Abstract
We have earlier described a haemagglutination-based assay for on-site detection of antibodies to HIV using whole blood. The reagent in this assay comprises of monovalent Fab fragment of an anti-human RBC antibody fused to immunodominant antigens of HIV-1 and HIV-2. In the present work, we describe a rational and systematic method for directed evolution of scFv and Fab antihuman RBC antibody fragments. Based on homology modeling and germline sequence alignments of antibodies, target residues in the anti-RBC MAb B6 sequence were identified for mutagenesis. A combinatorial library of 107 clones was constructed and subjected to selection on whole RBC under competitive binding conditions to identify several phage-displayed B6 scFv clones with improved binding as determined in an agglutination assay. Selected VL and VH sequences were shuffled to generate a second generation phage-displayed Fab library which on panning yielded Fab clones with several fold better binding than wild type. The mutants with better binding also displayed more Fab molecules per phage particle indicating improved in vivo folding which was also confirmed by their increased periplasmic secretion compared to the wild type. The mutant Fab molecules also showed superior characteristics in large scale production by in vitro folding of LC and Fd. The biophysical measurements involving thermal and chemical denaturation and renaturation kinetics clearly showed that two of the mutant Fab molecules possessed significantly improved characteristics as compared to the wild type B6 Fab. Structural modelling revealed that B6 Fab mutants had increased hydrogen bonding resulting in increased stability. Our approach provides a novel and useful strategy to obtain recombinant antibodies with improved characteristics.
Key words: phage display, antibody maturation, Fab, antibody folding, scFv, agglutination
Introduction
Antibodies claim an unparalleled position in the current proteomics era and a large number of antibodies for diagnostic, therapeutic and research applications are being isolated. Synthetic antibody libraries1,2 have made it possible to isolate antibodies against any antigen including self antigens. These libraries are mostly single chain Fv (scFv)-based, though some are Fab based, and generally involve phage display technology for isolation of specific antibody molecules.3,4 However, of the antibodies isolated using these libraries and identified as invaluable for many diverse applications, only a few have gone ahead for potential commercial applications.5 Bottlenecks in the widespread application of the recombinant scFv fragments are the average expression behaviour, poor yields in E. coli and limited stability of the functional molecules. Fab fragments are closer in structure to the native antibody molecule and are superior to scFv fragments in both folding and stability. Fab fragments are better molecules for in vitro purposes and in applications of antibody fragments where the size of the molecule is not detrimental.
We have developed a haemagglutination based assay for on site detection of antibodies to HIV-1 and HIV-2 using whole blood6 from finger prick. The reagent used in this assay comprises monovalent Fab fragment of anti-human RBC antibody fused to immunodominant antigens of HIV-1 and HIV-2. Addition of this reagent to a drop of blood results in coating of RBC in the blood with the reagent and if this blood contains anti-HIV antibodies (as in the case of an HIV-infected individual), they will cross-link the reagent coated RBCs to give agglutination which can be seen with naked eye and is similar to that observed with blood-grouping reagent. This test gives results in less than two minutes and has potential utility in infrastructure-starved developing and underdeveloped nations of the world.
The specificity of the test depends on the HIV antigens used in the reagent while the sensitivity of the test depends upon both the binding of reagent to the RBCs and of anti- HIV antibodies to the reagent coated RBCs. The titre and quality of anti-HIV antibodies in patient's sera is variable and cannot be controlled; however the binding of reagent to the RBCs can be increased by improving the binding characteristics of the anti-RBC antibodies used in the reagent. Simultaneously, for the reagent to be commercially sustainable, the production cost of the reagent should be low; for this the folding yield of antibody fusion protein needs to be high. Also, for the reagent to have widespread utility, it needs to be stable to extremes of temperature encountered in different geographical locations.
Fab fragment of an anti-human RBC antibody B6 (B6 Fab), is one of several fusion proteins used in the above mentioned HIV-diagnostic assay. B6 is a murine monoclonal antibody that was isolated from mice immunised with O Rh D-human RBCs and binds to all human RBCs irrespective of the major and minor blood groups. To enhance the sensitivity of the assay and improve its commercial productivity, we undertook the task of improving the binding characteristics, folding yield and thermodynamic stability of B6 Fab. In this paper, we describe a rational and systematic approach to identify inappropriate residues in the VH and VL sequence, their mutagenesis and an efficient strategy of competitive selection on whole cells to isolate variants of B6 antibody with improved binding, folding and stability.
