Abstract
AIM: To establish if PTCH1a transcriptional regulation region (TRR) is methylated in gastric cancer and its influence in gastric tumorigenesis.
METHODS: The CpG islands in PTCH1a TRR were analyzed by Methyl Primer Express v1.0 software. The region from -643 to -355 bp (the transcription initiation site of PTCH1a was designated as 0) that contained 19 CpG sites was chosen for bisulfite-sequencing PCR (BSP) and methylation-specific PCR (MSP) detection. The gastric cancer cell line AGS was treated with 5-aza-2’-deoxycytidine (5-Aza-dC; 1 μmol/L) for 3 d. Alterations in PTCH1a TRR methylation in treated AGS cells was measured through BSP clone sequences, and their PTCH1 expression was measured by quantitative RT-PCR. The cell cycle and apoptosis were observed with flow cytometry through propidium iodide (PI) staining or annexin V/PI double staining. The prevalence of PTCH1a TRR methylation was investigated in 170 gastric cancer tissue samples and the adjacent normal tissues by MSP. The correlation of PTCH1a TRR methylation with PTCH1 expression or with patients’ clinical features was analyzed.
RESULTS: Methylation of PTCH1a TRR was observed in AGS cells and a subset of gastric cancer tissues (32%, 55/170), while no methylation amplification products were observed in any normal tissues by MSP. The methylation of PTCH1a TRR was correlated negatively with PTCH1 expression (Spearman’s r = -0.380, P = 0.000). However, methylation of PTCH1a TRR was not related to the gastric cancer patients’ clinical features, such as sex, age of onset, clinical stage, lymph node metastasis or histological grade. The methylation of PTCH1a TRR in AGS cells was almost converted to non-methylation after 5-Aza-dC treatment, which increased PTCH1 expression (5.3 ± 2.5 times; n = 3) and apoptosis rate (3.0 ± 0.26 times; P < 0.05; n = 3).
CONCLUSION: Methylation of PTCH1a TRR is present in a subset of gastric cancers and correlated negatively with PTCH1 expression. This may be an early event in gastric tumorigenesis and a new treatment target.
Keywords: Carcinogenesis, Methylation, Hedgehog signaling pathway, Methylation, PTCH1, Stomach neoplasms
INTRODUCTION
The hedgehog (HH) pathway plays a critical role in embryonic development, tissue polarity and carcinogenesis. In the HH pathway, Sonic HH binds to the receptor PTCH1, which is encoded by the PTCH1 gene. This liberates the Smoothened protein, which allows glioma-associated oncogene homolog 1 zinc finger protein (GLI) and MYCN transcription factors to turn on target genes, including the PTCH1 gene itself, in a negative feedback loop as a tumor suppressor gene. More recently, abnormal activation of the HH pathway has been reported in subsets of human basal cell carcinoma[1], medulloblastoma[2], pancreatic cancer[3–5], lung cancer[6], prostate cancer[7] and gastrointestinal cancer[8–10].
Gastric cancer is one of the most common cancers worldwide, and has high mortality. Patients with gastric cancer usually present at late stages and have a poor prognosis. Loss-of-function mutation of PTCH1 gene participates in the abnormal activation of the HH pathway, which occurs frequently in some cases of human basal cell carcinoma[11] and medulloblastoma[12], but it has never been observed in gastric cancer[13]. Loss-of-function of tumor suppressor gene is also known to result from methylation of the transcriptional regulation region (TRR). Recently, several studies have argued that PTCH1 TRR methylation is involved in tumorgenesis[14–17]; however, none has been reported in gastric cancer. Previous studies have shown that the PTCH1 gene has three major isoforms in the first exon, PTCH1a, PTCH1b and PTCH1c that code for different N-sequence PTCH1 proteins, PTCH1-l, PTCH1-m and PTCH1-s, respectively, and expression of each is regulated by its own independent TRR[18].
The present study analyzed the methylation of PTCH1a TRR in gastric cancer cell line AGS and some gastric cancer tissue samples. We showed that methylation of PTCH1a TRR took place in a subset of gastric cancers, and was correlated negatively with PTCH1 gene expression. It was not related to the patients’ clinical features of gastric cancer, which suggested that the methylation of PTCH1a TRR might be an early event in gastric tumorigenesis.
MATERIALS AND METHODS
Gastric cancer patients’ tissue samples and cell line
All the tissue samples were obtained from Shanghai Xinhua Hospital with hospital ethics board approval. One hundred and seventy gastric cancer tissue samples were collected from radical gastrectomy, to analyze the methylation of the PTCH1 gene, and its expression. All patients gave informed consent for their specimens to be studied. The tumor and adjacent macroscopically normal tissue samples were preserved in liquid nitrogen immediately after being resected. Only the samples in which the proportion of tumor cells was > 70% and adjacent normal tissues with no inflammation or tumor infiltration were selected. Patients’ clinical features were recorded, including sex, age of onset, clinical stage, lymph node metastasis, and histological grade. Gastric cancer cell line AGS was obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA) and cultured under recommended conditions.
