Abstract
For gene products that must be present in cells at defined concentrations, expression levels must be tightly controlled to ensure robustness against environmental, genetic, and developmental noise. By studying the regulation of the concentration-sensitive Drosophila melanogaster Hox gene Ultrabithorax (Ubx), we found that Ubx enhancer activities respond to both increases in Ubx levels and genetic background. Large, transient increases in Ubx levels are capable of silencing all enhancer input into Ubx transcription, resulting in the complete silencing of this gene. Small increases in Ubx levels, brought about by duplications of the Ubx locus, cause sporadic silencing of subsets of Ubx enhancers. Ubx enhancer silencing can also be induced by outcrossing laboratory stocks to D. melanogaster strains established from wild flies from around the world. These results suggest that enhancer activities are not rigidly determined, but instead are sensitive to genetic background. Together, these findings suggest that enhancer silencing may be used to maintain gene product levels within the correct range in response to natural genetic variation.
Author Summary
Gene expression is generally governed by cis-regulatory elements, also called enhancers. For genes whose expression levels must be tightly controlled, enhancer activities must be tightly regulated. In this work, we show that enhancers that control the expression of the Hox gene Ultrabithorax (Ubx) in Drosophila are regulated by a negative autoregulatory feedback mechanism. Negative autoregulation can be triggered by less than a two-fold increase in Ubx levels or by varying the genetic background. Together, these data reveal that enhancer activities are not always hardwired, but instead may be sensitive to genetic and environmental variation and, in some cases, to the amount of gene product they regulate. The finding that enhancers are sensitive to genetic background suggests that the regulation of gene expression is more plastic than previously thought and has important implications for how transcription is controlled in vivo.
Introduction
The transcriptional control of gene expression in eukaryotes is governed by cis-regulatory elements, also known as enhancers, that integrate cell-type and temporal information by binding combinations of transcription factors. Genes that exhibit complex expression patterns are typically controlled by multiple cis-regulatory elements, some of which have overlapping, partially redundant activities [1],[2],[3],[4]. Current estimates suggest that from 10 to 80% of the non-coding DNA of higher eukaryotes is devoted to gene regulation [5],[6],[7], raising the question of how all of this regulatory information is integrated to generate accurate and stereotyped patterns of gene expression in space and time. A third dimension of gene regulation is quantity, which is especially relevant for genes that must be expressed within a narrow range of levels. One possible solution is that enhancers are precisely tuned to generate the appropriate level of transcription that is required in each cell. However, the precision that this type of mechanism demands seems difficult to achieve and especially vulnerable to genetic, environmental, and developmental noise. An alternative solution is that feedback or other regulatory mechanisms exist that modulate enhancer activities in response to the levels of gene product. Although feedback autoregulation is a well-known motif in transcriptional networks [8], mechanisms that might be used to tune expression levels are not well understood. This problem is particularly challenging for genes that have multiple, partially redundant regulatory inputs.
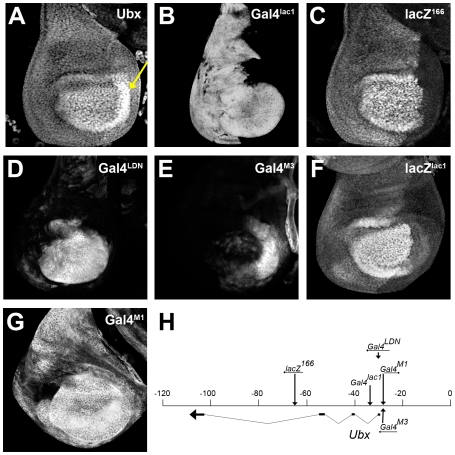
We have begun to study this problem in the fruit fly, Drosophila melanogaster, by analyzing the mechanisms that control the expression of the Hox gene Ultrabithorax (Ubx) in the haltere–a dorsal appendage on the third thoracic segment (T3) that helps the fly balance during flight [9]. Although Ubx protein is detected in all cells of the developing haltere imaginal disc, its pattern of expression is not uniform [10] (Figure 1A). Subsets of the complex regulatory input into the Ubx locus can be monitored by examining the expression patterns of Ubx enhancer traps, which exhibit different, overlapping subsets of the Ubx expression pattern (Figure 1). Ubx-Gal4lac1, for example, (monitored with UAS-GFP) is expressed uniformly throughout the anterior (A) compartment of the haltere disc, but only in the distal portion of the posterior (P) compartment (Figure 1B). In contrast, Ubx-Gal4LDN is expressed in distal regions (in both the A and P compartments) but is not expressed proximally (Figure 1D).
Figure 1. Ubx enhancer traps.
(A) Haltere disc stained for Ubx protein. Note the higher levels in the center of the disc and in the P compartment (arrow). (B–G) Patterns of Ubx enhancer trap expression in wild type haltere discs. The Gal4 inserts were monitored using a UAS-GFP transgene. (H) Map of the Ubx locus showing the location of the Ubx enhancer traps as described previously [28],[29],[30].
Results/Discussion
Ubx negative autoregulation
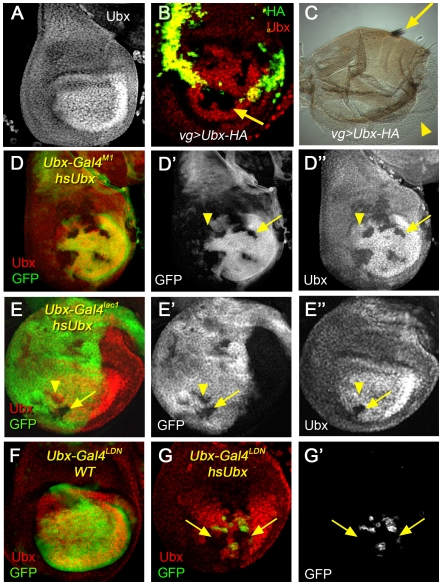
Somewhat paradoxically, transient ectopic expression of Ubx, induced either by heat shock or Gal4-mediated expression, resulted in Ubx loss-of-function transfomations that can be visualized both in the adult (as haltere to wing transformations; [11]) and in 3rd instar haltere imaginal discs (as groups of cells that showed a reduction or complete loss of Ubx protein) [12] (Figure 2). Thus, a transient pulse of high Ubx protein levels can lead to the complete and heritable silencing of all Ubx expression, implying that Ubx is being silenced by its own gene product.
Figure 2. Ubx enhancer silencing in response to hs-Ubx.