Results
Rational design of B6 mutants for selection of improved antibody molecules.
B6 genes were cloned as LC and Fd sequences7 using RNA isolated from B6 hybridoma. With the aim of improving the binding, folding and stability of B6 Fab for its better performance as a diagnostic reagent, we outlined a strategy for introducing directed mutations in B6 VL and B6 VH. Previous studies on B6 Fab had shown that random/spiked mutagenesis of CDR3 and Error-prone PCR based random mutagenesis of B6 VL and B6 VH led to a very high number of non-functional clones and weak activity clones (data not shown), indicating the need to focus on specific regions of B6 through site directed mutagenesis rather than randomly mutating the entire variable domains of B6.
Constant and variable domains from both heavy and light chains of immunoglobulins have proved to have similar structures, which is known as the ‘immunoglobulin fold.’ Also the CDR loops, which are attached to the essentially invariant β-sheet framework of the variable domains, assume only a limited range of conformations, except for the heavy chain CDR3. This enables one to predict and model with high degree of accuracy the main chain conformation of an antibody with known sequence, based upon alignment with antibody structures available in the protein database. Known as homology modelling, this method allows comparison of the model of an antibody with available structures in database to identify residues, which do not fit into the modelled structure and the potential mutations which could lead to a more stable conformation of the modelled molecule.
The germline sequence database of various antibody classes provides information about the characteristics of antibodies, the signature sequences which help in defining the framework and CDR regions in an Fv sequence, and allows comparison of an antibody sequence to identify residues that are ambiguous and are not generally found at those specific positions in antibody sequences.
A bi-directional approach based on sequence comparison and structure refinement was employed to analyse B6 Fv and identify candidate residues, which on mutagenesis would lead to improvement in folding, activity and thermodynamic stability of B6. Based on the homology model of B6 Fv fragment and a comparison of B6 Fv sequence with the closest murine germline sequences as described in Methods (Suppl. Table S1), a total of 15 residues, 7 in VL and 8 in VH, were delineated for mutagenesis (Fig. 1) and their replacement residues were also identified (Table 1). These changes were introduced as one set of mutations keeping the possibility of a wild type residue or a mutated residue at each position. A total of ∼6 × 105 combinations defined the complete set of possible mutations. Accordingly, a phage display library of mutant B6 scFv clones was made. Using oligonucleotides which were degenerate to encode the wild or mutant residue at each target position and wild type B6 scFv as template, a library of mutant scFv genes was obtained by PCR as described in Methods and this was cloned into a gIIIp-display based phagemid vector, pVCB6 scFv10633042. This vector carries B6 scFv gene having a stop codon in CDR3 of VL, and does not express a full-length scFv-gIIIp fusion protein. Replacement of this scFv gene by the mutant scFv library allows formation of scFv-gIIIp fusion protein. Hence, wild type clones arising from the vector in the mutant library are eliminated.
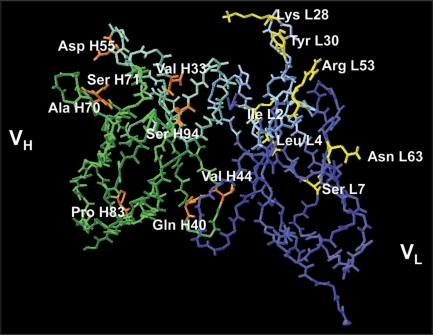
Figure 1.
Model of wild type B6 scFv showing the residues selected for mutagenesis. The VL chain is shown in blue and the VH in green with their CDRs in lighter shades of blue and green respectively. The residues selected for mutagenesis are colored yellow (of VL) and orange (of VH).
Table 1.