DNA/RNA isolation
Frozen tissue in liquid nitrogen was pulverized for subsequent DNA isolation using the Blood and Cell Culture DNA kit (Qiagen, Hilden, Germany) or RNA isolation with TRIzol (Gibco-BRL, Glasgow, UK) according to the protocols of the manufacturers.
Relative quantitative (RQ) RT-PCR
Total RNA (2.5 μg) was treated with DNAase RQ1 (Promega, Madison, WI, USA) to remove trace amounts of genomic DNA contamination, and converted to cDNA using the oligo (dT) primer system (TaKaRa, Dalian, China), in a total volume of 50 μL. Aliquots of the reaction mixture were used for quantitative PCR amplification with ABI7500 (Applied Biosystems, Foster City, CA, USA) using SYBR Premix EX TagTM (TakaRa). PCR was run for 30 cycles of denaturation at 95°C for 5 s, annealing at 55°C for 20 s, and elongation at 72°C for 20 s. Gene expression was quantified by the comparative CT method, with normalizing CT values to the housekeeping gene β-actin. After amplification, melting curve analysis was performed to ensure the products’ specificity. The RQ value of PTCH1 expression in the samples was calculated in comparison with a calibrator (the expression level of pooled adjacent normal tissue samples). To ensure experimental accuracy, all reactions were performed in triplicate. The primer sequences for the gene amplification are shown in Table 1.
Table 1.
Primers and size of PCR products
| Methods | Primers | Sequence | Length (bp) |
| QRT-PCR | PTCH1 | 5'-TGTGCGCTGTCTTCCTTCTG-3' | 119 |
| 5'-ACGGCACTGAGCTTGATTC-3' | |||
| β-actin | 5'-GCCATCCTGCGTCG-3' | 260 | |
| 5'-TGGGCACCGGAACCGCT-3' | |||
| BSP | 5'-GGGAGTATTTGGGTGGTATATT-3' | 351 | |
| 5'-AAAAAACTACAAAAAAACACCACCTTTC-3' | |||
| MSP | Methylation | 5'-GAGGGATCGATACGAATTC-3' | 143 |
| 5'-GAAAACGCGAAAAAACTAAA-3' | |||
| Non-methylation | 5'-GAGGGATTGATATGAATTT-3' | 143 | |
| 5'-AAAAACACAAAAAAACTAAA-3' |
QRT-PCR: Quantitative PCR.
Methylation-specific PCR (MSP) and bisulfite-sequencing PCR (BSP)
Bisulfite modification, MSP and BSP were performed as described before[19,20]. The primers of MSP and BSP are shown in Table 1. One microgram of genomic DNA was treated using the EZ DNA Methylation Kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s instructions. For MSP, 1 μL modified DNA was amplified using MSP primers that specifically recognized the methylated or unmethylated DNA after bisulfite conversion. CpGenome Universal Methylated DNA (S7821) and CpGenome Universal Unmethylated DNA (S7822) (Chemicon Company, Temecula, CA, USA) were used as control DNA for methylated and unmethylated detection, respectively. Amplification products were visualized by UV illumination on 3% low-range ultra-agarose gel (Bio-Rad Laboratories, Hercules, CA, USA) that contained ethidium bromide. For BSP clone sequence analysis, the PCR products were subcloned into a pMD-18-T vector (TaKaRa). Ten clones were sequenced for cell line AGS and some gastric cancer tissues.
5-Aza-2’-deoxycytidine (5-Aza-dC) treatment
Cells were plated at a density of 3 × 104 cells/cm2 in a six-well plate on day 0. The demethylating agent 5-Aza-dC (Sigma-Aldrich, Deisenheim, Germany) was added on days 1, 2 and 3 to maintain its concentration as 1 μmol/L in fresh medium. Cells were harvested on day 4 for RNA and DNA extraction. Control cells were incubated without the addition of 5-Aza-dC.
Analysis of cell cycle and apoptosis by flow cytometry
About 1 × 106 AGS cells were centrifuged at 1000 r/min for 5 min to remove the culture solution. Cell cycle was measured by propidium iodide (PI) staining (final concentration 100 μg/mL, 0.01 mol/L PBS, pH 7.4; R&D System, Abingdon, UK) and flow cytometry (Becton Dickinson, Fullerton, CA, USA). Meanwhile, cell apoptosis rate was measured by annexin V/PI double staining (R&D Systems) and flow cytometry.
