, PS2M_Mod-idT, and PS2M-idT (300 μM), and washed exactly as the loaded cells used above. RBCs from all 3 exposures supported parasite growth equally well (Fig. S1B), confirming that the growth inhibitory effect observed with aptamer encapsulated RBCs is not simply the result of selective adhesion of residual amounts of aptamer to the external surface of loaded RBCs.
Fig. 4.

Heme-binding aptamers inhibit parasite growth compared with control oligonucleotides. * P value = 0.04; ** P value <0.001.
Similarly, oligonucleotide leakage from preloaded erythrocytes into the media and a subsequent decrease in invasion efficiency by the mechanisms described above is not a significant mechanism for the observed growth inhibition. By calculation, if RBCs at 0.5% hematocrit in a 200-μL culture are loaded with 300 μM nucleic acid at 60–80% efficiency, and complete leakage of encapsulated nucleic acid into the media is assumed, then the maximum attainable media nucleic acid concentration would be 0.9–1.2 μM. Therefore, we directly added control oligonucleotides and aptamers (0–2.5 μM) into both early and late stage parasite cultures. However, up to 2.5 μM added nucleic acid had no major impact on growth (Fig. 5A), and more importantly, the observed growth pattern did not recapitulate the dramatic differential toxicity observed between control oligonucleotides and heme-binding aptamers shown in Fig. 4. Altogether, these data demonstrate conclusively that the selective inhibitory effect of the heme-binding aptamers on parasite growth is entirely dependent upon interaction of the parasite with the RBC-encapsulated aptamers.
Fig. 5.
(A) Growth assays conducted in the presence of control oligonucleotides and selected aptamers added directly to complete media. The horizontal line indicates the extent of growth expected if this mechanism could account for the observed parasite toxicity. (B) Growth assays to examine the parasite stage affected by heme-binding aptamers preloaded into RBCs.
To gain further insight into the parasite stage(s) impacted by the aptamers, growth in sham, PS2M_Mod-idT and PS26-idT loaded erythrocytes was measured. At 24 h post invasion (ring/trophozoite stage), the total parasitemia in cultures containing sham-loaded and PS2M_Mod-idT-loaded erythrocytes increased to approximately 3% and 10% after the first and second invasion cycles, respectively, while minimal parasite expansion occurred in PS26-idT loaded erythrocyte cultures, even after 2 invasion cycles (Fig. 5B). Viable parasites progressing through the typical developmental stages are present in the latter culture, and this is likely a reflection of the fact that a fraction of RBCs are either unloaded or suboptimally loaded during the HL/R procedure. The heme-binding aptamers, therefore, are inducing their differential toxicity early in the parasite's intraerythrocytic developmental cycle, between invasion and ring/trophozoite stages. This is consistent with existing data on the timing of when the parasite begins to take up components from the red cell cytosol. While Hb degradation and hemozoin formation are maximal in the trophozoite and schizont stages of parasite development (26), there is substantial and growing evidence that these processes occur to a significant extent during the earlier ring stages in parasite development. Previous microscopy data indicate that early ring stage parasites are actively sampling the red cell cytosol, as demonstrated by their ability to take up non-toxic fluorescent and biotinylated dextrans from preloaded RBCs (23, 34); similarly, ultrastructural and biochemical studies provide convincing evidence for hemozoin formation in ring-stage parasites (26, 37–39). More recent work in this area describes 4 distinct pathways used by P. falciparum to take up RBC cytosolic components, namely: (i) the “big gulp” (BG); (ii) small Hb vacuoles (SHV); (iii) cytostomal tubes; and (iv) phagotrophic uptake (40). The BG and SHV mechanisms, in particular, are active during the early ring stages and could account for uptake of concentrations of preloaded aptamers inhibitory to hemozoin formation occurring during ring-stage parasites. Importantly, the SHV uptake mechanism begins at a very early time, and although it is a relatively low volume pathway, it must be noted that the steady-state aptamer concentration present in the cytosol of preloaded RBCs in which growth inhibition is observed is sufficiently high to prevent heme detoxification by polymerization to hemozoin, potentially resulting in early parasite toxicity. This is expected to be even more prominent once the higher volume BG pathway becomes active.