Amino acid positions mutated in the B6scFv phage display library
| Position | Residue in WT | Codon | Residue incorporated | Codon incorporated |
| L2 (FR1) | I | ATT | V | GTT |
| L4 (FR1) | L | CTG | M | ATG |
| L7 (FR1) | S | TCT | T | ACT |
| L28 (CDR1) | K | AAA | K/N/D/E | (A/G)A(A/C) |
| L30 (CDR1) | Y | TAC | Y/N | (T/A)AC |
| L53 (CDR2) | R | CGT | R/N/H/S | (C/A) (G/A)T |
| L63 (FR3) | N | AAT | N/S | A(A/G)T |
| H33 (CDR1) | V | GTT | V/Y/N/D/F/I | (G/T/A)(T/A)T |
| H40 (FR2) | Q | CAG | Q/H/R/K/N/S | (C/A)(A/G)(G/T) |
| H44 (FR2) | V | GTC | V/G | G(T/G)C |
| H55 (CDR2) | D | GAT | D/G/N/M | (G/A)(A/G)T |
| H70 (FR3) | A | GCT | A/T | (G/A)CT |
| H71 (FR3) | S | TCA | S/A/L/V | (T/G)(C/T)A |
| H83 (FR3) | P | CCC | P/T | (C/A)CC |
| H94 (FR3) | S | AGC | S/R | (A/C)GC |
Total combinations of DNA sequences= 589824 (∼6 × 105)
To ensure representation of all possible combinations of mutations, a library of 1 × 107 clones was obtained in E. coli TG1 and was used to produce phage particles using helper phage AGM13. AGM13 is a derivative of M13K07 and contains a trypsin cleavage site in gIIIp. Treatment of AGM13 phages with trypsin renders them non-infectious. The library phages display B6 scFv in VL-linker-VH orientation followed by cmyc tag and full-length gIIIp. The cmyc decapeptide tag has an internal trypsin cleavage site. Treatment of library phage particles with trypsin renders all helper phage particles and phagemid particles not displaying an scFv molecule, non-infectious. Only those phagemid particles which display a scFv fused to phagemid encoded gIIIp remain infectious. Also, action of trypsin within the cmyc sequence cleaves the scFv from gIIIp, thereby restoring full-infectivity to the phage particle.
Competitive panning and selection of improved B6.
The mutant B6 library was analysed for its variability and display characteristics. Phages were produced from 96 clones of the initial unscreened library and were tested for agglutination activity. As shown in Figure 2A, 30% clones gave agglutination in the unselected library. The clones were also sequenced to check for the randomness of mutant sequences in the library. In selection experiments B6 Fab protein was used as competitor to block binding to RBC of library phages displaying B6 scFv, having affinity equal to or less than wild type B6 scFv. 50 ng of recombinant B6 Fab protein in 200 µl completely inhibited agglutination of 5 × 106 RBCs by 109 B6 scFv phages.
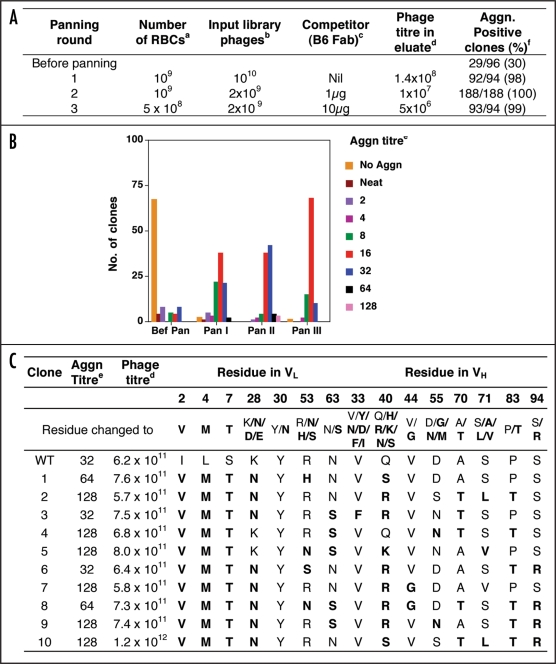
Figure 2.
Results of all the panning rounds for the first generation library of B6 scFv displaying phages. (A) Conditions used in the panning rounds. (B) Agglutination titre of clones tested from each round of panning. (C) Agglutination titre, Phage titre and sequence of VL and VH at the mutagenised positions in ten clones selected after panning. (a) Number of Red Blood Cells used in panning. (b) Number of phages used in panning. (c) Amount of B6 Fab protein (wild type) used in panning as competitor for binding to RBCs. (d) Phage titre determined as ampicillin resistant transductants. (e) Agglutination titre recorded as the maximum dilution of sample that gave visible agglutination (end point). (f) Percentage of clones in each panning round that gave agglutination.