(A) Wild type haltere disc stained for Ubx protein. Note the higher levels in the distal region. (B) Haltere disc in which an HA-tagged Ubx protein was expressed via the vg-Gal4 driver, which is transiently expressed in all haltere cells. The disc was stained for HA (green) and Ubx (red). At this stage, the vg-Gal4 driver is active along the dorso-ventral boundary (strong green and yellow stain). Groups of cells that do not stain for Ubx (arrow) are observed. (C) Adult haltere from a vg>Ubx fly showing a transformation from haltere to wing. Both wing margin (arrow) and wing blade (arrowhead) tissue is observed. (D,E) Ubx-Gal4M1 (D) and Ubx-Gal4lac1 (E) haltere discs that were given a transient pulse of Ubx expression by heat shock during the 2nd instar, stained for GFP (green, to monitor enhancer trap activities) and Ubx (red). Some cells no longer express the enhancer traps and Ubx (arrows). Some cells no longer express the enhancer traps, but still express Ubx (arrowheads). (F) Wild type Ubx-Gal4LDN haltere disc stained for GFP (green, to monitor the enhancer trap) and Ubx (red). (G) A Ubx-Gal4LDN haltere disc that was given a transient pulse of Ubx expression by heat shock during the 2nd instar, stained for Ubx (red) and GFP (green, to monitor the enhancer trap). Silencing of both Ubx and the enhancer trap are observed (arrows). Surrounding the Ubx silenced cells, some cells have reduced Ubx levels but still express the enhancer trap.
Transient pulses of ectopic Ubx also resulted in the stable silencing of Ubx enhancer traps, including Ubx-Gal4lac1, Ubx-Gal4M1, Ubx-Gal4LDN, and Ubx-lacZ166 (Figure 2 and Table S1). When the absence of Ubx protein was observed, these cells also had no enhancer trap expression (Figure 2). However, in many cases enhancer trap silencing was observed in cells that had normal Ubx protein levels (Figure 2). In these cases we suggest that only the enhancers captured by the enhancer trap were silenced, and that other, partially redundant, enhancers in the Ubx locus remained active, resulting in an apparently normal pattern of Ubx expression. We also find, consistent with previous results [12], that the patches of Ubx-silenced cells in the haltere are clonal events and that the Polycomb system of epigenetic regulators is required for silencing (Figure S1 and Figure S2).
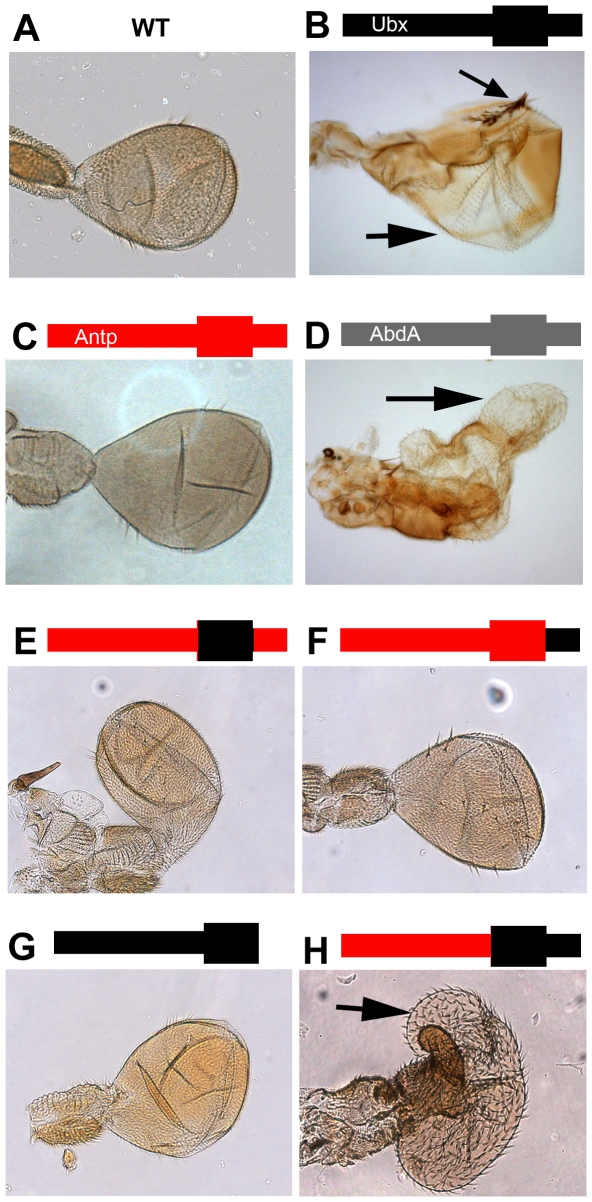
To obtain initial mechanistic insights into Ubx autoregulatory silencing, we carried out experiments that suggest it requires specific DNA binding by Ubx. For these experiments, we monitored the ability of chimeric Hox proteins to induce haltere-to-wing transformations when expressed via the vg-Gal4 driver. Although the more anterior Hox protein Antennapedia (Antp) was unable to induce Ubx silencing, transient overexpression of Antp-Ubx chimeric proteins revealed that the Ubx homeodomain and adjacent C-terminal sequences were both necessary and together sufficient to induce robust Ubx silencing (Figure 3). These findings suggest that Ubx protein, and not Ubx mRNA, is responsible for the induction of silencing. Further, as both the homeodomain and adjacent sequences are implicated in Ubx specificity and DNA binding [13],[14],[15], these results suggest that Ubx triggers silencing by binding to Ubx-specific cis-regulatory elements. Consistently, the Hox protein Abdominal-A (Abd-A), which is very similar to Ubx in both domains, also induced Ubx silencing when transiently expressed during haltere development (Figure 3).
Figure 3. Ubx Silencing requires the Ubx homeodomain and C-terminus.
(A) Wild type haltere. (B) vg-Gal4 UAS-Ubx halteres show haltere to wing transformations due to Ubx silencing. (C) vg-Gal4 UAS-Antp halteres fail to produce any haltere to wing transformations. (D) vg-Gal4 UAS-AbdA halteres show haltere to wing transformations that are indistinguishable from those seen with UAS-Ubx. AbdA and Ubx have very similar homeodomains and also share the UbdA motif in the C-terminus, consistent with these domains playing a critical role in silencing. (E) vg-Gal4 UAS-AUA (Antp N-terminus, Ubx homeodomain, Antp C-terminus) halteres show no transformation to wing in 8/10 samples and mild transformations in 2/10 samples. (F) vg-Gal4 UAS-AAU (Antp N-terminus, Antp homeodomain, Ubx C-terminus) halteres show no haltere to wing transformations. (G) vg-Gal4 UAS-UU* (Ubx N-terminus, Ubx homeodomain, deletion of the C-terminus) halteres show no haltere to wing transformations. (H) vg-Gal4 UAS-AUU (Antp N-terminus, Ubx homeodomain, Ubx C-terminus) halteres show haltere to wing transformations indistinguishable from those seen with UAS-Ubx.