At first glance, there appear to be inherent differences between the heme-binding aptamers and chloroquine with respect to the temporal onset of parasite toxicity and their IC50 values, but a possible explanation may relate to the differences between the mechanisms by which these compounds become bioavailable to the parasite. Typically, chloroquine is added directly to parasite culture medium, and in sensitive strains will kill trophozoite stage parasites with an IC50 ranging from 2–30 nM (41, 42), in contrast with the approximate 150 μM observed for preloaded heme-binding aptamers. Because it is an amphipathic compound, chloroquine readily permeates RBC and parasite membranes to enter and accumulate in the acidic Hb digestive compartments of the parasite by: (i) protonation to become positively charged and membrane impermeable (43); and (ii) binding to heme released during Hb degradation (41). Thus, chloroquine exerts its toxic effects only when these 2 mechanisms combine to produce sufficiently high concentrations of drug inhibitory to hemozoin formation. Additionally, several studies have shown that parasites exposed to nanomolar concentrations will accumulate this drug to between 1,800–2,400 fold above media concentrations leading to concentrations in the micromolar range in the acidic digestive compartments (44). Thus, while chloroquine is toxic at media concentrations in the nanomolar range, at the site of action the drug has accumulated to micromolar levels, comparable with the concentrations at which heme-binding aptamers are inducing their toxicity. Since the aptamers are preloaded at a fixed concentration and directly sampled at this loaded concentration, levels compatible with inhibiting hemozoin formation are immediately present thus resulting in the onset of toxicity at earlier parasite developmental stages. Our data demonstrating aptamer-mediated inhibition of hemozoin formation by a model lipid-catalyzed system and particularly by parasite extracts, and the growth inhibitory effects of these aptamers when taken together with previously published studies as discussed above, can be combined into a model whereby heme-binding aptamers co-localize with the sites of hemozoin formation, with concurrent inhibition of this process.
While the heme-binding aptamers inhibit hemozoin formation in vitro and induce parasite toxicity in a manner consistent with inhibition of this process in situ, we cannot presently fully exclude additional toxicity mechanisms. As the heme-binding aptamers induce toxicity relatively early during parasite development in the RBC, it is possible that invasion defects may be contributing. We have conclusively ruled out inhibition resulting from disruption of merozoite-ligand interactions with RBC surface receptors, an important mechanism mediating the early stage of parasite invasion (45), and a proposed mechanism for oligonucleotide-mediated inhibition of parasite growth (35, 36). In the later stages of invasion, the merozoite invaginates into the RBC membrane and enters the RBC such that it remains completely enveloped in a portion of the RBC membrane (the parasitophorous vacuole) that maintains physical separation of the parasite from the RBC cytosol (45, 46) until the earlier described mechanisms facilitating communication between the parasite and RBC cytosol become active. Studies aimed at more precisely understanding the role that inhibition of this phase of invasion might play in the observed toxicity of these heme-binding aptamers are being pursued. Similarly, since heme is a critical cofactor for many cellular redox reactions, such as oxidative phosphorylation and energy production, we are also examining whether these aptamers interfere significantly with other aspects of cellular metabolism.
In conclusion, we have demonstrated that heme-binding DNA aptamers can specifically inhibit the essential P. falciparum heme detoxification pathway in vitro in assays using both a model lipid-catalyzed system and parasite-derived lysates retaining the native hemozoin formation components. Additionally, when preloaded into RBCs, these aptamers induce parasite toxicity in a manner consistent with inhibiting this process in early stage parasites. Here, we have focused on modulating a biophysical process specific to P. falciparum, but this strategy has the potential to be more generally applicable for specifically and robustly regulating the biological activity of a much wider range of physiologically important parasite-specific pathways. Therefore, aptamers may soon play an integral role in expanding the currently limited approaches available for elucidating biological mechanisms in a pathogen of immense global importance.
