Abstract
The application of a 1H NMR spectroscopy based screening method for determining the use of two widely available analgesics (acetaminophen and ibuprofen) in epidemiologic studies has been investigated. We used samples and data from the cross-sectional INTERMAP Study involving participants from Japan (n=1,145), China (n=839), UK (n=501) and USA (n=2,195). An orthogonal projection to latent structures discriminant analysis (OPLS-DA) algorithm with an incorporated Monte Carlo re-sampling function was applied to the NMR dataset to determine which spectra contained analgesic metabolites. OPLS-DA pre-processing parameters (normalization, bin width, scaling and input parameters) were assessed systematically to identify an optimal acetaminophen prediction model. Subsets of INTERMAP spectra were examined to verify and validate the presence/absence of acetaminophen/ibuprofen based on known chemical shift and coupling patterns. The optimized and validated acetaminophen model correctly predicted 98.2% and the ibuprofen model correctly predicted 99.0 % of the urine specimens containing these drug metabolites. The acetaminophen and ibuprofen models were subsequently used to predict the presence/absence of these drug metabolites for the remaining INTERMAP specimens. The acetaminophen model identified 415 of 8,436 spectra containing acetaminophen metabolites while the ibuprofen model identified 245 of 8,604 spectra containing ibuprofen metabolites from the global dataset. The NMR-based metabolic screening strategy provides a new objective approach for evaluation of self-reported medication data and is extendable to other aspects of population xenometabolome profiling.
INTRODUCTION
High resolution 1H NMR spectroscopy is an efficient technique for empirical drug metabolite detection and has been successfully applied to investigate drug metabolism in both animals1–3 and healthy volunteers4–7. The development of 1H NMR spectroscopy for high throughput metabolic screening of urine8 and plasma9 in human populations, and for the analysis of population risk factors in so-called metabolome-wide association studies10–12, offers the potential for objective assessment of drug intake in epidemiological studies. Here we are concerned with the detection of xenobiotic substances in urine spectra that can be used to validate self-reported medication data acquired from the participants. At present, there are a lack of suitable techniques with which to validate self-reported data particularly with respect to over-the-counter (OTC) medicines. However, scaling up the method for use in large-scale epidemiologic studies presents a number of challenges. Visual confirmation of the presence or absence of drug metabolites in thousands of individual 1H NMR biofluid spectra is impractical due to the time constraints, and significant peak overlap from both endogenous and exogenous compounds can render the identification of drug metabolite peaks problematic (Supplementary Figure 1 and Supplementary Figure 2).
We have previously shown that Statistical Total Correlation Spectroscopy (STOCSY)13 provides an excellent method for identifying structural and pathway connectivities for drug metabolites and reaction products14–16. We have also shown the use of STOCSY for identification of drug metabolite signals connectivities in human population studies 17 and indeed have shown the extension of this approach using NMR-MS statistical heterospectroscopy18 to the detection of novel metabolites and ionization patterns in epidemiologic samples19. In the present study, a semi-automated computer-based prediction model is developed for the identification of urinary metabolites of two common OTC analgesics, acetaminophen and ibuprofen in urine samples taken from the general population. These OTC drugs were selected because of their widespread usage and extensive knowledge on their metabolism20–26 and their urinary excretion profiles4, 7, 23, 27–30.
Pre-processing parameters such as bin size, scaling, normalization and input parameters can affect the predictability of Orthogonal Projection to Latent Structures Discriminant Analysis (OPLS-DA) models. In this study, these parameters were assessed systematically to identify an optimal prediction model. Normalization is usually performed on each spectrum to compensate for the concentration difference between urine samples31. Pattern recognition of NMR-based metabolite data was initially performed using quantitated integrals of specific spectral peaks32, 33. However, in regions of the NMR spectra where there is substantial peak overlap, this approach is not ideal as it is not easily automated for application to large sample sets. Scaling, in addition to normalization, can be applied to give different weighting to the data variables31, 34. When the data variables are mean-centred, by subtracting the respective mean for each data variable, this tends to over emphasize NMR data variables with large intensity. Scaling methods such as unit-variance, achieved by dividing each variable by the standard deviation, thus gives all NMR data variables an equal weight irrespective of intensity, whilst pareto scaling, by dividing each variable by the square root of the standard deviation, gives greater weight to the NMR data variables with larger intensity but is not as extreme as using un-scaled data31, 34.
Binning can be used both to reduce the size or dimensionality of data and to accommodate small differences in the peak shift caused by pH variation and to ensure all samples included in pattern recognition analyses are corrected for such variation31, 35. Bin size of between 0.01 to 0.04 ppm provides a good compromise between spectral resolution and positional variation of resonances, although more recently other approaches such as full resolution36, 37 and dynamic binning38 of NMR spectra have been successfully applied. However, for development of chemometric classification models, it is not essential for the use of full resolution spectra and indeed the use of full resolution spectra is much more important for biomarker identification. Various bin regions including full digitized spectral region or selected drug metabolite-containing regions will also be tested in the prediction models. Having validated the prediction models, they were then used to predict the presence or absence of acetaminophen and ibuprofen related metabolites in the 1H NMR urine spectra acquired from the INTERMAP Study, a large-scale epidemiological investigation of the relation of diet and life style factors to blood pressure 39.
METHODS
INTERMAP Study design
Urine specimens from 4,680 men and women were sourced from the INTERMAP Study, involving 17 population samples ages 40–59 in China, Japan, UK and USA39, 40. In brief, each individual attended a specified clinic centre on four occasions; the first 2 and last 2 visits were on adjacent days with an average of 3 weeks between the two pairs of visits. Data collection included four indepth 24-h dietary recalls, two 7-day daily alcohol intake assessments by interview, measurement of blood pressure on 8 occasions (2 per visit), smoking status, and other variables. At the first and third visits, a timed 24-h urinary collection was initiated in the clinic and was completed the following day in the clinic, according to a standardized protocol. Information on intake of medications from self-completed questionnaires was acquired at the first and third clinic visits.
Preparation and 1H NMR analysis of urine specimens
Boric acid was utilized as a preservative (at ca 5g/L) and incorporated in the urine collection bottles. Specimens were mixed thoroughly and total volume recorded. Multiple aliquots of urine specimens were prepared, frozen and transported on dry ice to the central laboratory in Leuven, Belgium and stored in both −40 °C and −80 °C freezers. The aliquots used for this study were those stored at −40 °C for up to a maximum of 8 years prior to 1H NMR analysis. Our in-house data and other studies 41 show that urine specimens stored at −40 °C show minimal changes in the metabolic profiles for up to 6 years. Urine specimens were thawed completely before mixing 500 µL of urine with 250 µL of phosphate buffer (0.2M) for the stabilization of urinary pH 7.4 (± 0.5), and 75 µL of sodium 3-trimethylsilyl-(2,2,3,3-2H4)-1-propionate (TSP) in D2O (final concentration 0.1mg/mL) solution for chemical shift referencing of TSP (δ 0.0). The D2O provided a lock signal for the NMR spectrometer. Each urine specimen was mixed, placed in a 96-well plate, and left to stand for 10 minutes before centrifuging at 1,500 g for 10 minutes to remove any precipitates prior to analysis. The remaining urine specimen was refrozen.