Immunohistochemistry
Sections of 3 μm were dried for 30 min at 72°C, deparaffinized in xylene, and rehydrated in a decreasing ethanol series. Endogenous peroxidase activity was blocked with 3% H2O2 in methanol for 30 min and endogenous biotin with a blocking kit (SP-2001; Vector Laboratories, Burlingame, CA, USA). Antigen retrieval was performed by autoclaving for 10 min at 120°C in 10 mmol/L citrate buffer, pH 6.0. Sections were blocked for 30 min in a protein block (X0909; Dako, Carpinteria, CA, USA), and incubated overnight at 4°C with diluted goat polyclonal antibody directed against human PTCH1 protein (sc-6149, 1:100; Santa Cruz Biotechnology, Santa Cruz, CA, USA). After washing the sections with PBS that contained 0.1% Tween 20, biotinylated secondary antibodies were added for 30 min at room temperature. After extensive rinsing and incubation with avidin-biotin, immunoperoxidase antibody staining was visualized with the 3,3’ diaminobenzidine system (Nichirei, Tokyo, Japan), and the sections were counterstained with Mayer’s hematoxylin. The application of primary antibody to tissue sections was omitted in negative controls.
Statistical analysis
Statistical analysis was carried out using SPSS version 14.0 (SPSS, Chicago, IL, USA). Differences were considered statistically significant at P < 0.05. The nonparametric correlations of PTCH1 expression with methylation were analyzed with Spearman’s test. Differences in the clinicopathological parameters between positive and negative PTCH1a TRR methylation were determined with the χ2 test.
RESULTS
PTCH1a TRR methylation and PTCH1 gene expression in gastric cancer cell line AGS
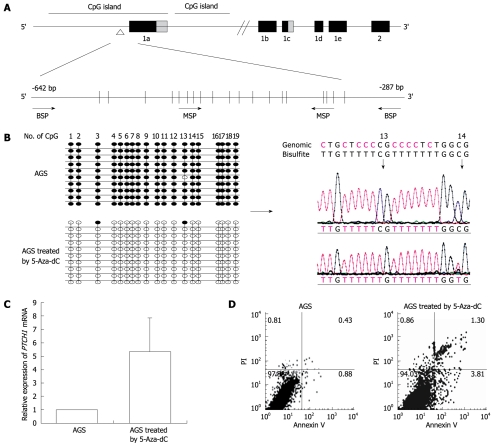
According to National Center for Biotechnology Information, PTCH1 gene has three mRNA transcripts. We analyzed the CpG island at -3950 bp upstream and +2050 bp downstream from the transcription initiation site of PTCH1a (designated as 0) by the methylation analysis software Methyl Primer Express v1.0 (Applied Biosystems). As shown in Figure 1A, two CpG islands exist in this region. One is -1139 to +860 bp and the other is +875 to +1692 bp. The region from -643 to -355 bp in the first CpG island that contained 19 CpG sites was chosen for the BSP and MSP primer amplifications. The MSP up-primer contained the fifth to eighth CpG sites, while the down-primer contained the sixteenth to eighteenth CpG sites. The methylation level of PTCH1a TRR in gastric cancer cell line AGS treated with 5-Aza-dC after 72 h was measured through BSP clone sequences. As shown in Figure 1B, almost all the CpG sites were methylated in the untreated cells, while almost all of them were converted to unmethylated after treatment with 5-Aza-dC. The RQ value of PTCH1 expression increased by 5.3 ± 2.5 times (P < 0.05, n = 3) (Figure 1C). The cell cycle had no significant alteration after treatment (data not shown), by PI staining. However, as shown in Figure 1D, the apoptosis rate increased significantly by 3.0 ± 0.26 times (P < 0.05, n = 3) by annexin V/PI double staining. These results indicated that the PTCH1a TRR was highly methylated in AGS cells, and became unmethylated after 5-Aza-dC treatment, which substantially increased PTCH1 expression and induced more apoptosis.
Figure 1.
Analysis of methylation and expression of PTCH1a in gastric cancer cell line AGS. A: Illustration of PTCH1a TRR and topology of MSP and BSP primers. BSP detection region contained 19 CpG sites, and MSP up-primer and down-primer contained four and three CpG sites, respectively. The detection amplicon is indicated by the empty triangle; B: Alteration of PTCH1a TRR methylation in gastric cancer cell line AGS treated with 5-Aza-dC. Genomic DNA from untreated AGS cells and those treated with 5-Aza-dC (1 μmol/L) were analyzed by BSP at day 4. The left column indicates alterations of the 19 CpG sites contained in the BSP amplicon through 10 cloned sequences after 5-Aza-dC treatment. The right column displays part of the sequence of the methylated and unmethylated clones. Black dot, methylated; white dot, unmethylated; C: Alteration of PTCH1 expression in gastric cancer cell line AGS treated with 5-Aza-dC. PTCH1 gene expression in AGS cells treated with 5-Aza-dC (1 μmol/L) at day 4 was detected by real-time PCR relative to untreated AGS cells. Expression of PTCH1 gene was enhanced significantly in the treated AGS cells compared with untreated (P < 0.01, n = 3, independent tests). Box, mean; bar, SD; D: Analysis of apoptosis of AGS cells treated with 5-Aza-dC. The rate of apoptosis in AGS cells treated with 5-Aza-dC (1 μmol/L) at day 4 was significantly higher compared to untreated cells (P < 0.01, n = 3). The representative analysis of AGS cell apoptosis by annexin V/PI method is shown.