Materials and Methods
General reagents were analytical grade. Aqueous solutions were prepared in distilled, deionized water (Millipore) made nuclease free by treatment with 0.1% diethylpyrocarbonate (DEPC) (Sigma-Aldrich) at 37 °C overnight followed by autoclaving. Freshly prepared hemin (Sigma-Aldrich) stock solutions were prepared in DMSO, and their concentrations were determined in the same solvent using an extinction coefficient at 404 nm of 170 mM−1cm−1. Oligonucleotides (Integrated DNA Technologies) were dissolved in DEPC-H2O to make mM stocks that were kept frozen at −20 °C then thawed on ice when needed. Chloroquine diphosphate (Sigma-Aldrich) stocks were prepared in DEPC-H2O, and made fresh each day.
Spectroscopic Studies.
Absorption spectra were recorded in either 100 mM sodium acetate, 5 mM potassium chloride, 1 mM magnesium chloride, pH 5, or 20 mM HEPES, 120 mM sodium chloride, 5 mM potassium chloride, 1 mM magnesium chloride, 1 mM calcium chloride, pH 7 (SHMCK) buffers on a Cary 300 Bio UV-Visible spectrophotometer equipped with a Peltier (Varian Inc.) for temperature control and sample stirring as needed. Titrations were carried out by difference UV-Visible spectroscopy in a reaction volume of 2.5 mL SHMCK in stirred cuvettes at 25 °C. Heme (500 μM DMSO stock) was added simultaneously in 0.2-μM aliquots to reference (buffer only) and sample (2 μM aptamer) cuvettes, with 5-min equilibration time between additions. The ΔA408 nm was plotted versus [heme], and fitted to Eq. 1, where m0 = log{[heme] μM}, m1 = minimum A408 nm, m2 = maximum A408 nm, m3 = Hill coefficient, and m4 = apparent KD using Kaleidagraph version 3.6.4 for MacOS X (Synergy).
Hemozoin Inhibition Assays.
1-Mono-oleoylglycerol (MOG) (Sigma-Aldrich) was suspended to a final concentration of 150 μg/mL in 100 mM sodium acetate, 5 mM potassium chloride, 1 mM magnesium chloride, pH 5, buffer by sonicating and vortexing. The resulting emulsion was quickly transferred to untreated polystyrene round bottom 96-well plates (200 μL per well) (Corning) using a multichannel pipette. Turbidity measurements at 600 nm in a 96-well plate reader (Molecular Devices) were made to ascertain that similar MOG concentrations were deposited to each well. Hemin was added from a DMSO stock to a final concentration of approximately 50 μM per well. Aptamer and control oligonucleotides were added to obtain final concentrations of 0, 10, 25, 49, 96, and 188 μM. On each plate, a negative control was included in which MOG was omitted, along with a chloroquine (100 μM) positive control for hemozoin inhibition. Similar plates were prepared containing 0, 10, 25, 50, 75, and 200 μM chloroquine. The final DMSO concentration in all reactions was less than 2%, and did not interfere with hemozoin formation. Each reaction condition was done in triplicate or quadruplicate. Completed reaction mixtures were sealed to minimize evaporation, and agitated at medium speeds for 24 h at ambient temperature. At the end of the incubation period, 50 μL of a 70:30 200 mM HEPES, pH 7:pyridine solution was added and the plate shaken at medium speeds at room temperature for 10–15 min to allow selective dissolution of non-polymerized heme (47), and formation of the pyridine complex. After sitting for an additional 15 min to allow any suspended hemozoin to settle, 50 μL from each well were transferred to a flat bottom 96-well plate containing 150 μL 70:30 200 mM HEPES, pH 7:pyridine, and the absorbance measured at 405 nm. IC50 values were obtained by plotting A405 nm versus log {aptamer or chloroquine concentrations} and fitting to Eq. 2, where m0 = log{[aptamer] μM} or log {[chloroquine] μM}, m1 = minimum A405 nm, m2 = maximum A405 nm, m3 = Hill coefficient, and m4 = IC50, using Kaleidagraph.