Ubx enhancer silencing triggered by additional copies of the Ubx+ gene
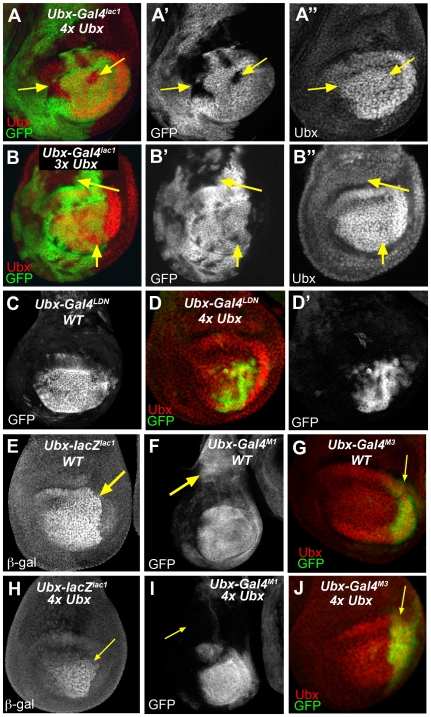
We next tested whether more subtle increases in Ubx levels could also induce silencing. For these experiments, we monitored the expression of Ubx lacZ or Gal4 enhancer traps in flies that had extra copies of the wild type Ubx locus. Ubx-Gal4lac1 and Ubx-Gal4LDN were silenced in groups of haltere cells of 3x Ubx+ and 4x Ubx+ flies (100% of 4x Ubx+ haltere discs had at least one group of silenced cells) (Figure 4A–4D; Table S1). In these haltere discs, probably because the flies had multiple copies of Ubx+, the pattern of Ubx protein was invariably wild type (Figure 4A, 4B, 4D). Interestingly, the amount of silencing induced by 4 copies of Ubx was significantly decreased when one of these copies encoded a non-functional Ubx protein (the Ubx9–22 allele; data not shown). This result supports the idea that Ubx protein, not Ubx mRNA, is the inducer of silencing in response to extra copies of the Ubx locus.
Figure 4. Ubx enhnacer trap silencing in response to increasing Ubx+ dose.
(A) Ubx-Gal4lac1 is silenced in groups of cells by 4 copies of the Ubx+ locus (arrows), but Ubx protein levels are normal. (B) Ubx-Gal4lac1 is silenced in groups of cells by 3 copies of the Ubx+ locus (arrows), but Ubx protein levels are normal. (C) Wild type haltere expression pattern of Ubx-Gal4LDN. (D) Ubx-Gal4LDN is partially silenced by 4 copies of the Ubx+ locus. (E–G) Wild type haltere expression patterns of Ubx-lacZlac1 (E), Ubx-Gal4M1 (F), and Ubx-Gal4M3 (G). (H–J) Ubx-lacZlac1 (H) and Ubx-Gal4M1 (I), but not Ubx-Gal4M3 (J), are partially silenced by 4 copies of Ubx+. Note that for Ubx-lacZlac1 and Ubx-Gal4M1, silencing does not occur in random clones, but instead is manifest by a loss of expression in proximal regions of the disc (arrows).
Ubx-Gal4M1 and Ubx-lacZlac1 responded differently to 4x Ubx+: instead of being silenced in clones, these enhancer traps were no longer expressed in proximal regions of the haltere disc, but distal expression remained unchanged (Figure 4E, 4F). For Ubx-lacZ166, the levels were strongly reduced in 4x Ubx+ flies compared to 2x Ubx+ flies (Table S1). Note, however, that Ubx-lacZ166 can be completely silenced in clones in response to hs-Ubx (Figure S3 and Table S1). Finally, the expression of Ubx-Gal4M3 did not change in the presence of four copies of the Ubx+ locus (Figure 4G and Table S1). Taken together, these results allow us to make three important conclusions. First, silencing is occurring at the level of Ubx enhancers, not entire Ubx alleles, because different Ubx enhancer traps respond in different ways. Second, silencing can be triggered by the presence of only one or two additional Ubx+ loci, suggesting that less than doubling Ubx levels is sufficient to silence some enhancers. Third, although all Ubx enhancers can be silenced by high Ubx levels, lower Ubx levels result in a range of responses that depend on which enhancer trap, and therefore which subset of Ubx enhancers, is being monitored. Thus, we conclude that different Ubx enhancers are sensitive to different levels of Ubx protein. We also generated flies to monitor two different enhancer trap insertions into the Ubx locus (Ubx-lacZ166 and Ubx-Gal4lac1) at the same time. When silencing was triggered by heat shock-induced Ubx, we observed silencing of both enhancer traps, but at different frequencies: Ubx-Gal4lac1 was silenced to a greater extent than Ubx-lacZ166 (Figure S3). This finding provides additional support for the idea that individual enhancer traps, and thus different subsets of Ubx enhancers, respond differently to the same increase in Ubx levels.
Haltere size and Ubx levels are buffered in response to increased Ubx+ copy number
The above results show that epigenetic autoregulatory silencing of Ubx enhancers occurs in response to elevated Ubx levels. Interestingly, increasing the dose of Ubx+ results in smaller halteres [16], but this size change does not scale linearly with the number of Ubx+ genes. Haltere size is similar to wild type in flies with 3x Ubx+ or 4x Ubx+, while in flies with 6 copies of Ubx+, haltere size is greatly reduced (Figure 5A and Figure S4A). These results suggest that haltere size is buffered against increasing doses of the Ubx+ gene. A similar buffering can be observed when Ubx protein levels are quantified in haltere discs from animals with different numbers of Ubx+ genes. When one copy of Ubx is inactivated (1x Ubx+), Ubx protein levels are nearly halved (Figure S4A). However, when the Ubx+ complement is doubled (4x Ubx+) or tripled (6x Ubx+) only 39% and 60% increases in Ubx protein levels were detected, respectively (Figure S4A). The less-than-expected increases in Ubx levels seen in Ubx duplications is not because they fail to express wild type levels, as they are sufficient to fully rescue a Ubx null mutation, both phenotypically [17],[18] and with respect to Ubx protein levels (data not shown). Together with the results described above, we suggest that the buffering of Ubx levels and haltere size is due, at least in part, to the epigenetic silencing of Ubx enhancers in response to higher than normal doses of Ubx+.
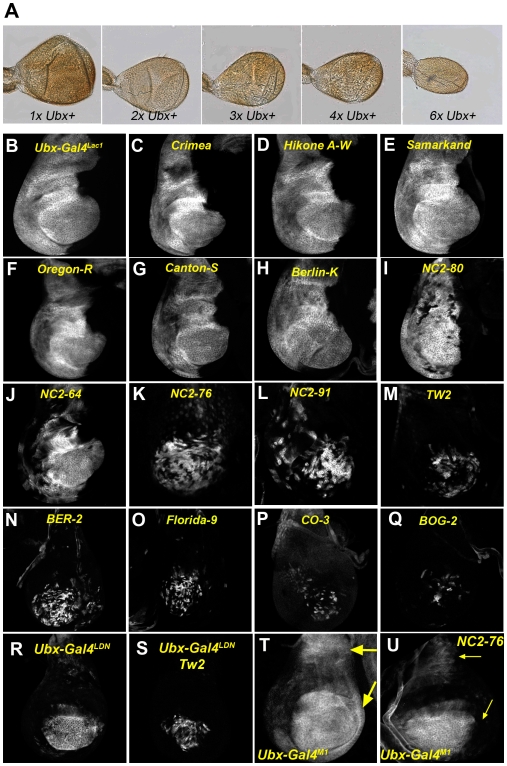
Figure 5. Ubx enhancer silencing in response to natural genetic variation.