1H-NMR spectra of the urine specimens were obtained using a Bruker (Bruker Biospin, Rheinstetten, Germany) Avance 600 spectrometer operating at a 1H frequency of 600.29 MHz in flow-injection mode. Specimens were automatically delivered to the spectrometer using a Gilson 215 robot incorporated into the BEST (Bruker Efficient Sample Transfer) system. A standard one-dimensional pulse sequence (recycle delay - 90° - t1 - 90° - tm - 90° acquisition; XWIN-NMR 3.5) was used. Water suppression was achieved with a saturation pulse during the recycle delay (2 s) and mixing time (tm, 100 ms) defines t1. For each specimen, 64 free induction decays (FIDs) were collected into 32K data points using a spectral width of 20 ppm and the total repetition time was 4.8 s. FIDs were multiplied by an exponential weighting function corresponding to a line broadening of 0.3 Hz and data were zero-filled to 64k data points prior to Fourier transformation.
A small population (ca 0.5 % of urine specimens analyzed) of the urine spectra were found to be sub-standard e.g. due to extreme dilution or poor solvent suppression for one or both of the urine specimens obtained for 50 participants and these were omitted from the analysis, leaving a total of 4,630 participants (n=9,260 urine spectra) available for this study.
Processing of 1H-NMR spectra
Baseline correction, phasing and referencing to TSP were achieved automatically using an in-house routine written in MATLAB® 7.0.1 (MathWorks, Natick, MA). Each spectrum was collected into either 0.04 or 0.01 ppm spectral regions. Reduction of spectral data into these bin sizes allowed optimal computing efficiency without jeopardizing the ability to detect the presence of drug metabolites by pattern recognition techniques. The spectral regions containing water and urea resonances (δ 4.5 – 6.4) were eliminated to remove variation in water suppression and in the integral of the urea signal due to partial cross-saturation with water via deuterium solvent exchangeable protons. The remaining bucketed data, for the 0.04 (n = 177 regions) and 0.01 ppm (n = 710 regions) bin sizes, were normalized using three different approaches prior to data analysis (see analysis strategy below).
Strategy for mathematical modeling of the acetaminophen data
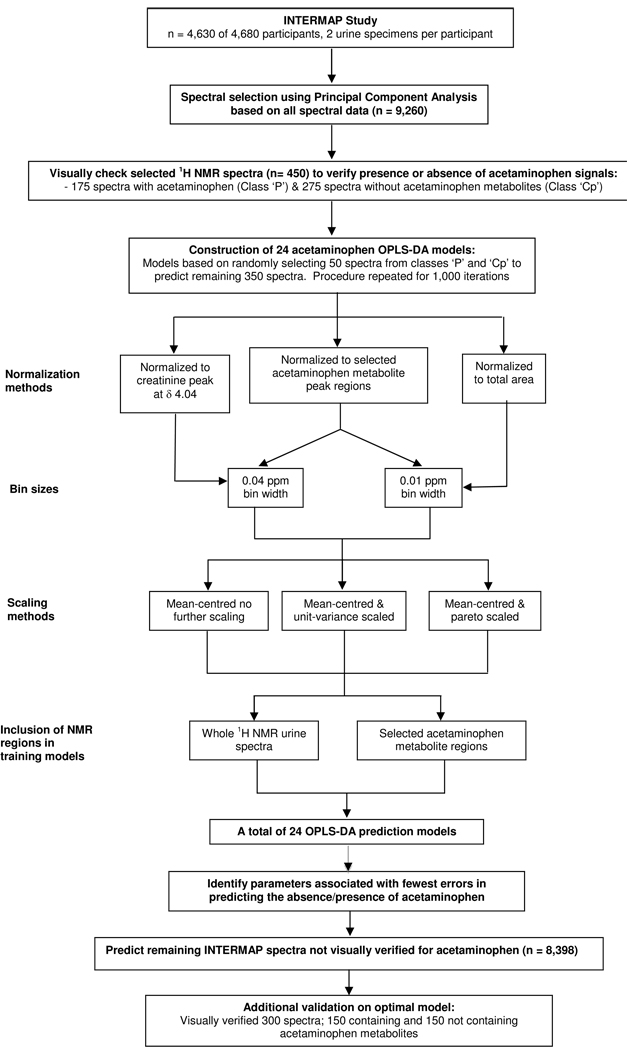
A schematic diagram of the study design for generation of acetaminophen prediction models is shown in Figure 1. To establish optimal parameters and to ensure the best prediction model for spectra containing acetaminophen metabolites, 24 prediction models were constructed based on permutations of pre-processing parameters.
Method of normalization, where three normalization methods were assessed: normalization to a) the −CH2 creatinine peak at δ 4.04 since many chemical assays are expressed as a ratio to creatinine excretion, b) selected acetaminophen metabolite regions at δ 1.84 –1.88, 2.13 –2.25 and 7.13 – 7.49 and c) total area of the spectrum from δ 0.5 – 9.5, (excluding the region containing the residual water and urea resonances).
Width of spectral frequency bin corresponding to 0.04 and 0.01 ppm were assessed for models normalized to selected acetaminophen regions. Models analyzed using the 0.01ppm width were marginally better with respect to predictive ability for the acetaminophen models, thus, the 0.01 ppm bin width was used for the data normalized to total spectral area. However, the bin width for creatinine was set to 0.04 ppm since this bin width approximates the creatinine singlet.
Scaling methods. Models were generated using mean-centred and unit-variance or pareto scaling or the application of no secondary scaling (un-scaled) data.
The input parameters used in the model construction were as follows. Models were constructed using either the complete digitally reduced spectrum (excluding water and urea resonances) or only selected acetaminophen metabolite regions (δ 1.84 –1.88, 2.13 –2.25 and 7.13 – 7.49); acetaminophen glucuronide resonances present in the regions δ 3.62 and 3.88 were not included due to significant overlap with other resonances, e.g., glucose signals making it difficult to distinguish acetaminophen metabolites from endogenous and other exogenous resonances.
Table 1 summarizes each of the models indicating the contribution of parameters with respect to the prediction model.
Figure 1.
Schematic diagram showing the study design for the generation of prediction models for acetaminophen.
Table 1.