PTCH1a TRR methylation in gastric cancer tissues
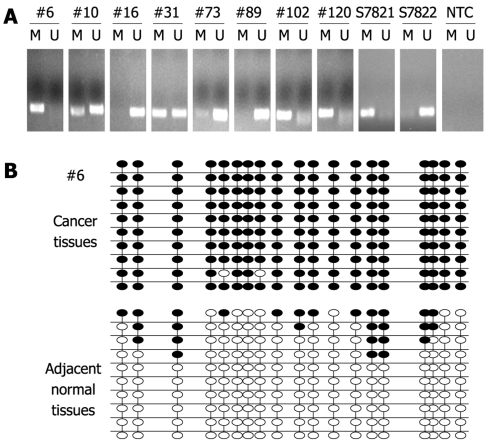
In order to investigate the prevalence of PTCH1 TRR methylation in gastric cancer tissues, detection of PTCH1a TRR methylation in 170 gastric cancer tissues was performed by MSP. If the methylation amplification products appeared after electrophoresis in the investigated sample, PTCH1 TRR was predicted to be methylated. The prevalence of PTCH1a TRR methylation was 32% (55/170) in gastric cancer tissues, while no methylation amplification products were observed in any normal tissues (data not shown). Part of the representative MSP amplification products electrophoretogram is shown in Figure 2A. To further confirm the fact of PTCH1a TRR methylation, we chose gastric cancer tissue sample #6 with a positive methylation amplification product, and the pool of adjacent normal tissues (n = 12) for BSP clone sequencing. As shown in Figure 2B, almost all of the 19 CpG sites in 10 clones exhibited methylation in cancer tissues, while very few CpG sites in adjacent normal tissues did. These results demonstrated that the methylation of PTCH1a TRR did exist in a subset of gastric cancer tissues.
Figure 2.
Methylation of PTCH1a TRR in gastric cancer tissues. A: Methylation of PTCH1a TRR in gastric cancer tissues (n = 170) using MSP. MSP results from eight representative patients (#) are shown. The DNA bands in lanes labeled with M represent the products amplified with the methylation-specific primers, while DNA bands labeled with U represent the products amplified with the non-methylation-specific primers. CpGenome Universal Methylated DNA (S7821) and the CpGenome Universal Unmethylated DNA (s7822) were used as controls for methylation and non-methylation. Water was used as non-template control (NTC); B: Genomic DNA of gastric cancer tissues and its corresponding normal tissues from a representative patient (#6) was analyzed by BSP. Methylation patterns of the 19 CpG sites contained in the BSP amplicon through 10 clone sequence analyses in cancer tissues, and corresponding normal tissues are shown. Black dot, methylation; white dot, non-methylation.
Correlation between methylation of PTCH1a TRR and PTCH1 expression in gastric cancer tissues
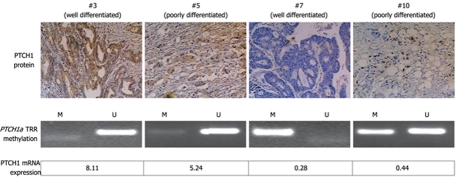
We analyzed the correlation between methylation of PTCH1a TRR and PTCH1 expression. As shown in Figure 3, there was a significant difference in PTCH1 mRNA expression between methylated and unmethylated gastric cancer tissues. High expression had a negative correlation with high methylation (Spearman’s r = -0.380; P = 0.000). To further determine this negative correlation, the PTCH1 protein was examined in four representative samples by immunohistochemistry. As shown in Figure 4, two samples (#3 and #5) with visible unmethylated products by MSP were positive for PTCH1 protein and had higher RQ value of PTCH1 mRNA expression, while another two samples (#7 and #10) with visible methylated products by MSP were negative for PTCH1 protein and had lower RQ value of PTCH1 mRNA expression. Samples #3 and #7 were well-differentiated, while #5 and #10 were poorly differentiated. Notably, this was further proof that a subset of gastric cancer tissues were characterized by methylation of PTCH1a TRR, along with lower expression of the PTCH1 gene.
Figure 3.
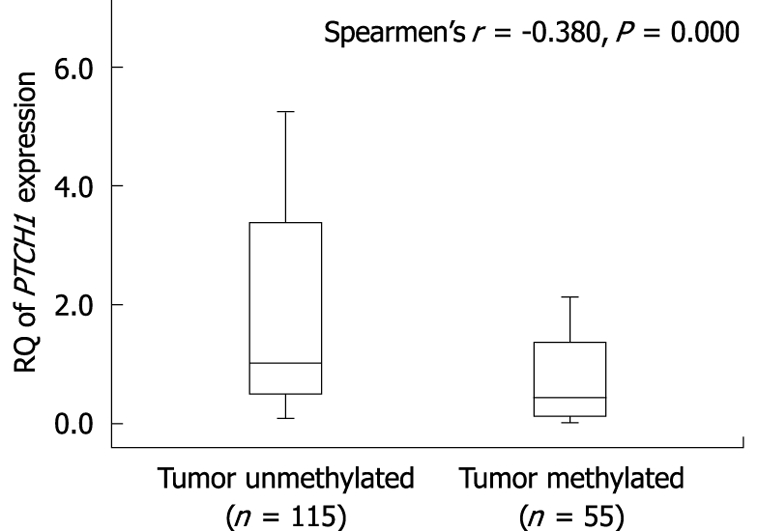
Correlation between methylation of PTCH1a gene TRR and expression of PTCH1 in gastric cancer tissues. Box plot illustrating the loss of PTCH1 gene expression in relation to the methylation of PTCH1a gene TRR in human gastric cancer tissues (n = 170). The Y axis indicates the RQ value of PTCH1 gene mRNA expression was calculated in comparison with a calibrator (the expression level of pooled adjacent normal tissue samples). Horizontal lines: Group medians; Boxes: 25%-75% quartiles; Vertical lines: Range, peak and minimum.