(A) Halteres decrease in size with increasing Ubx+ copy number. UbxDf(109)/+(1xUbx+); Wild Type (2xUbx+); Dp(P5)/+(3xUbx+); Dp(P10)2x/+(4xUbx+); Dp(P10)2x/+; Dp(P5)/Dp(P5) (6xUbx+). (B–U) All images show haltere discs stained for enhancer trap expression. (B–Q) Ubx-Gal4lac1 driven UAS-GFP reporter expression in the lab stock (B) and outcrossed to various wild type stocks (C–Q). Stocks beginning with “NC2” were collected in North Carolina. Other wild type stocks were obtained from the Bloomington Stock Center. See Table S1 and Table S2 for a complete summary of these results. (C–H) Outcrossing to these stocks does not cause Ubx-Gal4lac1 silencing. (I–L) Outcrossing to these wild type stocks causes mild to moderate Ubx-Gal4lac1 silencing. (M–Q) Outcrossing to these wild type stocks causes moderate to strong Ubx-Gal4lac1 silencing. (R,S) Ubx-Gal4LDN in the lab background (R) and in F1 progeny when crossed to Tw2 (S). Strong clonal silencing is observed. (T,U) Ubx-Gal4M1 in the lab background (T) and in F1 progeny when crossed to NC2-76 (U). Loss of proximal expression (arrows) is observed.
Ubx enhancer silencing induced by genetic variation
In wild type animals, we hypothesized that enhancer silencing may be used to ensure uniform Ubx levels in response to naturally occurring genetic variation in the cis- and trans-regulation of Ubx expression. We tested this idea by out-crossing our laboratory Ubx-Gal4lac1 flies to 32 D. melanogaster strains established from wild populations around the world. In our lab stock, less than 5% of haltere discs showed any evidence of Ubx-Gal4lac1 silencing. However, when outcrossed to wild D. melanogaster strains, we frequently observed silencing of Ubx-Gal4lac1 in haltere discs of the F1 generations (Figure 5 and Table S2). Although the frequency of silencing varied between wild stocks, it was consistent for each wild stock in a statistically significant manner (Figure 6). Of the 32 stocks crossed to Ubx-Gal4lac1, 14 resulted in no detectable silencing in the F1 generation, 6 showed weak silencing in the F1 generation, and 12 showed strong silencing in the F1 generation (Figure 5 and Table S2). Because the amount of silencing can, in some cases, approach 100% (e.g. Tw2 F1), while 4x Ubx+ resulted in ∼20–30% silencing (Figure 6), we suggest that differences beyond Ubx levels contribute to silencing in these F1 outcrosses. Genetic variation may, for example, result in differences in the levels or activities of the trans-regulators of Ubx. Silencing was also observed when Ubx-lacZlac1 and Ubx-Gal4LDN were outcrossed to wild populations, demonstrating that this effect is not limited to Ubx-Gal4lac1 (Figure 5R–5U and Table S1). Despite the silencing of Ubx enhancer traps, the pattern and levels of Ubx protein were similar in the wild stocks, our laboratory stocks, and in their F1 progeny (Figure S4B). We ruled out that the lack of enhancer trap expression in these outcrosses was due to a failure to initiate expression by carrying out a lineage tracing experiment, which demonstrates that Ubx-Gal4lac1 was expressed prior to silencing (see Materials and Methods). We also ruled out that transposon instability (e.g. hybrid dysgenesis [19]) was responsible for the loss of enhancer trap expression using several criteria (see Materials and Methods). Most importantly, silencing occurred at the same frequency when the male or female parent was from the wild (non-laboratory) stock and the amount of enhancer trap DNA, measured by qPCR, was unchanged between the parental and F2 generations. Further, silencing of enhancer traps in other genes, including Distalless-Gal4, homothorax-lacZ, and teashirt-lacZ was not observed by crossing these insertions to the same wild strains (data not shown).
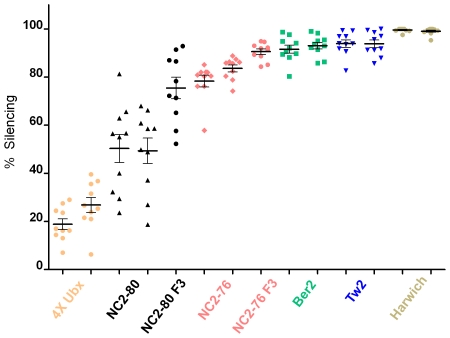
Figure 6. Quantification of silencing.
Each point records the % silencing of the Ubx-Gal4lac1 enhancer trap for a single haltere disc. % silencing is defined as the amount of non-stained tissue relative to wild type controls measured in parallel (see Materials and Methods for details). Unless otherwise indicated, all measurements were of haltere discs from F1 animals grown under non-crowded conditions produced by crossing our laboratory Ubx-Gal4lac1 stock to the indicated genetic backgrounds (4× Ubx, orange circles; NC2-80, black triangles; NC2-76, pink diamonds; Ber2, green squares; Tw2, blue triangles; Harwich, tan circles). Silencing was measured in two independent sets of crosses, separated in time by two weeks, and are graphed in neighboring columns. The thick black bars correspond to averages and the thinner bars show the standard error of the mean. For each cross, a minimum of 10 haltere discs, from 10 different animals, were scored. An analysis of variance (ANOVA) shows that the differences among the five wild type genotypes (NC2-80, NC2-76, Ber2, Tw2 and Harwich) in % silencing were highly significant (t ratio = 9.4, p = 0.0007) with 83% of the variance among lines, and no differences between replicates. Also graphed is the % silencing measured in 10 independent haltere discs resulting from the continued introgression (F3 generations) of Ubx-Gal4lac1 into the NC2-80 background (black circles) and into the NC2-76 background (pink circles). The average % silencing increased in the F3 generations compared to the F1 generations.
We postulate that silencing induced in these outcrosses may be due to an incompatibility between the trans-acting factors (largely derived from the wild stocks) and cis-regulatory elements (linked to the monitored Ubx locus of the laboratory stock) controlling Ubx expression. In support of this idea, when Ubx-Gal4lac1 was further introgressed into weakly or strongly silencing wild stocks, which effectively increases the genetic complement from the wild strain background, an increase in the severity of silencing was observed when compared to the F1 generation (Figure 6 and Figure S5). We also never observed the complete absence of Ubx protein or haltere-to-wing transformations in any of these outcrosses, arguing that only a subset of enhancer inputs into Ubx is silenced in response to genetic variation. Consistently, individual enhancer traps responded differently when crossed to the same wild strains (Table S1).