Summary: Pre-processing parameters for each acetaminophen prediction model.
| Model name |
Type of scaling |
Normalization method |
Width of bin (ppm) |
Chemical shift regions included in training sets |
Number of chemical shift regions used in modeling |
|---|---|---|---|---|---|
| PCT1 | MC + none | Total spectral area* | 0.01 | Acetaminophen metabolite regions only*** |
55 |
| PCT2 | MC + UV | ||||
| PCT3 | MC + Par | ||||
| PCT4 | MC + none | Total spectral area* | 0.01 | Full spectrum | 710 |
| PCT5 | MC + UV | ||||
| PCT6 | MC + Par | ||||
| PCT7 | MC + none | Acetaminophen metabolites regions*** |
0.04 | Acetaminophen metabolite regions only*** |
16 |
| PCT8 | MC + UV | ||||
| PCT9 | MC + Par | ||||
| PCT10 | MC + none | Acetaminophen metabolites regions*** |
0.01 | Acetaminophen metabolite regions only*** |
55 |
| PCT11 | MC + UV | ||||
| PCT12 | MC + Par | ||||
| PCT13 | MC + none | Acetaminophen metabolites regions*** |
0.01 | Full spectrum | 710 |
| PCT14 | MC + UV | ||||
| PCT15 | MC + Par | ||||
| PCT16 | MC + none | Acetaminophen metabolites regions*** |
0.04 | Full spectrum | 177 |
| PCT17 | MC + UV | ||||
| PCT18 | MC + Par | ||||
| PCT19 | MC + none | Creatinine peak at 4.04 ppm** |
0.04 | Acetaminophen metabolite regions only*** |
16 |
| PCT20 | MC + UV | ||||
| PCT21 | MC + Par | ||||
| PCT22 | MC + none | Creatinine peak at 4.04 ppm** |
0.04 | Full spectrum excluding creatinine at 4.04 ppm |
174 |
| PCT23 | MC + UV | ||||
| PCT24 | MC + Par | ||||
Key: MC = mean-centred un-scaled data; UV = unit-variance scaled; Par = pareto scaled
For models normalized to total spectral region, only bin size of 0.01 ppm were used
For models normalized to creatinine peak at 4.04 ppm, only bin size of 0.04 ppm were used
Acetaminophen metabolite regions included: δ1.84 –11.88. 2.13–2.25 & 7.13 –7.49
Spectra selection for constructing acetaminophen prediction models
A principal components analysis (PCA) model was initially constructed using all 9,260 1H NMR urine spectra from the INTERMAP Study. These spectra were digitized, reduced into spectral regions of 0.01 ppm width and then normalized to total area of the 1H NMR urine spectra. Loadings of this PCA model identified acetaminophen metabolites as one of the main influences on spectra positions in principal components 3 and 5 (PC 3 and PC 5), where PC 1, PC 2 and PC 4 were mainly dominated by high concentrations of glucose, ethanol and trimethylamine-N-oxide (TMAO) metabolite resonances respectively. Two groups relating to acetaminophen status could be visualized in the dataset from the scores plot (see Supplementary Figure 3), one group containing acetaminophen metabolite resonances (group A, n = 605 spectra), the other spectra not containing acetaminophen metabolites (group B, n = 8,655). We randomly selected 175 spectra from group A (28.9 % of group A) and 275 spectra from group B (3.2 % of group B) based on the scores of the constructed PCA. A higher percentage of spectra from group A than from group B were included in order to obtain a set with sufficient spectra containing acetaminophen metabolites for statistical analysis. These 450 selected spectra were chosen as representative of groups A and B (see Supplementary Figure 3), and were subsequently examined visually to confirm presence or absence of acetaminophen metabolite signals. For 170 of the 175 (97.1 %) spectra selected from group A, presence of acetaminophen metabolites was confirmed whilst 270 of the 275 spectra (98.2 %) selected from group B did not have acetaminophen metabolites as verified visually. The 170 spectra from group A and the 5 spectra from group B (n = 175) were grouped together and are spectra containing acetaminophen, referred to as ‘P’ class. The remaining 275 spectra, 5 spectra from group A and the 270 spectra from group B not containing acetaminophen resonances are referred to as ‘Cp’ class. These 450 spectra (i.e. 175 ‘P’ class and 275 ‘Cp’ class) were subsequently used to construct the 24 prediction models using various pre-processing parameters (see validation of acetaminophen prediction models later). These 450 spectra corresponded to the 1H NMR urine spectra for 412 individuals and a further 19 individuals from whom both urinary spectra were used.
To confirm that the selected spectra from ‘P’ and ‘Cp’ classes were representative of groups A and B, we performed MANOVA, to compare a randomly selected group of spectra of equivalent size to class ‘P’ and ‘Cp’ respectively from the whole INTERMAP dataset using a computational algorithm. MANOVA results showed no significant differences between randomly selected spectra and those selected as representative of groups A and B.
Validation of acetaminophen prediction models
A total of 24 OPLS-DA prediction models, each assessing different pre-processing parameters including normalization, scaling, binning and input of parameters, were constructed (models PCT1 to PCT24 in Table 1). For each OPLS-DA model, 100 spectra were randomly selected, 50 from class ‘P’ (acetaminophen group) and 50 from class ‘Cp’ (group without acetaminophen metabolites). These 100 spectra were used to predict the remaining 125 spectra from ‘P’ class and 225 spectra from ‘Cp’ class. This process of randomly selecting 100 spectra to predict the remaining 350 spectra from the 450 visually verified spectra was repeated 1,000 times using Monte Carlo re-sampling 42, 43. The re-sampling procedure allowed calculation of mean and median number of errors as well as the overall percentage error rates for each of the 24 OPLS-DA models to be assessed. Predictive ability of the constructed model using different pre-processing parameters in the OPLS-DA model was also assessed. These 24 different OPLS-DA models were each re-sampled for 1,000 iterations (PCT1 to PCT24), all using one predictive and one orthogonal component, for modeling the discrimination between the two classes of urine spectra. A Y-predicted value of 0.5 was used as cut off value between the two classes, i.e., spectra falling below 0.5 assigned to the ‘Cp’; those over 0.5 assigned to the ‘P’ class, following criteria described by Keun et al 44. The model with the lowest overall mean error rates was considered as the optimal model. However, when more than one model showed an identical low overall mean error rates, the mean number of errors in predicting spectra from ‘P’ and ‘Cp’ class was also considered in order to determine the model with an optimal prediction capacity. The optimized parameters were then applied to evaluate presence or absence of acetaminophen in the remaining 4,218 individuals (n = 8,436 urine spectra) for the INTERMAP urine spectra where the presence or absence of acetaminophen in the urine spectra were not visually verified. A further subset (n = 300 of 8,436) of urine spectra were visually verified to further confirm the predictive ability of the optimal model.
Ibuprofen Metabolite Model
To further exemplify this method for drug metabolite screening, the same parameters used for constructing the optimal acetaminophen model were applied for detecting the presence or absence of ibuprofen metabolite signals in the urinary spectra. A similar strategy for selecting samples with or without ibuprofen metabolites was undertaken. Forty percent (n = 104) of spectra identified as containing ibuprofen were chosen based on the scores of the global PCA model from PC 8, n = 260 (40 % instead of 30 % as in the case of acetaminophen since there were relatively fewer spectra containing ibuprofen than acetaminophen metabolites, to ensure sufficient sample size for constructing the prediction model). Visual examination of these selected spectra confirmed that 100 of the 104 spectra contained ibuprofen metabolites (from here on, these 100 spectra are referred to as ‘I’ class). A further 3.0 % (n = 266) of the spectra were selected from the remainder of the 9,000 spectra and all 266 spectra were confirmed not to have ibuprofen metabolites. These were grouped together with the 4 spectra from the 104 spectra found not to display ibuprofen metabolites (from here on, referred to as ‘Ci’ class) giving a total of 270 spectra that did not contain ibuprofen metabolites. Thus, the complete dataset consisted of 370 spectra, from which 50 spectra were randomly selected from the ‘Ci’ class and another 50 spectra from the ‘I’ class. These combined 100 spectra were used to predict the presence or absence of ibuprofen metabolites from the remaining 270 spectra. A Monte Carlo re-sampling strategy (n=1,000 iterations) was applied to assess the accuracy of the OPLS-DA model in predicting the presence or absence of ibuprofen metabolites (known as IBU1). This particular set of 370 spectra corresponded to the 1H NMR urinary spectra for 328 individuals and a further 21 individuals from whom both urinary spectra were used (n = 370 spectra, 328 individuals). Once IBU1 had been validated using the 370 spectra, it was subsequently used to evaluate the spectra from the remaining 4,302 individuals (excluding 328 individuals, 656 spectra) where their spectra had not been verified visually for the presence or absence of ibuprofen metabolites. All data analyses were performed using in-house Matlab ® 7.3.0 (MathWorks, Natick, MA) routines.