Hemozoin Formation Catalyzed by Parasite Lysates.
Late ring-early trophozoite stage parasite cultures were collected by centrifugation, and intact parasites harvested by saponin lysis as previously described (48). Parasite pellets were stored at −80 °C until needed. Lysates were prepared by thawing pellets on ice, resuspending in 15 mL ice cold PBS in 50 mL Falcon tubes, and sonicating on ice (4 × 10-s pulses at power setting = 4 on a Misonix 3000 sonicator). This suspension was transferred to polypropylene tubes and centrifuged at 35,000 × g for 35 min at 4 °C. The supernatant was discarded and the pellet resuspended in 12 mL of 300 mM sodium acetate, 5 mM potassium chloride, 1 mM magnesium chloride, pH 5, buffer. This suspension was dispensed in 200-μL aliquots into 96-well round bottom plates with heme present at a final concentration of 50 μM. The control oligonucleotides (PS2R and PS2M_Mod) and heme-binding aptamers (PS2M, PS21, PS26, OKA_26–3, and OKA_26–5) were added to their respective wells in triplicate at 100 μM final concentration. Control wells with buffered heme (no lysate) and heme-containing lysate only (no oligonucleotides) were also included in triplicate. Plates were incubated at 37 °C with shaking for 12 h before measuring hemozoin formation as above.
Evaluation of Stability of Modified Oligonucleotides.
Three oligonucleotides, PS2R-idT, PS2M_Mod-idT, and PS2M-idT were incubated at 37 °C in 10% FBS in SHMCK. Aliquots were analyzed by HPLC (Agilent) on a 250 × 2.1 mm, 5 μM, 300 Å Columbus C18 column (Phenomenex) using 150 mM triethylamine acetate, pH 7 (Solvent A) and acetonitrile (Solvent B) as mobile phases. The column was eluted with 5% B for 5 min, a gradient from 5% to 25% B over 25 min, then back to 5% B at 32 min. The flow rate was 0.2 mL/min, and products were detected at 254 nm. A calibration curve generated using the appropriate authentic oligonucleotide was used to determine the amount of oligonucleotide recovered after RBC lysis.
Loading RBCs with Oligonucleotides.
RBCs were loaded with oligonucleotides using the hypotonic lysis with resealing (HL/R) method. This was carried out as previously described (23), except that the magnesium chloride and ATP concentrations were 5 mM, and a 5-min incubation step added after RBC suspension was returned to isotonicity. Resealed cells were washed sequentially with 2 × 12 mL PBS, 1 × 12 mL RPMI 1640 wash, and 1 × 12 mL complete RPMI 1640. This media was removed, and cells resuspended in fresh complete RPMI 1640 to 2% hematocrit, and used immediately in growth assays as needed. All manipulations were carried out under sterile conditions.
Parasite Cultures and Determination of Parasitemia.
3D7 parasites were maintained in RPMI 1640 media (minus glutamine and sodium bicarbonate) (Invitrogen) supplemented with 25 mM sodium bicarbonate (Sigma-Aldrich), 100 μM hypoxanthine (Sigma-Aldrich), 2.5 g/L Albumax II (Invitrogen), and 10 μg/mL gentamicin at 2% hematocrit in incubators maintained at 90% N2, 5% CO2, 5% O2, and 37 °C. Synchronization was achieved by treating parasite pellets with 5% sorbitol for 5–10 min at 37 °C. Routine parasitemia and staging were determined by Giemsa staining. High throughput parasitemia measurements used FACS (LSR II, BD Biosciences) and 3D7 parasites stably transfected with the PfGNr plasmid [Malaria Research and Reference Reagent Resource (MR4)], and expressing GFP. Assays to assess the impact of aptamers on parasite growth were done in 96-well plate format at 0.5% hematocrit in media containing between 250–400 μg/mL geneticin sulfate, and inoculated at initial parasitemias of 0.4%, 0.8%, and 1.6% of synchronous ring or late stage parasites as needed, in 200-μL cultures. If the final parasitemia attained did not increase proportionally with the seeding parasitemia, the experiment was repeated. Each condition was tested in triplicate, and experiments with control oligonucleotide- and aptamer-loaded cells were done at the same time on the same plate. No attempt was made to separate parasitized from non-parasitized RBCs since the inoculating culture did not change the overall percent of uninfected cells in the final culture significantly. Cultures were incubated for about 72 h before diluting to 0.25% hematocrit, fixing for 30–60 min in 1% paraformadehyde at room temperature and FACS analysis.