Together, these results demonstrate that Ubx enhancer silencing is triggered when Ubx is present at higher than normal levels. When Ubx concentration is especially high (when Ubx is ectopically expressed via Gal4 or heat-shock promoters) all enhancer input into Ubx can be silenced, resulting in the complete absence of Ubx expression and haltere-to-wing transformations. Although such high levels of Ubx are not physiological, we also find that Ubx enhancer silencing can be triggered by additional copies of Ubx+, which in principle results in less than double the amount of Ubx protein. In this case, we find that the expression of some Ubx enhancer traps is clonally silenced (e.g. Ubx-Gal4lac1), while the expression of other enhancer traps (e.g. Ubx-lacZ166) is reduced. Thus, different Ubx enhancers are differentially sensitive to negative autoregulation; some are shut off by relatively low Ubx levels, while others require high Ubx levels to be silenced.
Enhancer silencing and natural genetic variation
Most remarkably, we found that enhancer silencing can occur simply by varying the genetic background. In Drosophila melanogaster, due in part to its large population size, the frequency of DNA polymorphisms between individuals in the wild is estimated to be as high as 1 in 100 basepairs [20]. Due to these polymorphisms, we imagine that different strains of D. melanogaster, when kept in isolation from each other, may have subtly different ways of regulating Ubx. These may be due to strain-specific differences in the Ubx cis-regulatory elements, in the trans regulators of Ubx expression, or both. Consistent with this idea, it is of interest that gene expression levels, when assayed across entire genomes, show a lot of variability in natural populations [21],[22],[23],[24],[25]. Although we find that the final Ubx expression pattern and levels are very similar between lab and wild D. melanogaster strains, when two strains are bred together genetic differences may result in fluctuations in the initial Ubx levels. The silencing system described here may function to compensate for these fluctuations and thus ensure that the correct Ubx levels are produced throughout the haltere.
Plasticity of enhancer activities
In the crosses to wild D. melanogaster strains, we found that the expression of genetically marked Ubx alleles varied tremendously, depending on the genetic background. Extrapolating from these results suggests that there is a lot of previously undetected variability in enhancer activities at the Ubx locus in wild files that would not have been detected using traditional assays. Thus, these results challenge the standard view that a given transcriptional enhancer integrates the same inputs and produces the same outputs, regardless of genetic background. Instead, due to natural genetic variation, the activity of a particular enhancer may vary widely between individuals in wild populations. Additionally, our results show that the activity of an enhancer can even vary among the cells within its expression domain (e.g. the haltere) in a single individual. We suggest that plasticity in enhancer activities is essential to compensate for genetic and perhaps environmental variation. Moreover, given that many genes may have multiple, partially redundant enhancers, enhancer silencing may be essential to buffer gene expression levels so that they remain within a narrow, biologically tolerable range. On the other hand, small differences in enhancer activities in flies in the wild may serve as a potential source of phenotypic variation that can be acted upon by natural selection. Since population genetic theory predicts that selection differentials of a small fraction of a percent are seen in natural populations with the effective population size of Drosophila [20], it is plausible that this variation is functionally significant, perhaps through a subtle influence of haltere morphology on flight performance.
Materials and Methods
Genetic variation experiments
The NC2 stocks were obtained from Greg Gibson (N.C. State University); all other wild stocks were obtained from the Bloomington Stock Center (Table S2).
To show that the lack of expression in these outcrosses was not due to a failure to initiate enhancer trap expression in the wild backgrounds, we carried out a lineage tracing experiment. The genotype of the stock was: Ubx-Gal4lac1 UAS-flp; actin>stop>GFP. The combination of UAS-flp and actin>stop>GFP records the history (i.e. marks the lineage) of Gal4 expression. When outcrossed to wild backgrounds, GFP expression was not silenced (in contrast to when the direct UAS-GFP readout was monitored). Together, these results suggest that Ubx-Gal4lac1 was initially activated but then silenced.
Hybrid dysgenesis was ruled out as a reason for loss of expression from P transposons by the following tests: 1) silencing occurs equally well, regardless of the direction the cross was set up, 2) silencing occurs equally well at 18° and 25°C (while hybrid dysgenesis is suppressed at 18°C), 3) silencing was not observed for some other transposon insertions (inside or outside of the Ubx locus) when crossed to the same wild stocks, 4) the miniwhite gene associated with the P element insertions did not lead to a variegated eye phenotype as would be expected for somatic transposon excision, and 5) quantitative PCR analysis confirmed that the amount of transposon DNA was the same in the parent (unsilenced) and F2 (silenced) generations. Finally, enhancer trap expression can be recovered when back-crossed into the laboratory stock background.
Quantification of Ubx protein levels
To measure Ubx protein levels in different genetic backgrounds, we stained haltere discs obtained from uncrowded yw (2x Ubx+), yw;If/Cyo;TM2/TM6B (1x Ubx+), yw;If/Cyo;DpP5/TM6B (3x Ubx+), yw;DpP10x2/CyoGFP;MKRS/TM6B (4x Ubx+), yw; DpP10x2/CyoGFP;DpP5/DpP5 (6x Ubx+),Hikone-R, Berlin-K, NC2-76, NC2-80, yw x NC2-76 F1s, Tw2, yw x Tw2 F1s, Florida-9, Reids-2, and Harwich wandering larvae with anti-Ubx (FP3.38) and a fluorescent secondary antibody. Stainings and confocal imaging were done identically and in parallel for ≥8 haltere discs from each genotype. The pixel intensities in identically sized regions of the distal anterior compartments were measured using Adobe Photoshop. This region was quantified because it is a relatively large area that expresses Ubx at uniform levels and gives rise to the main body of the haltere (the same portion measured in Figure 5A and Figure S4A). Similar trends were observed when average pixel intensities for the entire distal haltere were measured. The average intensities for each wild population differed by no more than 16%, suggesting that final Ubx levels are very similar despite differences in genetic background and silencing.
Quantification of Ubx reporter silencing
To quantify the extent of silencing of the Ubx-Gal4lac1 reporter in response to Ubx+ copy number and outcrosses to wild populations, third instar haltere discs were dissected from wandering larvae of yw122; DpP10x2/CyoGFP; Ubx-Gal4lac1UAS-GFP/TM6B (4xUbx+), and the GFP positive, F1 progeny of yw122; If/Cyo; Ubx-Gal4lac1UAS-GFP/TM6B crossed with NC2-80, NC2-76, Ber-2, Tw-2, and Harwich. GFP positive F3 progeny of yw122; If/Cyo; Ubx-Gal4lac1UAS-GFP/TM6B crossed with NC2-80 and NC2-76 were also dissected. For the outcrosses, we always used females from the wild populations. Haltere discs were fixed, mounted, and imaged for GFP and DAPI on a confocal microscope. Images were made binary in ImageJ. The GFP expressing area relative to the total disc area was measured for each disc, and this value was subtracted from the average GFP expressing area (relative to total disc size) of yw122; If/Cyo; Ubx-Gal4lac1UAS-GFP/TM6B haltere discs to yield a ‘% silencing’ value for each disc.