RESULTS
Assessment of predictive ability for acetaminophen models
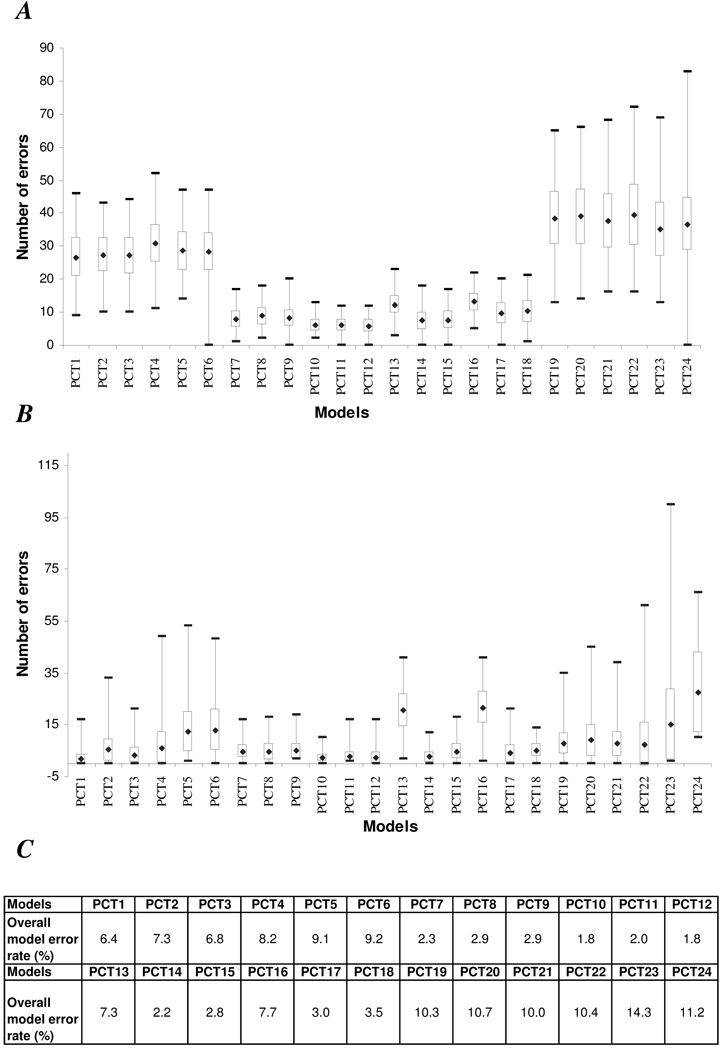
Distribution of numbers of errors (based on 1,000 re-sampling iterations) for the 24 acetaminophen OPLS-DA prediction models for spectra with acetaminophen metabolites is shown in Figure 2 (A); for spectra without acetaminophen metabolites in Figure 2 (B). Models PCT7 to PCT18 (exception for models PCT13 and PCT16) had lower overall mean error rates (< 5 %) in predicting class membership of the spectra in classes ‘P’ and ‘Cp’ (Figure 2 (C)). These models all shared a common normalization feature, i.e., chemical shift regions included in the modeling were normalized to the sum of acetaminophen metabolite regions at δ 1.84 – 1.87, 2.12 –2.23 and 7.12 – 7.48 rather than the whole spectrum. This observation suggests that this method of normalization had a greater impact on predictive ability of the model than bin width or scaling method. Data normalized to the specific peaks of interest allowed the models to focus on more subtle variation in acetaminophen and its metabolites. Thus these models were more predictive than models normalized to total spectral area (PCT1 to PCT6), or to the creatinine peak (PCT19 to PCT24). Additionally, when comparing models PCT7 to PCT18, it was noted that for models based on the same bin size and pre-processing parameters, those with only selected acetaminophen regions tended to have a lower number of errors than models that included the full spectral region, e.g. PCT10 vs PCT13. With respect to the impact of bin size, models using the smaller bin size (i.e., 0.01 ppm per bin for PCT10 to PCT15) tended to show lower overall mean error rates than those constructed using the 0.04 ppm bin size (PCT7 to PCT9 and PCT16 to PCT18) (Figure 2). The scaling method generally had little impact on the overall mean error, except for models PCT13 and PCT16, where mean-centred unscaled data showed a 2 to 3 fold higher overall error rate than models with unit-variance scaling (model PCT14 and PCT17) or with pareto scaling (model PCT15 and PCT18). This suggests that the type of scaling has little impact on the predictive ability of the model if appropriate pre-processing parameters are used. Figure 2 also clearly shows that the normalized models that included selected acetaminophen regions only (PCT10 for mean-centred unscaled data, PCT11 for mean-centred and unit-variance scaled data, and PCT12 for mean-centred and pareto scaled data) not only showed low number of mean errors but also demonstrated a narrow distribution of errors based on Monte Carlo re-sampling of the 100 spectra (n = 1,000 times). Models PCT19 to PCT24, based on normalization to the creatinine peak, tended to have higher overall number of mean errors (> 10 %) in comparison to models normalized to total spectral area or to acetaminophen metabolite regions. The relative width of the error bars for models PCT19 to PCT24 was also larger, particularly for predicting spectra containing acetaminophen metabolites, suggesting that creatinine excretion is variable and metabolism of acetaminophen is generally not dependent on renal function in the population at large.
Figure 2.
Box-plot of minimum and maximum number of errors in predicting 1H NMR urine spectra, also the mean number of errors ± one standard deviation, after 1000 re-sampling iterations for (A) spectra containing acetaminophen related metabolites (n = 175); (B) spectra not containing acetaminophen related metabolites (n = 275); (C) percentage of overall model error rates, calculated based on the mean number of spectra incorrectly predicted for spectra with and without acetaminophen metabolites.