Figure 4.
Correlation of PTCH1 mRNA and protein expression with methylation of PTCH1a TRR. Expression of PTCH1 genes, as well as methylation of PTCH1a TRR are displayed in four representative gastric cancer tissue samples (#3, #5, #7 and #10). PTCH1 protein expression was detected by immunohistochemical staining (original magnification, × 100). Methylation of PTCH1a TRR was detected by MSP. The RQ value of PTCH1 gene mRNA expression was calculated in comparison with a calibrator (the expression level of pooled adjacent normal tissue samples). The well-differentiated tissue sample #3 and the poorly differentiated tissue sample #5 showed non-methylated products by MSP, positive expression of PTCH1 protein, and higher RQ value of PTCH1 mRNA expression. The well-differentiated tissue sample #7 and poorly differentiated tissue sample #10 showed methylated products by MSP, negative expression of PTCH1 protein, and lower RQ value of PTCH1 mRNA expression.
Relationship between PTCH1a TRR methylation in gastric cancer tissues and clinical features
We analyzed statistically the relationship between the methylation of PTCH1a TRR in gastric cancer tissues and clinical features. As shown in Table 2, there was no correlation between the methylation of PTCH1a TRR and clinical features, including sex, age of onset, clinical stage, lymph node metastasis, and histological grade. These data suggest that methylation of PTCH1a TRR is an early event in gastric tumorigenesis.
Table 2.
Clinical features in relation to methylation of PTCH1a TRR in gastric cancer
| Variable |
PTCH1methylation |
|||
| n | Positive | Negative | P value | |
| Total | 170 | 55 | 115 | |
| Clinicopathological parameters | ||||
| Sex | ||||
| Male | 89 | 26 | 63 | 0.359 |
| Female | 81 | 29 | 52 | |
| Age of onset (yr) | ||||
| < 50 | 68 | 22 | 46 | 1.000 |
| ≥ 50 | 102 | 33 | 69 | |
| Clinical stage | ||||
| pT1 | 75 | 25 | 50 | 0.808 |
| pT2-4 | 95 | 30 | 65 | |
| Lymph node metastasis | ||||
| pN0 | 55 | 16 | 39 | 0.530 |
| pN1-3 | 115 | 39 | 76 | |
| Histological grade | ||||
| Well and moderately differentiated | 63 | 19 | 44 | 0.735 |
| Poorly differentiated | 107 | 36 | 71 | |
DISCUSSION
The methylation of tumor suppressor gene plays an important role in the tumorigenesis of gastric cancer. PTCH1 gene is a known tumor suppressor gene in the HH pathway. Loss of function mutation and epigenetic regulation has been found in many kinds of tumors[3,6,13,14].
PTCH1 gene has three main isoforms of the first exon, PTCH1a, PTCH1b, and PTCH1c, which code for different N-sequence PTCH1 proteins PTCH1-l, PTCH1-m and PTCH1-s, respectively[18]. Each expression is regulated by its own independent TRR. Although it has been reported that PTCH1-l and PTCH1-m have the same effect on inducing apoptosis and suppressing GLI-mediated transcription, only the methylation analysis of PTCH1b TRR has been reported by some research groups. Cretnik et al[17] have reported that methylation of PTCH1b TRR (-1593 bp, transcription initiation site of PTCH1b as 0) occurs in ovarian tumors (dermoids and fibromas) compared to healthy controls, but not in basal cell carcinoma. Wolf et al[14] have demonstrated methylation of PTCH1b TRR (-776 to +1238 bp, transcription initiation site of PTCH1b as 0) in breast cancer cell lines and tissues, which has a negative correlation with PTCH1 expression. However, Pritchard et al[15] have found that there is no methylation of PTCH1b TRR (-983 bp, transcription initiation site of mRNA1b as 0) in primary medulloblastoma.