Supplementary Material
Acknowledgments.
This work was supported by an National Institutes of Health Postdoctoral Fellowship (J.C.N.), Howard Hughes Medical Institute (J.L.D), and the Aldo DeBenedictis Fund (M.A.M).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906370106/DCSupplemental.
References
- 1.Snow RW, et al. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. World Malaria Report 2008. Switzerland: Geneva; 2008. [Google Scholar]
- 3.Franco-Paredes C, Santos-Preciado JI. Problem pathogens: Prevention of malaria in travelers. Lancet Infect Dis. 2006;6:139–149. doi: 10.1016/S1473-3099(06)70410-8. [DOI] [PubMed] [Google Scholar]
- 4.Hyde JE. Drug-resistant malaria. Trends Parasitol. 2005;21:494–498. doi: 10.1016/j.pt.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Koning-Ward TF, Janse CJ, Waters AP. The development of genetic tools for dissecting the biology of malaria parasites. Annu Rev Microbiol. 2000;54:157–185. doi: 10.1146/annurev.micro.54.1.157. [DOI] [PubMed] [Google Scholar]
- 6.Dasaradhi PVN, et al. A role of falcipain-2, principal cysteine proteases of Plasmodium falciparum in merozoite egression. Biochem Biophys Res Commun. 2005;336:1062–1068. doi: 10.1016/j.bbrc.2005.08.213. [DOI] [PubMed] [Google Scholar]
- 7.Malhotra P, et al. Double-stranded RNA-mediated gene silencing of cysteine proteases (falcipain-1 and-2) of Plasmodium falciparum. Mol Microbiol. 2002;45:1245–1254. doi: 10.1046/j.1365-2958.2002.03105.x. [DOI] [PubMed] [Google Scholar]
- 8.McRobert L, McConkey GA. RNA interference (RNAi) inhibits growth of Plasmodium falciparum. Mol Biochem Parasit. 2002;119:273–278. doi: 10.1016/s0166-6851(01)00429-7. [DOI] [PubMed] [Google Scholar]
- 9.Gissot M, et al. PfMyb1, a Plasmodium falciparum transcription factor, is required for intra-erythrocytic growth and controls key genes for cell cycle regulation. J Mol Biol. 2005;346:29–42. doi: 10.1016/j.jmb.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 10.Siau A, et al. Whole-transcriptome analysis of Plasmodium falciparum field isolates: Identification of new pathogenicity factors. J Infect Dis. 2007;196:1603–1612. doi: 10.1086/522012. [DOI] [PubMed] [Google Scholar]
- 11.Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: Genomic gleanings. Cell. 2003;115:771–785. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- 12.Ullu E, Tschudi C, Chakraborty T. RNA interference in protozoan parasites. Cell Microbiol. 2004;6:509–519. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- 13.Meissner M, et al. Tetracycline analogue-regulated transgene expression in Plasmodium falciparum blood stages using Toxoplasma gondii transactivators. Proc Natl Acad Sci USA. 2005;102:2980–2985. doi: 10.1073/pnas.0500112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armstrong CM, Goldberg DE. An FKBP destabilization domain modulates protein levels in Plasmodium falciparum. Nat Methods. 2007;4:1007–1009. doi: 10.1038/nmeth1132. [DOI] [PubMed] [Google Scholar]
- 15.Agop-Nersesian C, Pfahler J, Lanzer M, Meissner M. Functional expression of ribozymes in Apicomplexa: Towards exogenous control of gene expression by inducible RNA-cleavage. Int J Parasitol. 2008;38:673–681. doi: 10.1016/j.ijpara.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 16.Greenbaum DC, et al. A role for the protease falcipain 1 in host cell invasion by the human malaria parasite. Science. 2002;298:2002–2006. doi: 10.1126/science.1077426. [DOI] [PubMed] [Google Scholar]