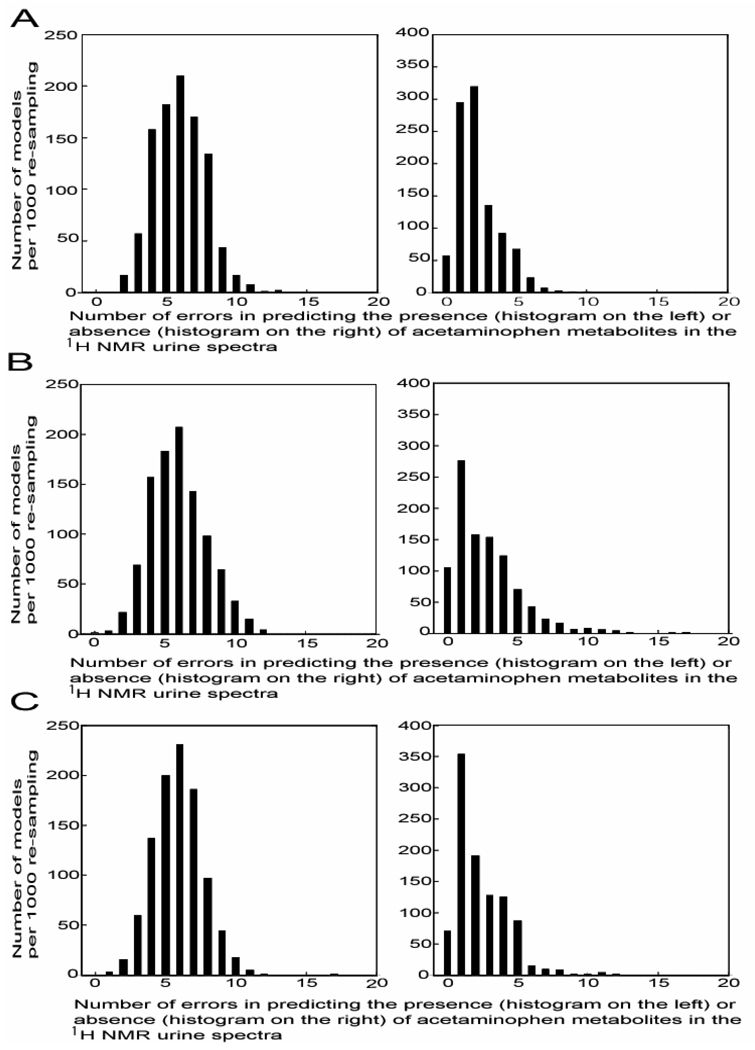
Overall statistics for PCT10 (mean-centred data) and PCT12 (unit variance scaled data) were the same, with an overall model error rate of 1.8 %; 95 % confidence interval (CI) for number of errors for class ‘P’ was 3 to 9 errors per 125 spectra, 0 to 5 errors per 225 spectra for class ‘Cp’ (data not shown). However, maximum and minimum distribution error rates were smaller for PCT10 than PCT12 for predicting both spectra with and without acetaminophen, based on the 1,000 iteration re-sampling procedure (Figure 2). The distribution of number of errors for prediction for models PCT10 to PCT12 after re-sampling iterations (n = 1,000) is shown in Figure 3 and Supplementary Figure 4 and Supplementary Figure 5 for all other models. In general, model PCT10 incorrectly predicted ≤ 6 (which corresponds to the median number of errors for model PCT10 when predicting class ‘P’) of the 125 spectra from class ‘P’ in 624 of the 1,000 re-sampling iterations. For the remaining 376 iterations, model PCT10 incorrectly predicted ≤ 13 spectra from class ‘P’. As for spectra from class ‘Cp’, model PCT10 incorrectly predicted ≤ 2 spectra (which corresponds to the median number of errors for model PCT10 when predicting class ‘Cp’) in 671 of the 1,000 re-sampling iterations, whilst for the remaining 329 iterations, model PCT10 incorrectly predict ≤ 10 spectra from class ‘Cp’ . Thus, the PCT10 model was considered the optimal model for predicting the presence or absence of acetaminophen in the urine spectra and was subsequently used to predict the presence or absence of aceaminophen metabolites in the remaining urine spectra of the INTERMAP Study (n = 8,436) not visually verified.
Figure 3.
Histograms of number of errors in predicting specimens with acetaminophen related metabolites (red bars) and specimens without acetaminophen related metabolites (blue bars) in each of the 1000 re-samplings for spectra with the presence or absence of acetaminophen visually verified for (A) model PCT10, based on mean-centred data; (B) model PCT11, based on unit-variance scaling and (C) model PCT12, based on pareto scaling. Results for all other models, see Supplementary Figure 4 and Supplementary Figure 5.
Ibuprofen Metabolite Prediction Models
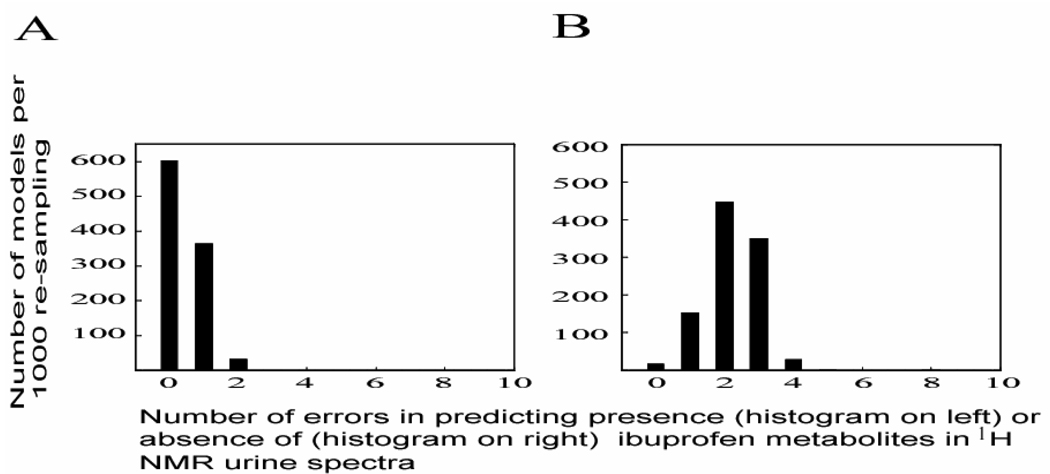
Based on the optimal validated acetaminophen prediction model (i.e. model PCT10), presence or absence of ibuprofen and its metabolites in the 1H NMR urine spectra was assessed using 23 equal chemical shift regions across the spectral region containing ibuprofen metabolites i.e. δ 0.87 -0.90, 1.05 -1.09, 1.21 - 1.23, 1.38 -1.43 and 1.51 – 1.55 at a width of 0.01 ppm, and data were normalized to these selected ibuprofen regions. Ibuprofen metabolite peaks in the regions δ 3.64 and 3.98 were excluded from the multivariate data analysis due to potential interference by other endogenous metabolites e.g., glucose and hippurate resonances. Aromatic spectral regions for ibuprofen metabolites (δ 7.22 – 7.33) were also excluded due to potential interference from both gut microbial (e.g. p-cresol sulfate, phenylacetylglutamine, Supplementary Figure 2) and exogenous acetaminophen glucuronide metabolites. Validation of the ibuprofen prediction model (IBU1) was performed using 370 spectra. Random selection of 100 spectra was repeated 1,000 times for mean-centred data as described for acetaminophen modeling. Model IBU1 (mean-centred un-scaled data normalized to selected ibuprofen metabolites region, at a bin width of 0.01 ppm for selected ibuprofen metabolite regions only) showed a low overall model error rate of 1.0 %; maximum number of errors for predicting spectra with and without ibuprofen metabolites was ≤ 2 and ≤ 4 respectively (Figure 4). Model IBU1 correctly predicted all 50 spectra for 604 of the 1,000 re-sampling iterations and incorrectly predicted ≤ 2 of 50 spectra from class ‘I’ for the remaining 396 iterations. For spectra from class ‘Ci’, the model incorrectly predicted ≤ 2 of the 220 spectra (which corresponds to the median number of errors for model IBU1) in 617 of the 1,000 re-sampling iterations, whist for the remaining 383 iterations, model IBU1 incorrectly predicted ≤ 4 spectra. Due to its high sensitivity and specificity in determining the presence or absence of ibuprofen metabolites, parameters used in this model were deemed appropriate for predicting presence or absence of ibuprofen metabolites for the remaining INTERMAP spectra (n = 8,604, after excluding 656 spectra which corresponds to 328 individuals where either one of the two spectra acquired for each participant was included in the construction of model IBU1).