The conditions used for the selection of better binders from the library are shown in Figure 2A. The first round of panning was done without any selection pressure in order to merely select for functional clones and maintain maximum diversity for subsequent panning rounds. For this, 1010 library phages were allowed to bind to 109 RBCs in absence of competitor and bound phages were eluted with trypsin. This round led to enrichment of scFv displaying phages and eliminated sequences with deletions or unfavourable mutations that led to loss of display or loss of activity of the displayed scFv. As seen in Figure 2A, the 1st round of panning led to complete elimination of non-functional clones. However, a large number of clones had agglutination activity less than the wt clone, which showed an agglutination titre of 32 (Fig. 2B). The eluted phages were amplified by infecting TG1 cells along with helper phage. In the 2nd and the 3rd round of panning, the ratio of input phages and RBC was changed and B6 Fab protein as competitor was added at 1 µg and 10 µg, respectively (Fig. 2A). The 2nd and 3rd round of panning successfully eliminated weak binders and more than 50% clones after 2nd round had agglutination activity equal to or higher than wild type. However, the 3rd round of panning did not lead to any further increase in the percentage of high activity clones. It has been reported that repeated rounds of panning often lead to loss of high-activity clones. Therefore, no more panning rounds were carried out.
Based upon the agglutination titres obtained for individual clones analysed from eluate obtained after each round of panning, nine clones of Pan II and one clone of Pan III which showed up to four times higher agglutination titre than wild type were selected. Phages were prepared in larger volume for these clones, and tested for agglutination activity, phage titre and DNA sequence of scFv to identify the mutations in the scFv sequence. The phage numbers in culture supernatant for all the ten clones were similar while the agglutination activity showed a 2–4-fold improvement over the wild type (Fig. 2C). Sequence analysis showed that while certain positions could tolerate more than one type of residue, other positions were more resistant to changes. L28 present in the solvent exposed region of CDR-L1 of B6 was either a positively charged residue or a neutral residue in all the clones selected indicating that this residue is involved in electrostatic interaction with the antigen and a positive charge or a hydrogen bond donor is absolutely essential as none of the clones analysed in any panning round had a negatively charged residue at this position (data not shown). L30, also present in CDR-L1, is a tyrosine residue in wild type B6. All the clones obtained after the selection had tyrosine at this position. This indicates that the tyrosine residue makes important contribution to the paratope and is essential for maintaining binding of B6.
In the heavy chain sequence, H33 present in CDR-H1 was Valine in all the clones except one. Valine is also present in the wild type antibody sequence suggesting that it makes important contact with the antigen and is essential for B6 binding to its antigen on RBC. Similarly, H44 present in FR2 is Valine as is also the case in wild type B6 indicating that this residue is important for activity of B6. The other positions in heavy chain were more tolerant to mutations.
Generation of a secondary library using chain shuffling and selection of improved B6 Fab.
Fab fragment of an antibody has superior folding characteristics and higher thermodynamic stability compared to an scFv fragment. Also, B6 is used in the Fab format as a diagnostic reagent for HIV. Therefore, the improved B6 mutants obtained as scFv were converted into Fab to confirm the improvement in a different quaternary structure. To increase the diversity of sequences during this conversion, the 8VL and 10VH domains (Fig. 2C) were shuffled to generate eighty LC-Fd combinations and used for panning on human RBCs (Fig. 3A). Only 12.7% clones in the unselected Fab library showed agglutination activity (Fig. 3A), indicating that several combinations of LC and Fd resulting from chain shuffling were non-productive and did not yield functional Fab molecules. Therefore, to enrich for binders from the pool of non-binders, the 1st round of selection (Pan I) was performed under non-competitive conditions. A 6-fold enrichment was obtained and more than 80% clones showed agglutination activity. Also, majority of clones showed activity better than wild type B6 Fab displaying phages (Fig. 3B). A 2nd round of panning (Pan II) in presence of 1 µg B6 Fab competitor, completely eliminated clones which either showed no agglutination (Fig. 3A) or agglutination less than or equal to wild type and were predominant after the 1st round of selection. Almost all the Pan II clones analysed showed agglutination activity 2–64-fold better than wild type. Twenty-two clones from Pan I and Pan II, which showed 32–64-fold higher activity than wild type, were selected for further analysis (Fig. 3C). Phages were prepared in 5 ml culture volume and tested for agglutination activity, phage titre and DNA sequence to identify the mutations responsible for the observed improvement in activity.