Heat-shock induced Ubx overexpression
Larvae bearing the hs-UbxIa22 transgene [26] were heat-shocked at 37°C for 15–20 minutes 3 or 4 days after egg laying. Larvae were dissected at least 48 hours after heat shock to allow for total dissipation of exogenous Ubx. hs-UbxIa22 larvae that were not heat shocked showed no Ubx silencing. Neutral clones were induced using the same heat shock regime in flies of the genotype yw hsflp; FRT 42D Ub-GFP/FRT 42D; hs-UbxIa22/+.
Ubx enhancer traps and duplications
Ubx-Gal4lac1 [27]; Ubx-lacZlac1 [28]; Ubx-Gal4LDN [29]; Ubx-Gal4M1 [29]; Ubx-lacZ166 [30]; and Ubx-Gal4M3 [29]. Although these lines are hypomorphic mutations of the Ubx locus, this is unlikely to contribute to our results because decreased production of Ubx would, if anything, cause an underestimate of the amount of silencing that occurs at the Ubx locus.
3x Ubx+ flies contain a tandem duplication of the Ubx locus (Dp(3;3)P5).
4x Ubx+ flies contain a tandem duplication of a transpositon of the Ubx locus onto the 2nd chromosome (Dp(3;2)P10). Further increases in Ubx+ copy number were created by combining these duplications [16]. Ubx9–22 expresses a non-functional Ubx protein due to a ∼1500 bp deletion that removes a splice acceptor site and part of the Ubx homeodomain-encoding exon [31].
Before crossing to enhancer traps, Ubx duplications were introduced into stocks containing marked chromosomes that do not cause silencing (yw hsflp; If/cyo; Dp(P5)/Tm6B and yw hsflp; Dp(3;2)P10x2/CyoGFP; MKRS/Tm6B).
To monitor silencing of Ubx-lacZ166 and Ubx-Gal4lac1 simultaneously (Figure S3), flies of the genotype, Dp(3;2)P10x2/heat shock-Ubx; Ubx-lacZ166/Ubx-Gal4lac1 UAS-GFP were given a 15 min. heat shock at 37°C 48 to 96 hrs after egg laying. Imaginal discs were dissected at wandering stage and stained for Ubx, βgal, and GFP. Silencing was not observed in flies of the same genotype without heat shock.
PcG mutations
FRT101 ph504
FRT2A PcXT109
FRT42D Su(Z)2l.b8
FRT82B ScmD1
FRT42D PclD5
Of these mutations, when analyzed in loss-of-function clones, all but Pcl resulted in repression of Ubx in the haltere (due to derepression of more posterior Hox genes; data not shown) and therefore could not be used to assess their role in silencing.
Other lines used
UAS-GFP Ubx-Gal4lac1/TM6B
UAS-GFP (X); Ubx-Gal4LDN/TM6B
UAS-GFP (X); Ubx-Gal4M1/TM6B
FRT 82B UbxDf(109)/TM6B
hs-UbxIa22/TM6B [26]
Ubx9–22/TM6B
vg-Gal4 UAS-GFP
vg-Gal4 UAS-GFP UAS-flp act>cd2>Gal4
UAS-UbxHA
FRT42D Ub-GFP
FRT42D Ub-GFP; hs-UbxIa22/Tm6B
FRT42D
UAS-GFP; FRT42D arm-lacZ; Ubx-Gal4Lac1
hs-Gal4
Antp-Ubx chimeras
(Previously described by [14]
UAS-Antp
UAS-AUA
UAS-UU* (* refers to a stop codon inserted immediately following the homeodomain)
UAS-AAU
UAS-AUU
Quantitative PCR
Whole-fly genomic DNA was isolated from the lab stock containing the Ubx-Gal4lac1 enhancer trap (yw122; If/CyoGFP; Ubx-Gal4lac1 UAS-GFP/TM6B) and the GFP+ F2 progeny of the Ubx-Gal4lac1 stock crossed to strains Tw2, NC2-76, and NC2-80. Silencing was confirmed to be occurring in these crosses. The F2 progeny were generated by crossing Gal4lac1UAS-GFP F1 males to wild population females, precluding the possibility of recombination between chromosomes of the lab and wild genotypes. Primers were designed to amplify ∼200 bp in the Gal4 and UAS transgenes to determine their relative abundance in each genotype. A ∼200 bp sequence in the 5′UTR of homothorax was amplified to normalize for different amounts of template DNA. PCR amplification was performed in triplicate using Applied Biosystems 7300 Real Time PCR System, and SYBR Green PCR Master Mix. Product dissociation curves were examined to ensure that each primer set only amplified a single product. CT values and amplification curves were consistent with an equal abundance of the Gal4 and UAS sequences in all genotypes.
Antibody staining
Standard protocols were used with the following primary antibodies:
Rabbit anti-β-Gal 1:10,000 (Cappel)
Mouse anti-En 1:10 (Hybridoma Bank)
Mouse anti-Ubx 1:20
Rat anti-HA 1:100
Supporting Information
Neutral clones respect the borders of Ubx silencing. (A,B) Two examples of haltere discs with neutral clones (marked by the absence of GFP) and Ubx silencing (induced by hs-Ubx). In (A), there is no crossing between the neutral clones and Ubx-silenced patches. In (B), although most of the neutral clones respect the Ubx-silenced patches, there are two small exceptions (arrows). Ubx- silenced patches are outlined in yellow and the neutral clones are outlined in blue. The exceptions observed in these experiments are likely due to multiple neutral clones that were scored as a single clone because they fused during growth.
(9.78 MB TIF)
PcG functions are required for Ubx autoregulatory silencing. (A) Wing disc with Pcl- clones (absence of GFP) stained for Ubx (red) and GFP. Ubx expression is observed in pouch clones. (B) Haltere disc with Pcl- clones (absence of GFP) stained for Ubx (red) and GFP. Ubx expression is unaffected by the absence of Pcl. Pcl was the only PcG gene we tested to show strong, autonomous Ubx derepression in the wing disc, and no affect on Ubx expression in the haltere disc; the PcG mutations Pc, Scm, ph, and Su(Z)2 could not be used for this experiment because they result in a loss of Ubx expression in the haltere, due to the derepression of more posterior Hox genes in these clones (data not shown). (C) A Ubx-Gal4lac1 haltere disc in which both silencing (by hs-Ubx) and Pcl- clones were induced. Pcl- tissue is outlined in yellow. Silencing of both Ubx and the enhancer trap are observed, but not in Pcl- tissue. Note that Pcl- clones only affect Ubx expression in the distal, “pouch” domain of the wing and haltere (Beuchle D, Struhl G, Muller J (2001) Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128: 993-1004).