Figure 4.
Histograms of overall distribution, number of errors for model IBU1, based on mean-centred data, for predicting (A) spectra with ibuprofen and (B) without ibuprofen related metabolites.
Prediction of acetaminophen and ibuprofen on non-verified INTERMAP data
The presence or absence of acetaminophen or ibuprofen for the remaining INTERMAP data, which were not visually verified (n = 8,436 for acetaminophen and n = 8,604 for ibuprofen), but were determined here using the optimized parameters as defined in models PCT10 and IBU1. Both models PCT10 and IBU1 were constructed based on: selected regions of interest, data normalized to the regions of interest and mean-centred without further scaling using a bin width size 0.01 ppm.
To predict the presence or absence of acetaminophen, the calculated OPLS-DA model (from here on, this prediction model is known as the PCT model) using one predictive component and one orthogonal component, described 80.1 % of the variation in X (R2X = 0.801), 87.0 % of the variation in response Y (R2Y = 0.870) and predicted 85.7 % (Q2Y = 0.857) of the variation in response Y, as determined by seven fold cross-validation. R2 is defined as the proportion of variance in the NMR data matrix (X) or class discrimination (Y) explained by the model and Q2 is defined as the proportion of variance in the data predictable by the model under cross validation. The high Q2Y value is thus a measure of the robustness of the model. Based on the distribution of Y-predicted scores of the model PCT, 415 (4.9 %) of the remaining 8,436 urine spectra were predicted as containing acetaminophen metabolites; the remaining 8,021 (95.1 %) urine spectra were predicted as not containing acetaminophen metabolites. To further estimate error rates for model PCT, 300 spectra were randomly selected from spectra not included in the construction of the 24 prediction models, 150 from the 415 spectra predicted as containing acetaminophen metabolites and another 150 spectra from the remaining 8,436 spectra predicted as without acetaminophen metabolites in the urinary spectra. Model PCT incorrectly predicted 19 of the 300 spectra (6.3 %), of which 12 spectra were incorrectly predicted to contain acetaminophen metabolites due to the presence of overlapping peaks in the aromatic regions, and the remaining 7 spectra were incorrectly predicted as not containing acetaminophen metabolites. Assuming binomial statistics (n = 300, p = 0.063) the estimated 95 % CI for the error rate was 3.5 – 9.1 %.
As for the Ibuprofen model (from here on designated as IBU) based on one predictive component and one orthogonal component, the model statistics were R2X = 0.887; R2Y = 0.924 and Q2Y = 0.918. The Y-predicted scores of model IBU showed 245 urine spectra (2.8 %) as containing ibuprofen metabolite resonances and the remaining 8,359 spectra (97.2 %) as not containing ibuprofen metabolites. To estimate the error rate for model IBU, a random selection of 300 spectra from the set of spectra not used to construct the model IBU1 were assessed. A total of 150 spectra were randomly selected from the 245 spectra predicted to contain ibuprofen metabolites and a further 150 spectra from the remaining 8,359 spectra predicted not to contain ibuprofen metabolites. The model IBU was found to correctly predict all 300 spectra.
DISCUSSION
This study presents a novel approach to the empirical assessment of self-administrating drug exposure and a means of testing the veracity of self-reported data via use of a high throughput analytical platform in an epidemiologic setting. The approach is exemplified in a large population study consisting of 4,630 participants each sampled twice. From a pharmaco-epidemiological point of view, the results give useful insights into the urinary prevalence of two commonly used non-prescription analgesics in human populations. The approach provides a rapid means of objective screening for xenobiotics excreted in the urine. However, the prevalence of analgesics is not the main focus of this paper and will not be discussed further here. These results will be reported elsewhere.
Pharmacy or general practitioner electronic databases are commonly used to determine medication exposure together with self-reported data on the usage of medicines45–49. Although there is good concordance between self-reported and electronic data, none of these methods fully captures actual drug exposure, particularly if the drug involved can be obtained OTC, if patients use multiple pharmacies or surgeries, or do not take prescribed medication. Thus, the extent of over- or under-reporting of OTC medications cannot be fully evaluated due to the lack of a suitable external gold standard. Our results illustrate that it is possible to reliably predict absence or presence of acetaminophen and ibuprofen related metabolites based on prediction models generated from a relatively small subset of specimens. This predicted information can be used as an indicator of validity of such self-reported data. The strategy applied here offers the advantage of not depending on recall accuracy, as is the case for self-reported data. This could potentially strengthen our ability to generate hypotheses in epidemiologic studies and help overcome potential biases introduced by the under- or over-reporting of drug use.
As with any analytical technology, sensitivity and limits of detection of the method need to be established for each metabolite assessed, as well as accuracy which may be reduced by interfering factors e.g., overlapping peaks within spectra. In this study, optimal prediction models were based on selected regions of the NMR spectra containing the metabolites of interest, since inclusion of full spectral data introduces factors that confounded the models, e.g., variation in the intensity of glucose resonances, particularly apparent between diabetic and non-diabetic participants. Additionally, normalization of data matrices to the selected regions of NMR spectra containing the metabolites of interest focused the models on drug related variation. The bin size width and types of scaling factors were less influential in determining predictive ability of models once an appropriate normalization method was in place. The prediction models were able to correctly predict > 98 % of the spectra that had been visually verified based on the overall error rate (median error rates of 6.0 % for spectra containing acetaminophen and 2.0 % for spectra not containing acetaminophen based on model PCT10; median error rates of 0 % for spectra containing ibuprofen and 2.0 % for spectra not containing ibuprofen based on model IBU1). The enhanced predictive ability of the ibuprofen model probably reflects the fact that ibuprofen metabolites are more defined and less overlapped in the urinary spectra than those from acetaminophen.