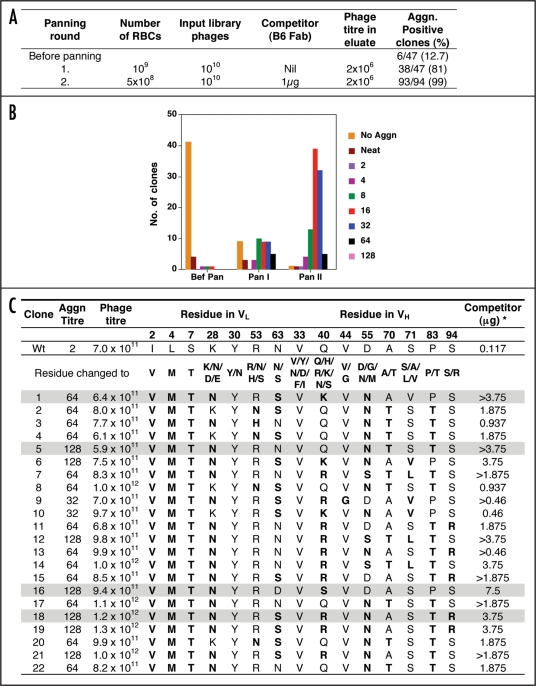
Figure 3.
Results of all the panning rounds for the second generation library of B6 Fab displaying phages. (A) Conditions used in the panning rounds. (B) Agglutination titre of clones tested from each round of panning. (C) Agglutination titre, Phage titre and sequence of VL and VH at the mutagenised positions in ten clones selected after panning. (a) to (f) Same as Figure 2. *Concentration of recombinant wild type B6 Fab required to compete out binding to RBCs of B6 Fab displaying phages in agglutination assay. The four mutant B6 Fab clones selected for subsequent studies are shaded in grey.
As shown in Figure 3C, all the clones had similar phage titres and showed agglutination activity 32–64-fold better than wild type. Sequence analysis of the clones showed that the Fab library panning led to narrowing down of the selected sequence diversity and optimum residue for the mutated positions could be identified. L30, H33 and H44 were conserved and could not be mutated to the other amino acids. L30 and H33 are CDR1 residues and could be involved in making important contacts with the antigen while H44 is present in FR2 and could be important for maintaining the binding activity of B6. All the clones had positive residue at L28 and L53. L28 is lysine in wild type and was mutated to Asparagine, Glutamate or Aspartate. None of the clones had glutamate or aspartate and majority of the clones contained asparagine in place of lysine. This indicates that L28 in CDR1 is involved in electrostatic interaction with a residue in the antigen and a negative charge at L28 leads to disruption of that interaction resulting in clones having reduced or no antigen binding. It is important to note that none of the binders analysed from Pan I, Pan II and Pan III eluate of the scFv library had Aspartate or Glutamate at L28 (data not shown) confirming the above conclusion. Similar to L28, L53 in CDR2 was also a basic residue in all the selected clones (Fig. 3C) indicating that a positive charge is important for interactions at this position. L63, which is part of FR3 could tolerate the presence of either asparagine or serine.
In VH, the first position mutated was H33, which is present in CDR1. All the selected clones had valine at this position as is present in wild type B6 indicating that valine is essential for binding and specificity of B6. Similarly valine at H44 in FR2 was also conserved in all the clones. At H40, arginine, glutamine or lysine were present in the clones indicating the importance of positive charge at this position. At other positions that were mutated in VH, more variations were observed.
Competition studies to identify the best binders.
To delineate the best binders among the 22 clones selected from B6 Fab library, a competition assay was used wherein increasing amount of B6 Fab (wt) protein was used to block the binding of a fixed number of phages to RBCs, thus resulting in inhibition of agglutination upon addition of anti-phage antibody. In this assay, higher concentrations of B6 Fab protein would be required to out-compete B6 Fab phage molecules with higher binding affinities. As seen in Figure 3C, while the wild type B6 Fab displaying phages were out-competed by less than 117 ng of B6 Fab protein, 4–70-fold higher concentration of B6 Fab protein was required to abolish agglutination by mutant clones indicating that these 22 clones were stronger binders than wild type B6. For eight clones B6 Fab protein in the range of 3.75 to 7.5 µg (30–70-fold excess over the amount required to out-compete wt binding) was required to abolish agglutination. This correlated well with the 16–64 fold better agglutination titre of these clones vis-à-vis wild type Fab displaying phages. Four clones were selected for further studies (Fig. 3C) and named as mut1, mut2, mut3 and mut4.
Since the antigen on the RBC surface to which B6 antibody binds is not known, the improvement in affinity of Fab could not be quantitatively estimated using SPR-based measurement methods. However, the competition in binding by soluble Fab in agglutination assay enabled delineation of mutants with improved affinity.