- 17.Arastu-Kapur S, et al. Identification of proteases that regulate erythrocyte rupture by the malaria parasite Plasmodium falciparum. Nat Chem Biol. 2008;4:203–213. doi: 10.1038/nchembio.70. [DOI] [PubMed] [Google Scholar]
- 18.Kato N, et al. Gene expression signatures and small-molecule compounds link a protein kinase to Plasmodium falciparum motility. Nat Chem Biol. 2008;4:347–356. doi: 10.1038/nchembio.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellington AD, Szostak JW. Selection in vitro ff single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;355:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 20.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment - RNA ligands to bacteriophage-T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 21.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 22.Famulok M, Blind M, Mayer G. Intramers as promising new tools in functional proteomics. Chem Biol. 2001;8:931–939. doi: 10.1016/s1074-5521(01)00070-9. [DOI] [PubMed] [Google Scholar]
- 23.Krogstad DJ, Schlesinger PH, Gluzman IY. Antimalarials increase vesicle pH in Plasmodium falciparum. J Cell Biol. 1985;101:2302–2309. doi: 10.1083/jcb.101.6.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yayon A, Cabantchik ZI, Ginsburg H. Identification of the acidic compartment of Plasmodium falciparum-infected human erythrocytes as the target of the antimalarial drug chloroquine. EMBO J. 1984;3:2695–2700. doi: 10.1002/j.1460-2075.1984.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klonis N, et al. Evaluation of pH during cytostomal endocytosis and vacuolar catabolism of haemoglobin in Plasmodium falciparum. Biochem J. 2007;407:343–354. doi: 10.1042/BJ20070934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Francis SE, Sullivan DJ, Goldberg DE. Hemoglobin metabolism in the malaria parasite Plasmodium falciparum. Annu Rev Microbiol. 1997;51:97–123. doi: 10.1146/annurev.micro.51.1.97. [DOI] [PubMed] [Google Scholar]
- 27.Li YF, Geyer CR, Sen D. Recognition of anionic porphyrins by DNA aptamers. Biochemistry. 1996;35:6911–6922. doi: 10.1021/bi960038h. [DOI] [PubMed] [Google Scholar]
- 28.Okazawa A, et al. In vitro selection of hematoporphyrin binding DNA aptamers. Bioorg Med Chem Lett. 2000;10:2653–2656. doi: 10.1016/s0960-894x(00)00540-0. [DOI] [PubMed] [Google Scholar]
- 29.Kelly JX, Winter R, Riscoe M, Peyton DH. A spectroscopic investigation of the binding interactions between 4,5-dihydroxyxanthone and heme. J Inorg Biochem. 2001;86:617–625. doi: 10.1016/s0162-0134(01)00217-3. [DOI] [PubMed] [Google Scholar]
- 30.Fitch CD, Cai GZ, Chen YF, Shoemaker JD. Involvement of lipids in ferriprotoporphyrin IX polymerization in malaria. Biochim Biophys Acta. 1999;1454:31–37. doi: 10.1016/s0925-4439(99)00017-4. [DOI] [PubMed] [Google Scholar]
- 31.Dass CR, Saravolac EG, Li Y, Sun LQ. Cellular uptake, distribution, and stability of 10–23 deoxyribozymes. Antisense Nucleic Acid Drug Dev. 2002;12:289–299. doi: 10.1089/108729002761381276. [DOI] [PubMed] [Google Scholar]
- 32.Ortigao JF, et al. Antisense effect of oligodeoxynucleotides with inverted terminal internucleotidic linkages: A minimal modification protecting against nucleolytic degradation. Antisense Res Dev. 1992;2:129–146. doi: 10.1089/ard.1992.2.129. [DOI] [PubMed] [Google Scholar]