Although NMR based approaches offer many advantages over other specific biochemical or enzymatic assays, the lack of sensitivity of NMR when compared to other spectroscopic methods presents a potential limitation in detecting low concentration of xenobiotics. NMR is more suitable for assessing the plasma concentration or urinary excretion of drugs that have been recently administered and are present at relatively high concentrations. For drugs administered at lower concentrations or with relatively low urinary excretion rates, alternative spectroscopic methods with higher sensitivity such as mass spectrometry may be more suitable. Although the half-life of the drug metabolites, the time of administration of the drug and the variation in metabolism between individuals due to disease states50–52 or ethnicities 53 were not considered here, the use of chemometric tools together with NMR can still provide useful means of assessing the compliance of medication intake. This study was performed with a focus on common analgesics, but the approach is generic and could be applied in a wide range of settings and with NMR could be extended in principle to pollutants or dietary products to give greater xenometabolome17 coverage. The method may be of particular relevance to the investigation of nutritional data e.g. to establish nutrient intake. Recent studies have successfully demonstrated the ability of NMR to detect different metabolic signatures, which are associated with altered dietary intake.54–61 NMR spectroscopy is an inherently quantitative technique as peak areas are proportional to the number of protons within the molecule and hence sample concentration. With the addition of an internal standard of known concentration and the use of deconvolution algorithms for partially overlapped spectral resonances 62, absolute concentration of the metabolites may be calculated. This ability to quantitate metabolite concentrations from NMR urinary spectra can equally be applied to calculate the excretion rate of a particular food or xenobiotic. Moreover, by integrating metabonomic and epidemiologic approaches, it is possible to improve the characterization of the use of xenobiotics in participants in epidemiologic studies and to assess the compliance of medication intake.
CONCLUSION
We have shown that the combined use of 1H NMR and pattern recognition approaches provide a reliable and an untargeted method for assessing self administered drug exposure in large scale population studies. This offers the possibility of acting as a non-selective high throughput platform for validating self-reported data on medication use, which may strengthen hypotheses generated from epidemiologic studies in relation to drug exposures and disease risks. It can also help correct bias introduced through reliance on self-reported data. This is particularly the case where the drug under investigation can be obtained OTC. Using this strategy, similar screening methods could be developed for a range of other high throughput analytical platforms such as liquid chromatography hyphenated with mass spectrometry.
Supplementary Material
Acknowledgements
We are grateful to the US National Heart, Lung, and Blood Institute, Bethesda, Maryland, MD, USA for their financial support of the project. The INTERMAP Study is supported by grant 2-RO1-HL50490, US NHLBI; and national and local agencies in the four countries, and the INTERMAP Metabonomics Study was supported by grant 5-RO1-HL71950-2 US NHLBI. It is a pleasure also to express appreciation to the many colleagues who collected and processed the INTERMAP data; for a listing of many of them, see Stamler et al 39. NMR spectra were preprocessed using MetaSpectra, a program developed in Matlab at Imperial College by O.C.
Abbreviations
- FID
Free Induction Decay
- NMR
Nuclear Magnetic Resonance
- OPLS-DA
Orthogonal Projection to Latent Structures Discriminant Analysis
- PCA
Principal Component Analysis
- SD
Standard Deviation
References
- 1.Nicholson JK, Wilson ID. Progress in Nuclear Magnetic Resonance Spectroscopy. 1989;21:449–501. [Google Scholar]
- 2.Prakash C, Chen W, Rossulek M, et al. Drug Metab Dispos. 2008;36:2064–2079. doi: 10.1124/dmd.108.022277. [DOI] [PubMed] [Google Scholar]
- 3.Prior MJ, Maxwell RJ, Griffiths JR. Biochem Pharmacol. 1990;39:857–863. doi: 10.1016/0006-2952(90)90200-5. [DOI] [PubMed] [Google Scholar]
- 4.Bales JR, Higham DP, Howe I, et al. Clin Chem. 1984;30:426–432. [PubMed] [Google Scholar]
- 5.Shockcor JP, Unger SE, Wilson ID, et al. Anal Chem. 1996;68:4431–4435. doi: 10.1021/ac9606463. [DOI] [PubMed] [Google Scholar]
- 6.Wade KE, Wilson ID, Troke JA, et al. J Pharm Biomed Anal. 1990;8:401–410. doi: 10.1016/0731-7085(90)80067-y. [DOI] [PubMed] [Google Scholar]
- 7.Bales JR, Nicholson JK, Sadler PJ. Clin Chem. 1985;31:757–762. [PubMed] [Google Scholar]
- 8.Dumas ME, Maibaum EC, Teague C, et al. Anal Chem. 2006;78:2199–2208. doi: 10.1021/ac0517085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maher AD, Crockford D, Toft H, et al. Anal Chem. 2008;80:7354–7362. doi: 10.1021/ac801053g. [DOI] [PubMed] [Google Scholar]
- 10.Nicholson JK, Holmes E, Elliott P. J Proteome Res. 2008;7:3637–3638. doi: 10.1021/pr8005099. [DOI] [PubMed] [Google Scholar]
- 11.Holmes E, Loo RL, Stamler J, et al. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes E, Wilson ID, Nicholson JK. Cell. 2008;134:714–717. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 13.Cloarec O, Dumas ME, Craig A, et al. Anal Chem. 2005;77:1282–1289. doi: 10.1021/ac048630x. [DOI] [PubMed] [Google Scholar]
- 14.Keun HC, Athersuch TJ, Beckonert O, et al. Anal Chem. 2008 doi: 10.1021/ac702040d. [DOI] [PubMed] [Google Scholar]
- 15.Cloarec O, Campbell A, Tseng LH, et al. Anal Chem. 2007;79:3304–3311. doi: 10.1021/ac061928y. [DOI] [PubMed] [Google Scholar]
- 16.Smith LM, Maher AD, Cloarec O, et al. Anal Chem. 2007 doi: 10.1021/ac0703754. [DOI] [PubMed] [Google Scholar]
- 17.Holmes E, Loo RL, Cloarec O, et al. Anal Chem. 2007;79:2629–2640. doi: 10.1021/ac062305n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crockford DJ, Holmes E, Lindon JC, et al. Anal Chem. 2006;78:363–371. doi: 10.1021/ac051444m. [DOI] [PubMed] [Google Scholar]