In the present study, we analyzed the methylation status of PTCH1a TRR. We selected the upstream regulation region of PTCH1a (-643 to +355 bp, transcription initiation site of mRNA1b as 0) as the target region to be analyzed because this region was in the CpG island that appeared nearest to the transcription initiation site of PTCH1a gene, according to the software analysis. In order to investigate the methylation status in a number of gastric cancer tissues, the BSP colon sequence method was used to identify the suitable CpG sites for the MSP primer design. We found that the upstream regulatory sequence of PTCH1a gene was methylated in the gastric cancer cell line and a subset of gastric cancer tissues, and this methylation correlated with low expression of the PTCH1 gene. Nagao et al[18] have reported that the expression of these three isoforms is regulated by GLI transcription factors, one in exon 1a and the other between exons 1a and 1b, in the vicinity of which methylation is found in ovarian tumors[17]. However, we found methylation at -441 bp upstream of the GLI binding site in exon 1, and the methylation of this target region was correlated negatively with PTCH1 gene expression. These results suggest that CpG island methylation in the TRR of PTCH1a gene plays a role in the regulation of not only PTCH1a transcription, but also downstream PTCH1b and PTCH1 transcription.
Several recent studies have demonstrated that activation of the HH signaling pathway is involved in gastric tumorigenesis[21–25]. However, PTCH1 gene expression has not been investigated extensively, especially in normal gastric tissues. Ma et al[26] have reported that PTCH1 mRNA expression was detected by hybridization in about 64% (63/99) of gastric cancer tissues but not in normal gastric tissues (0/18). Many other studies have shown by immunohistochemistry that PTCH1 gene expression was present in the fundic glandular epithelium of the stomach[8,24]. We found that adjacent normal tissues expressed PTCH1 gene, along with being unmethylated, which confirms that PTCH1 gene expression is present in normal gastric tissues.
Berman et al[13] have found that the PTCH1 gene was expressed in six human gastric cancer cell lines including AGS, which indicates that the Hedgehog signaling pathway is activated not by mutation, but by ligand expression. The expression of PTCH1a and PTCH1b genes was equally active in terms of suppressing GLI-mediated transcription, as a negative feedback for regulation of the HH signaling pathway, and induction of apoptosis[18]. We found that the demethylation reagent 5-Aza-dC reversed the methylation of PTCH1a gene, enhanced PTCH1 gene expression, and induced apoptosis. Our results implied that the enhanced expression of PTCH1 gene that resulted from demethylation strengthened the negative feedback function of PTCH1, which provided a new target for treating gastric cancer.
Previous studies have found that a high level of aberrant DNA methylation exists in Helicobacter pylori (H pylori)-infected gastric mucosa and is possibly associated with gastric cancer risk[27,28]. Others have shown that H pylori infection might affect the HH pathway that is involved in gastric carcinogenesis[29,30]. These results suggest that the methylation of PTCH1a TRR in gastric cancer may be triggered by H pylori infection in the early course of carcinogenesis. This will be studied in our laboratory in the future.
We demonstrated that methylation of PTCH1a TRR was present in a subset of gastric cancers. To the best of our knowledge, this phenomenon has not been observed or reported by any research group to date. Methylation of PTCH1 was correlated negatively with PTCH1 gene expression and was not related to clinical features of gastric cancer, which suggested that the methylation of PTCH1a TRR is an early event in gastric tumorigenesis. Downregulation of PTCH1a gene methylation may provide a new therapy for gastric cancer characterized by PTCH1a TRR methylation.
COMMENTS
Background
Abnormal activation of the hedgehog (HH) pathway has been reported in subsets of human basal cell carcinoma, medulloblastoma, pancreatic cancer, lung cancer, prostate cancer and gastrointestinal cancer. Although loss-of-function mutation in the PTCH1 gene participates in the abnormal activation of the HH pathway, several studies have argued that the PTCH1 transcriptional regulation region (TRR) methylation is involved in tumorigenesis. However, none has been reported in gastric cancer.
Research frontiers
Previous studies have shown that the PTCH1 gene has three major isoforms in the first exon, PTCH1a, PTCH1b and PTCH1c that code for different N-sequence PTCH1 proteins PTCH1-l, PTCH1-m and PTCH1-s, respectively. Expression of each is regulated by its own independent TRR. The present study analyzed the methylation of PTCH1a TRR in gastric cancer.
Innovations and breakthroughs
The present study showed that methylation of PTCH1a TRR is present in a subset of gastric cancers, and correlated negatively with PTCH1 gene expression. It was not related to the clinical features of gastric cancer, which suggests that methylation of PTCH1a TRR is an early event in gastric tumorigenesis. To the best of our knowledge, these phenomena have never been observed or reported by any research group to date.
Applications
By understanding that the methylation of PTCH1a TRR might be an early event in gastric tumorigenesis, this study may provide a new therapy for gastric cancer that is characterized by PTCH1a TRR methylation.
Terminology
The HH pathway plays a critical role in embryonic development, tissue polarity and carcinogenesis. In the HH pathway, the receptor PTCH1 is encoded by the PTCH1 gene as a tumor suppressor gene, and plays a role in negative feedback regulation of activation of the HH pathway. Methylation of TRR can induce the loss of gene expression. Thus, methylation of PTCH1a TRR might be an early event in gastric tumorigenesis.