- 33.Sun LQ, et al. Suppression of smooth muscle cell proliferation by a c-myc RNA-cleaving deoxyribozyme. J Biol Chem. 1999;274:17236–17241. doi: 10.1074/jbc.274.24.17236. [DOI] [PubMed] [Google Scholar]
- 34.Goodyer ID, et al. Characterization of macromolecular transport pathways in malaria-infected erythrocytes. Mol Biochem Parasit. 1997;87:13–28. doi: 10.1016/s0166-6851(97)00039-x. [DOI] [PubMed] [Google Scholar]
- 35.Kanagaratnam R, Misiura K, Rebowski G, Ramasamy R. Malaria merozoite surface protein antisense oligodeoxynucleotides lack antisense activity but function as polyanions to inhibit red cell invasion. Int J Biochem Cell B. 1998;30:979–985. doi: 10.1016/s1357-2725(98)00051-x. [DOI] [PubMed] [Google Scholar]
- 36.Ramasamy R, et al. Anti-sense oligodeoxynucleoside phosphorothioates nonspecifically inhibit invasion of red blood cells by malaria parasites. Biochem Biophys Res Commun. 1996;218:930–933. doi: 10.1006/bbrc.1996.0164. [DOI] [PubMed] [Google Scholar]
- 37.Asawamahasakda W, et al. Effects of antimalarials and protease inhibitors on plasmodial hemozoin production. Mol Biochem Parasit. 1994;67:183–191. doi: 10.1016/0166-6851(94)00128-6. [DOI] [PubMed] [Google Scholar]
- 38.Bannister LH, et al. Three-dimensional ultrastructure of the ring stage of Plasmodium falciparum: Evidence for export pathways. Microsc Microanal. 2004;10:551–562. doi: 10.1017/S1431927604040917. [DOI] [PubMed] [Google Scholar]
- 39.Orjih AU, Fitch CD. Hemozoin production by Plasmodium falciparum - Variation with strain and exposure to chloroquine. Biochim Biophys Acta. 1993;1157:270–274. doi: 10.1016/0304-4165(93)90109-l. [DOI] [PubMed] [Google Scholar]
- 40.Elliott DA, et al. Four distinct pathways of hemoglobin uptake in the malaria parasite Plasmodium falciparum. Proc Natl Acad Sci USA. 2008;105:2463–2468. doi: 10.1073/pnas.0711067105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bray PG, et al. Cellular uptake of chloroquine is dependent on binding to ferriprotoporphyrin IX and is independent of NHE activity in Plasmodium falciparum. J Cell Biol. 1999;145:363–376. doi: 10.1083/jcb.145.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gluzman IY, Schlesinger PH, Krogstad DJ. Inoculum effect with chloroquine and Plasmodium falciparum. Antimicrob Agents Chemother. 1987;31:32–36. doi: 10.1128/aac.31.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yayon A, Cabantchik ZI, Ginsburg H. Susceptibility of human malaria parasites to chloroquine is pH dependent. Proc Natl Acad Sci USA. 1985;82:2784–2788. doi: 10.1073/pnas.82.9.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bray PG, Hawley SR, Ward SA. 4-aminoquinoline resistance of Plasmodium falciparum: Insights from the study of amodiaquine uptake. Mol Pharmacol. 1996;50:1551–1558. [PubMed] [Google Scholar]
- 45.Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–766. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Sibley LD. Intracellular parasite invasion strategies. Science. 2004;304:248–253. doi: 10.1126/science.1094717. [DOI] [PubMed] [Google Scholar]
- 47.Ncokazi KK, Egan TJ. A colorimetric high-throughput beta-hematin inhibition screening assay for use in the search for antimalarial compounds. Anal Biochem. 2005;338:306–319. doi: 10.1016/j.ab.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 48.Menting JGT, Coppel RL. In: Malaria Methods and Protocols. Doolan D, editor. Totowa: Humana Press Inc.; 2002. pp. 587–605. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.