(7.73 MB TIF)
Simultaneous monitoring of silencing for two Ubx enhancer traps. (A,B) hs-Ubx/DpP10x2; UbxGal4lac1 UAS-GFP/UbxlacZ166 haltere disc from animals that were not given a heat shock (A) or were given a 15 min heat shock (B). The discs were stained for Ubx (blue), GFP (green), and βgal (red). Individual channels are shown as indicated. For (B), where silencing was observed, the outlines of the silenced clones are shown as follows: in the βgal channel (B') the outlines of Ubx (yellow outline) silenced clones are shown. In the GFP channel (B') the outlines of Ubx (yellow outline) silenced clones are shown. B' shows the GFP channel with the Ubx-lacZ166 (red outline) silenced clones. Note that the extent of silencing of Ubx-Gal4lac1 is greater than that of Ubx-lacZ166, and that Ubx-lacZ166 silencing is a subset of Ubx-Gal4lac1 silencing.
(3.13 MB TIF)
Quantification of haltere sizes and Ubx levels. (A) Quantifications of Ubx protein levels (blue bars) and haltere sizes (red bars) in genotypes with differing numbers of wild type Ubx+ alleles. Both measurements are shown relative to wild type (2x Ubx+). Note that neither measurement scales quantitatively with increases in Ubx+ dose, illustrating that these phenotypes are buffered. In contrast, one copy of Ubx+ shows a ∼60% reduction in Ubx protein levels and a ∼50% increase in haltere size compared to wild type (2x Ubx+). Error bars represent standard error of the mean. (B) Quantifications of Ubx levels in 8 different wild genetic backgrounds (Hikone-R, Berlin-K, NC2-80, NC2-76, Tw2, Florida-9, Reids-2, and Harwich) and two F1s (yw X NC2-76 and yw X Tw2) are all within ∼16% of those measured in yw. Moreover, this variation does not correlate with the degree of silencing (shown in the thumbnail images below the graph). For comparison, halving the dose of Ubx+ decreases Ubx levels by ∼40% (left-most bar). Error bars represent standard error of the mean.
(0.36 MB TIF)
Ubx silencing increases with introgression into wild genetic backgrounds. (A) Ubx-Gal4lac1 expression in the F1 progeny of a cross to the Tw2 wild type line. (B) Silencing increases when Ubx-Gal4lac1 is introgressed by backcrossing into the Tw2 line. Shown here is a haltere disc after 2 backcrosses (the F3 generation). (C) Ubx-Gal4lac1 expression in the F1 progeny of a cross to the NC2-80 wild type line. (D) Silencing increases when Ubx-Gal4lac1 is introgressed by backcrossing into the NC2-80 line. Shown here is a haltere disc after 2 backcrosses (the F3 generation).
(1.47 MB TIF)
Summary of Ubx enhancer traps and their responses to changes in Ubx levels and genetic variation
(0.08 MB DOC)
Summary of Ubx-Gal4lac1 silencing in F1 crosses to wild stocks
(0.07 MB DOC)
Acknowledgments
We thank G. Gibson, I. Dworkin, B. Gebelein, T. Jessell, L. Johnston, and D. Rogulja for helpful discussions and/or comments on the manuscript, G. Gibson for carrying out the ANOVA analysis, and L. Vosshall for hosting MAC during the final stages of experiments. We thank E. Sanchez-Herrero, W. Bender, G. Gibson, and J. Müller for providing fly stocks.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by NIH grants GM054510 and GM058575 to RSM. MAC was supported by NIH training grants DK07328 and GM008798. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Yuh CH, Davidson EH. Modular cis-regulatory organization of Endo16, a gut-specific gene of the sea urchin embryo. Development. 1996;122:1069–1082. doi: 10.1242/dev.122.4.1069. [DOI] [PubMed] [Google Scholar]
- 2.Keys DN, Lee BI, Di Gregorio A, Harafuji N, Detter JC, et al. A saturation screen for cis-acting regulatory DNA in the Hox genes of Ciona intestinalis. Proc Natl Acad Sci U S A. 2005;102:679–683. doi: 10.1073/pnas.0408952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson EH. New York: Academic Press; 2001. Genomic Regulatory Systems. p. 261. [Google Scholar]
- 4.Arnosti DN. Analysis and function of transcriptional regulatory elements: insights from Drosophila. Annu Rev Entomol. 2003;48:579–602. doi: 10.1146/annurev.ento.48.091801.112749. [DOI] [PubMed] [Google Scholar]
- 5.Cameron RA, Oliveri P, Wyllie J, Davidson EH. cis-Regulatory activity of randomly chosen genomic fragments from the sea urchin. Gene Expr Patterns. 2004;4:205–213. doi: 10.1016/j.modgep.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfeiffer B, Jenett A, Hammonds A, Ngo T, Misra S, et al. Tools for Neuroanatomy and Neurogenetics in Drosophila. Proc Natl Acad Sci U S A. In press. 2008 doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 9.Dickinson MH. Haltere-mediated equilibrium reflexes of the fruit fly, Drosophila melanogaster. Philos Trans R Soc Lond B Biol Sci. 1999;354:903–916. doi: 10.1098/rstb.1999.0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White RA, Wilcox M. Protein products of the bithorax complex in Drosophila. Cell. 1984;39:163–171. doi: 10.1016/0092-8674(84)90202-2. [DOI] [PubMed] [Google Scholar]
- 11.Irvine KD, Botas J, Jha S, Mann RS, Hogness DS. Negative autoregulation by Ultrabithorax controls the level and pattern of its expression. Development. 1993;117:387–399. doi: 10.1242/dev.117.1.387. [DOI] [PubMed] [Google Scholar]
- 12.Garaulet DL, Foronda D, Calleja M, Sanchez-Herrero E. Polycomb-dependent Ultrabithorax Hox gene silencing induced by high Ultrabithorax levels in Drosophila. Development. 2008;135:3219–3228. doi: 10.1242/dev.025809. [DOI] [PubMed] [Google Scholar]
- 13.Chan SK, Mann RS. The segment identity functions of Ultrabithorax are contained within its homeo domain and carboxy-terminal sequences. Genes Dev. 1993;7:796–811. doi: 10.1101/gad.7.5.796. [DOI] [PubMed] [Google Scholar]
- 14.Gebelein B, Culi J, Ryoo HD, Zhang W, Mann RS. Specificity of Distalless repression and limb primordia development by abdominal Hox proteins. Dev Cell. 2002;3:487–498. doi: 10.1016/s1534-5807(02)00257-5. [DOI] [PubMed] [Google Scholar]
- 15.Merabet S, Saadaoui M, Sambrani N, Hudry B, Pradel J, et al. A unique Extradenticle recruitment mode in the Drosophila Hox protein Ultrabithorax. Proc Natl Acad Sci U S A. 2007;104:16946–16951. doi: 10.1073/pnas.0705832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smolik-Utlaut SM. Dosage requirements of Ultrabithorax and bithoraxoid in the determination of segment identity in Drosophila melanogaster. Genetics. 1990;124:357–366. doi: 10.1093/genetics/124.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Struhl G. Splitting the bithorax complex of Drosophila. Nature. 1984;308:454–457. [Google Scholar]