Peer review
The authors detected the methylation of PTCH1a TRR in gastric cancer cell line AGS and 170 gastric cancer tissue samples and adjacent normal tissues, and analyzed the correlation of PTCH1a TRR methylation with PTCH1 expression and clinical features. They revealed that the methylation of PTCH1a TRR correlated negatively with PTCH1 expression. However, the methylation of PTCH1a TRR was not related to the clinical features of gastric cancer, such as sex, age of onset, clinical stage, lymph node metastasis, and histological grade. The results are interesting and may represent a molecular mechanism of gastric carcinogenesis.
Footnotes
Supported by City Hospital Trust Fund and the University of Birmingham Scientific Project Grant
Peer reviewer: Marco Romano, MD, Professor, Dipartimento di Internistica Clinica e Sperimentale-Gastroenterologia, II Policlinico, Edificio 3, II piano, Via Pansini 5, 80131 Napoli, Italy
S- Editor Tian L L- Editor Kerr C E- Editor Zheng XM
References
- 1.Tojo M, Mori T, Kiyosawa H, Honma Y, Tanno Y, Kanazawa KY, Yokoya S, Kaneko F, Wanaka A. Expression of sonic hedgehog signal transducers, patched and smoothened, in human basal cell carcinoma. Pathol Int. 1999;49:687–694. doi: 10.1046/j.1440-1827.1999.00938.x. [DOI] [PubMed] [Google Scholar]
- 2.Shahi MH, Lorente A, Castresana JS. Hedgehog signalling in medulloblastoma, glioblastoma and neuroblastoma. Oncol Rep. 2008;19:681–688. [PubMed] [Google Scholar]
- 3.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernández-del Castillo C, Yajnik V, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shao J, Zhang L, Gao J, Li Z, Chen Z. Aberrant expression of PTCH (patched gene) and Smo (smoothened gene) in human pancreatic cancerous tissues and its association with hyperglycemia. Pancreas. 2006;33:38–44. doi: 10.1097/01.mpa.0000222319.59360.21. [DOI] [PubMed] [Google Scholar]
- 5.Gao J, Li Z, Chen Z, Shao J, Zhang L, Xu G, Tu Z, Gong Y. Antisense Smo under the control of the PTCH1 promoter delivered by an adenoviral vector inhibits the growth of human pancreatic cancer. Gene Ther. 2006;13:1587–1594. doi: 10.1038/sj.gt.3302816. [DOI] [PubMed] [Google Scholar]
- 6.Chi S, Huang S, Li C, Zhang X, He N, Bhutani MS, Jones D, Castro CY, Logrono R, Haque A, et al. Activation of the hedgehog pathway in a subset of lung cancers. Cancer Lett. 2006;244:53–60. doi: 10.1016/j.canlet.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 7.Shaw A, Bushman W. Hedgehog signaling in the prostate. J Urol. 2007;177:832–838. doi: 10.1016/j.juro.2006.10.061. [DOI] [PubMed] [Google Scholar]
- 8.van den Brink GR. Hedgehog signaling in development and homeostasis of the gastrointestinal tract. Physiol Rev. 2007;87:1343–1375. doi: 10.1152/physrev.00054.2006. [DOI] [PubMed] [Google Scholar]
- 9.Yoshizaki A, Nakayama T, Naito S, Wen CY, Sekine I. Expressions of sonic hedgehog, patched, smoothened and Gli-1 in human intestinal stromal tumors and their correlation with prognosis. World J Gastroenterol. 2006;12:5687–5691. doi: 10.3748/wjg.v12.i35.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma XL, Sun HJ, Wang YS, Huang SH, Xie JW, Zhang HW. Study of Sonic hedgehog signaling pathway related molecules in gastric carcinoma. World J Gastroenterol. 2006;12:3965–3969. doi: 10.3748/wjg.v12.i25.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindström E, Shimokawa T, Toftgård R, Zaphiropoulos PG. PTCH mutations: distribution and analyses. Hum Mutat. 2006;27:215–219. doi: 10.1002/humu.20296. [DOI] [PubMed] [Google Scholar]
- 12.Frappart PO, Lee Y, Russell HR, Chalhoub N, Wang YD, Orii KE, Zhao J, Kondo N, Baker SJ, McKinnon PJ. Recurrent genomic alterations characterize medulloblastoma arising from DNA double-strand break repair deficiency. Proc Natl Acad Sci USA. 2009;106:1880–1885. doi: 10.1073/pnas.0806882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, Parker AR, Shimada Y, Eshleman JR, Watkins DN, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 14.Wolf I, Bose S, Desmond JC, Lin BT, Williamson EA, Karlan BY, Koeffler HP. Unmasking of epigenetically silenced genes reveals DNA promoter methylation and reduced expression of PTCH in breast cancer. Breast Cancer Res Treat. 2007;105:139–155. doi: 10.1007/s10549-006-9440-4. [DOI] [PubMed] [Google Scholar]