- 19.Crockford DJ, Maher AD, Ahmadi KR, et al. Anal Chem. 2008;80:6835–6844. doi: 10.1021/ac801075m. [DOI] [PubMed] [Google Scholar]
- 20.Forrest JA, Clements JA, Prescott LF. Clin Pharmacokinet. 1982;7:93–107. doi: 10.2165/00003088-198207020-00001. [DOI] [PubMed] [Google Scholar]
- 21.Blackledge HM, O'Farrell J, Minton NA, et al. Hum Exp Toxicol. 1991;10:159–165. doi: 10.1177/096032719101000302. [DOI] [PubMed] [Google Scholar]
- 22.Prescott LF. Br J Clin Pharmacol. 1980;10 Suppl 2:291S–298S. doi: 10.1111/j.1365-2125.1980.tb01812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson ID, Nicholson JK. Journal of Pharmaceutical and Biomedical Analysis. 1988;6:151–165. doi: 10.1016/0731-7085(88)80041-4. [DOI] [PubMed] [Google Scholar]
- 24.Davies NM. Clin Pharmacokinet. 1998;34:101–154. doi: 10.2165/00003088-199834020-00002. [DOI] [PubMed] [Google Scholar]
- 25.Lee EJ, Williams K, Day R, et al. Br J Clin Pharmacol. 1985;19:669–674. doi: 10.1111/j.1365-2125.1985.tb02694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Critchley JA, Nimmo GR, Gregson CA, et al. Br J Clin Pharmacol. 1986;22:649–657. doi: 10.1111/j.1365-2125.1986.tb02953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bales JR, Bell JD, Nicholson JK, et al. Magn Reson Med. 1988;6:300–306. doi: 10.1002/mrm.1910060308. [DOI] [PubMed] [Google Scholar]
- 28.Prescott LF. Am J Ther. 2000;7:143–147. [PubMed] [Google Scholar]
- 29.Mills RF, Adams SS, Cliffe EE, et al. Xenobiotica. 1973;3:589–598. doi: 10.3109/00498257309151547. [DOI] [PubMed] [Google Scholar]
- 30.Spraul M, Hofmann M, Dvortsak P, et al. Anal Chem. 1993;65:327–330. doi: 10.1021/ac00052a004. [DOI] [PubMed] [Google Scholar]
- 31.Craig A, Cloarec O, Holmes E, et al. Anal Chem. 2006;78:2262–2267. doi: 10.1021/ac0519312. [DOI] [PubMed] [Google Scholar]
- 32.Gartland KP, Beddell CR, Lindon JC, et al. Mol Pharmacol. 1991;39:629–642. [PubMed] [Google Scholar]
- 33.Gartland KP, Sanins SM, Nicholson JK, et al. NMR Biomed. 1990;3:166–172. doi: 10.1002/nbm.1940030404. [DOI] [PubMed] [Google Scholar]
- 34.Eriksson L, Johansson E, Kettaneh-Wold N, et al. Multi- and megavariate data analysis: Principles and applications. 1st. ed. Umetric Academy: Umea; 2001. [Google Scholar]
- 35.Holmes E, Foxall PJ, Nicholson JK, et al. Anal Biochem. 1994;220:284–296. doi: 10.1006/abio.1994.1339. [DOI] [PubMed] [Google Scholar]
- 36.Cloarec O, Dumas ME, Trygg J, et al. Anal Chem. 2005;77:517–526. doi: 10.1021/ac048803i. [DOI] [PubMed] [Google Scholar]
- 37.Coen M, Hong YS, Cloarec O, et al. Anal Chem. 2007;79:8956–8966. doi: 10.1021/ac0713961. [DOI] [PubMed] [Google Scholar]
- 38.Weljie AM, Newton J, Mercier P, et al. Anal Chem. 2006;78:4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 39.Stamler J, Elliott P, Dennis B, et al. J Hum Hypertens. 2003;17:591–608. doi: 10.1038/sj.jhh.1001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beevers DG, Stamler J. J Hum Hypertens. 2003;17:589–590. doi: 10.1038/sj.jhh.1001609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sperlingova I, Dabrowska L, Stransky V, et al. Anal Bioanal Chem. 2007;387:2419–2424. doi: 10.1007/s00216-006-0708-7. [DOI] [PubMed] [Google Scholar]
- 42.Shao J. Journal of the American Statistical Association. 1993;88:486–494. [Google Scholar]
- 43.Bylesjo M, Rantalainen M, Cloarec O, et al. Journal of Chemometrics. 2006;20:341–351. [Google Scholar]
- 44.Keun HC, Ebbels TMD, Antti H, et al. Analytica Chimica Acta. 2003;490:265–276. [Google Scholar]
- 45.Heerdink ER, Leufkens HG, Koppedraaijer C, et al. Pharm World Sci. 1995;17:20–24. doi: 10.1007/BF01875554. [DOI] [PubMed] [Google Scholar]
- 46.Klungel OH, de Boer A, Paes AH, et al. Pharm World Sci. 1999;21:217–220. doi: 10.1023/a:1008741321384. [DOI] [PubMed] [Google Scholar]
- 47.Curtis JR, Westfall AO, Allison J, et al. Pharmacoepidemiol Drug Saf. 2006;15:710–718. doi: 10.1002/pds.1226. [DOI] [PubMed] [Google Scholar]
- 48.Lau HS, de Boer A, Beuning KS, et al. J Clin Epidemiol. 1997;50:619–625. doi: 10.1016/s0895-4356(97)00040-1. [DOI] [PubMed] [Google Scholar]
- 49.Uiters E, van Dijk L, Deville W, et al. BMC Health Serv Res. 2006;6:115. doi: 10.1186/1472-6963-6-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thalhammer F, Horl WH. Clin Pharmacokinet. 2000;39:271–279. doi: 10.2165/00003088-200039040-00003. [DOI] [PubMed] [Google Scholar]
- 51.Rodvold KA. Clin Pharmacokinet. 1999;37:385–398. doi: 10.2165/00003088-199937050-00003. [DOI] [PubMed] [Google Scholar]
- 52.Kuipers M, Smulders R, Krauwinkel W, et al. J Pharmacol Sci. 2006;102:405–412. doi: 10.1254/jphs.fp0060311. [DOI] [PubMed] [Google Scholar]
- 53.Xie HG, Kim RB, Wood AJ, et al. Annu Rev Pharmacol Toxicol. 2001;41:815–850. doi: 10.1146/annurev.pharmtox.41.1.815. [DOI] [PubMed] [Google Scholar]
- 54.Bertram HC, Hoppe C, Petersen BO, et al. Br J Nutr. 2007;97:758–763. doi: 10.1017/S0007114507450322. [DOI] [PubMed] [Google Scholar]
- 55.Bertram HC, Malmendal A, Petersen BO, et al. Anal Chem. 2007;79:7110–7115. doi: 10.1021/ac070928a. [DOI] [PubMed] [Google Scholar]
- 56.Zuppi C, Messana I, Forni F, et al. Clinica Chimica Acta. 1998;278:75–79. doi: 10.1016/s0009-8981(98)00132-6. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Tang H, Nicholson JK, et al. J Agric Food Chem. 2005;53:191–196. doi: 10.1021/jf0403282. [DOI] [PubMed] [Google Scholar]
- 58.Solanky KS, Bailey NJ, Beckwith-Hall BM, et al. The Journal of Nutritional Biochemistry. 2005;16:236–244. doi: 10.1016/j.jnutbio.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 59.Daykin CA, Duynhoven JP, Groenewegen A, et al. J. Agric. Food Chem. 2005;53:1428–1434. doi: 10.1021/jf048439o. [DOI] [PubMed] [Google Scholar]
- 60.Stella C, Beckwith-Hall B, Cloarec O, et al. J. Proteome Res. 2006;5:2780–2788. doi: 10.1021/pr060265y. [DOI] [PubMed] [Google Scholar]
- 61.Rezzi S, Ramadan Z, Martin F-PJ, et al. J. Proteome Res. 2007;6:4469–4477. doi: 10.1021/pr070431h. [DOI] [PubMed] [Google Scholar]
- 62.Crockford DJ, Keun HC, Smith LM, et al. Anal Chem. 2005;77:4556–4562. doi: 10.1021/ac0503456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.