- 18.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 19.Castro JP, Carareto CM. Drosophila melanogaster P transposable elements: mechanisms of transposition and regulation. Genetica. 2004;121:107–118. doi: 10.1023/b:gene.0000040382.48039.a2. [DOI] [PubMed] [Google Scholar]
- 20.Aquadro CF, Bauer DuMont V, Reed FA. Genome-wide variation in the human and fruitfly: a comparison. Curr Opin Genet Dev. 2001;11:627–634. doi: 10.1016/s0959-437x(00)00245-8. [DOI] [PubMed] [Google Scholar]
- 21.Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 22.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rockman MV, Kruglyak L. Genetics of global gene expression. Nat Rev Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 24.Ronald J, Brem RB, Whittle J, Kruglyak L. Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet. 2005;1:e25. doi: 10.1371/journal.pgen.0010025. doi: 10.1371/journal.pgen.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raser JM, O'Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann RS, Hogness DS. Functional dissection of Ultrabithorax proteins in D. melanogaster. Cell. 1990;60:597–610. doi: 10.1016/0092-8674(90)90663-y. [DOI] [PubMed] [Google Scholar]
- 27.Pallavi SK, Shashidhara LS. Egfr/Ras pathway mediates interactions between peripodial and disc proper cells in Drosophila wing discs. Development. 2003;130:4931–4941. doi: 10.1242/dev.00719. [DOI] [PubMed] [Google Scholar]
- 28.Casares F, Bender W, Merriam J, Sanchez-Herrero E. Interactions of Drosophila Ultrabithorax regulatory regions with native and foreign promoters. Genetics. 1997;145:123–137. doi: 10.1093/genetics/145.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bender W, Hudson A. P element homing to the Drosophila bithorax complex. Development. 2000;127:3981–3992. doi: 10.1242/dev.127.18.3981. [DOI] [PubMed] [Google Scholar]
- 31.Weinzierl RO, Axton JM, Ghysen A, Akam ME. Ultrabithorax mutations in constant and variable regions of the protein coding sequence. Genes Dev. 1987;1:386–397. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Neutral clones respect the borders of Ubx silencing. (A,B) Two examples of haltere discs with neutral clones (marked by the absence of GFP) and Ubx silencing (induced by hs-Ubx). In (A), there is no crossing between the neutral clones and Ubx-silenced patches. In (B), although most of the neutral clones respect the Ubx-silenced patches, there are two small exceptions (arrows). Ubx- silenced patches are outlined in yellow and the neutral clones are outlined in blue. The exceptions observed in these experiments are likely due to multiple neutral clones that were scored as a single clone because they fused during growth.
(9.78 MB TIF)
PcG functions are required for Ubx autoregulatory silencing. (A) Wing disc with Pcl- clones (absence of GFP) stained for Ubx (red) and GFP. Ubx expression is observed in pouch clones. (B) Haltere disc with Pcl- clones (absence of GFP) stained for Ubx (red) and GFP. Ubx expression is unaffected by the absence of Pcl. Pcl was the only PcG gene we tested to show strong, autonomous Ubx derepression in the wing disc, and no affect on Ubx expression in the haltere disc; the PcG mutations Pc, Scm, ph, and Su(Z)2 could not be used for this experiment because they result in a loss of Ubx expression in the haltere, due to the derepression of more posterior Hox genes in these clones (data not shown). (C) A Ubx-Gal4lac1 haltere disc in which both silencing (by hs-Ubx) and Pcl- clones were induced. Pcl- tissue is outlined in yellow. Silencing of both Ubx and the enhancer trap are observed, but not in Pcl- tissue. Note that Pcl- clones only affect Ubx expression in the distal, “pouch” domain of the wing and haltere (Beuchle D, Struhl G, Muller J (2001) Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128: 993-1004).
(7.73 MB TIF)
Simultaneous monitoring of silencing for two Ubx enhancer traps. (A,B) hs-Ubx/DpP10x2; UbxGal4lac1 UAS-GFP/UbxlacZ166 haltere disc from animals that were not given a heat shock (A) or were given a 15 min heat shock (B). The discs were stained for Ubx (blue), GFP (green), and βgal (red). Individual channels are shown as indicated. For (B), where silencing was observed, the outlines of the silenced clones are shown as follows: in the βgal channel (B') the outlines of Ubx (yellow outline) silenced clones are shown. In the GFP channel (B') the outlines of Ubx (yellow outline) silenced clones are shown. B' shows the GFP channel with the Ubx-lacZ166 (red outline) silenced clones. Note that the extent of silencing of Ubx-Gal4lac1 is greater than that of Ubx-lacZ166, and that Ubx-lacZ166 silencing is a subset of Ubx-Gal4lac1 silencing.
(3.13 MB TIF)
Quantification of haltere sizes and Ubx levels. (A) Quantifications of Ubx protein levels (blue bars) and haltere sizes (red bars) in genotypes with differing numbers of wild type Ubx+ alleles. Both measurements are shown relative to wild type (2x Ubx+). Note that neither measurement scales quantitatively with increases in Ubx+ dose, illustrating that these phenotypes are buffered. In contrast, one copy of Ubx+ shows a ∼60% reduction in Ubx protein levels and a ∼50% increase in haltere size compared to wild type (2x Ubx+). Error bars represent standard error of the mean. (B) Quantifications of Ubx levels in 8 different wild genetic backgrounds (Hikone-R, Berlin-K, NC2-80, NC2-76, Tw2, Florida-9, Reids-2, and Harwich) and two F1s (yw X NC2-76 and yw X Tw2) are all within ∼16% of those measured in yw. Moreover, this variation does not correlate with the degree of silencing (shown in the thumbnail images below the graph). For comparison, halving the dose of Ubx+ decreases Ubx levels by ∼40% (left-most bar). Error bars represent standard error of the mean.
(0.36 MB TIF)
Ubx silencing increases with introgression into wild genetic backgrounds. (A) Ubx-Gal4lac1 expression in the F1 progeny of a cross to the Tw2 wild type line. (B) Silencing increases when Ubx-Gal4lac1 is introgressed by backcrossing into the Tw2 line. Shown here is a haltere disc after 2 backcrosses (the F3 generation). (C) Ubx-Gal4lac1 expression in the F1 progeny of a cross to the NC2-80 wild type line. (D) Silencing increases when Ubx-Gal4lac1 is introgressed by backcrossing into the NC2-80 line. Shown here is a haltere disc after 2 backcrosses (the F3 generation).
(1.47 MB TIF)
Summary of Ubx enhancer traps and their responses to changes in Ubx levels and genetic variation
(0.08 MB DOC)
Summary of Ubx-Gal4lac1 silencing in F1 crosses to wild stocks
(0.07 MB DOC)