- 15.Pritchard JI, Olson JM. Methylation of PTCH1, the Patched-1 gene, in a panel of primary medulloblastomas. Cancer Genet Cytogenet. 2008;180:47–50. doi: 10.1016/j.cancergencyto.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang JY, Xiao SD. Alteration of DNA methylation in gastrointestinal carcinogenesis. J Gastroenterol Hepatol. 2001;16:960–968. doi: 10.1046/j.1440-1746.2001.02554.x. [DOI] [PubMed] [Google Scholar]
- 17.Cretnik M, Musani V, Oreskovic S, Leovic D, Levanat S. The Patched gene is epigenetically regulated in ovarian dermoids and fibromas, but not in basocellular carcinomas. Int J Mol Med. 2007;19:875–883. [PubMed] [Google Scholar]
- 18.Nagao K, Toyoda M, Takeuchi-Inoue K, Fujii K, Yamada M, Miyashita T. Identification and characterization of multiple isoforms of a murine and human tumor suppressor, patched, having distinct first exons. Genomics. 2005;85:462–471. doi: 10.1016/j.ygeno.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki M, Anast J, Bassett W, Kawakami T, Sakuragi N, Dahiya R. Bisulfite conversion-specific and methylation-specific PCR: a sensitive technique for accurate evaluation of CpG methylation. Biochem Biophys Res Commun. 2003;309:305–309. doi: 10.1016/j.bbrc.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Li LC. Designing PCR primer for DNA methylation mapping. Methods Mol Biol. 2007;402:371–384. doi: 10.1007/978-1-59745-528-2_19. [DOI] [PubMed] [Google Scholar]
- 21.Yoo YA, Kang MH, Kim JS, Oh SC. Sonic hedgehog signaling promotes motility and invasiveness of gastric cancer cells through TGF-beta-mediated activation of the ALK5-Smad 3 pathway. Carcinogenesis. 2008;29:480–490. doi: 10.1093/carcin/bgm281. [DOI] [PubMed] [Google Scholar]
- 22.Yanai K, Nagai S, Wada J, Yamanaka N, Nakamura M, Torata N, Noshiro H, Tsuneyoshi M, Tanaka M, Katano M. Hedgehog signaling pathway is a possible therapeutic target for gastric cancer. J Surg Oncol. 2007;95:55–62. doi: 10.1002/jso.20606. [DOI] [PubMed] [Google Scholar]
- 23.Wang LH, Choi YL, Hua XY, Shin YK, Song YJ, Youn SJ, Yun HY, Park SM, Kim WJ, Kim HJ, et al. Increased expression of sonic hedgehog and altered methylation of its promoter region in gastric cancer and its related lesions. Mod Pathol. 2006;19:675–683. doi: 10.1038/modpathol.3800573. [DOI] [PubMed] [Google Scholar]
- 24.Fukaya M, Isohata N, Ohta H, Aoyagi K, Ochiya T, Saeki N, Yanagihara K, Nakanishi Y, Taniguchi H, Sakamoto H, et al. Hedgehog signal activation in gastric pit cell and in diffuse-type gastric cancer. Gastroenterology. 2006;131:14–29. doi: 10.1053/j.gastro.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Xie K, Abbruzzese JL. Developmental biology informs cancer: the emerging role of the hedgehog signaling pathway in upper gastrointestinal cancers. Cancer Cell. 2003;4:245–247. doi: 10.1016/s1535-6108(03)00246-0. [DOI] [PubMed] [Google Scholar]
- 26.Ma X, Chen K, Huang S, Zhang X, Adegboyega PA, Evers BM, Zhang H, Xie J. Frequent activation of the hedgehog pathway in advanced gastric adenocarcinomas. Carcinogenesis. 2005;26:1698–1705. doi: 10.1093/carcin/bgi130. [DOI] [PubMed] [Google Scholar]
- 27.Nakajima T, Yamashita S, Maekita T, Niwa T, Nakazawa K, Ushijima T. The presence of a methylation fingerprint of Helicobacter pylori infection in human gastric mucosae. Int J Cancer. 2009;124:905–910. doi: 10.1002/ijc.24018. [DOI] [PubMed] [Google Scholar]
- 28.Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M, et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12:989–995. doi: 10.1158/1078-0432.CCR-05-2096. [DOI] [PubMed] [Google Scholar]
- 29.Lee KM, Lee JS, Jung HS, Park DK, Park HS, Hahm KB. Late reactivation of sonic hedgehog by Helicobacter pylori results in population of gastric epithelial cells that are resistant to apoptosis: Implication for gastric carcinogenesis. Cancer Lett. 2009;12:Epub ahead of print. doi: 10.1016/j.canlet.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 30.Romano M, Ricci V, Zarrilli R. Mechanisms of disease: Helicobacter pylori-related gastric carcinogenesis--implications for chemoprevention. Nat Clin Pract Gastroenterol Hepatol. 2006;3:622–632. doi: 10.1038/ncpgasthep0634. [DOI] [PubMed] [Google Scholar]