Mutant density and folding characteristics.
The copy number of Fab molecules displayed on M13 phages was determined by densitometric scanning of immunoblots of a defined number of phages developed using anti-cmyc antibody. As shown in Figure 4(1), all the mutants showed a higher display number on phages than wild type B6 Fab. The display was highest for mut2 (40-fold) and mut3 (23-fold) followed by mut1 (7-fold) and mut4 (5-fold). This large improvement in the number of Fab molecules displayed on phage particles indicates that not only are the mutants better binders, they also have better folding characteristics and are less prone to aggregation resulting in their increased incorporation in the assembling phage particles. Moreover, this result indicates that the wild type phage preparation contains a predominant fraction of phages that do not display a Fab molecule.
Figure 4.
Biochemical properties of selected mutants. (1) Relative Display density of wild type and mutant B6 Fab on M13 phages. The number of molecules displayed per phage particle was calculated based on densitometric scanning of western blot of phages as described in Materials and Methods. (2) In vivo yield of B6 Fab. Mutant and wild type B6 Fab were expressed in E. coli and purified from periplasmic fraction obtained from 1 litre of culture using Ni-NTA affinity chromatography. The total protein obtained was determined using Bradford's Protein assay. (3) Agglutination activity of B6 Fab. In vivo expressed Mutant and wild type B6 Fab proteins purified using Ni-NTA affinity chromatography were tested in agglutination assay as described in Materials and Methods and agglutination titre was determined. For all the three panels, the maximum value obtained was normalised as 100 and all values were plotted as percentage.
The yield of Fab obtained from periplasm is a direct correlate of the in vivo folding properties of a molecule. To assess this, the mutant and the wild type Fab sequence were cloned into T7 promoter based periplasmic secretion vector having a (His)6 tag. BL21 (DE3) transformants of various clones were grown in minimal media at 30°C and expression was induced with IPTG. Periplasmic fraction of induced cultures was isolated and Fab was purified using affinity chromatography on Ni-NTA column. The purified protein was analysed for the total yield and activity of Fab. The yield of protein obtained after purification on Ni-NTA column was highest for mut3 followed by mut1, mut2 and mut4 [Fig. 4(2)]. All the mutants gave better yields in comparison to wild type indicating that they had improved folding in vivo and were less prone to misfolding and aggregation. Mut1 and mut3 showed better agglutination activity as compared to mut2, mut4 and wild type [Fig. 4(3)].
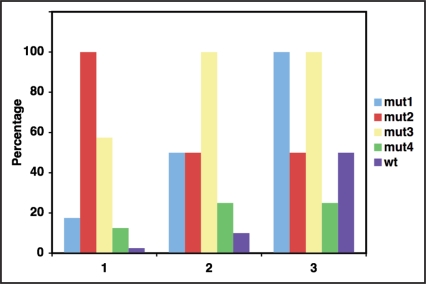
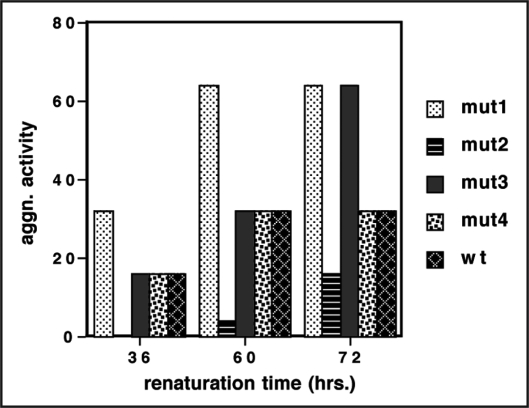
To make large amounts of Fab for commercial use, a protocol of in vitro refolding of denatured protein was followed. For this, the LC and Fd encoding sequences of mut1, mut2, mut3 and mut4 were cloned separately in T7 promoter driven cytoplasmic expression vector. The expressed LC and Fd were isolated as inclusion bodies, denatured in vitro, mixed in equimolar ratio and allowed to renature under redox buffer conditions as described in Methods. The refolding of the denatured LC and Fd and their association to form functional Fab was monitored at different time points in agglutination assay. As shown in Figure 5, refolding of mut1 was substantially faster than that of the other mutants and of wild type and it achieved more than 80% refolding within 36 hours and by 60 hours its refolding was complete. mut3 and mut4 exhibited slower refolding kinetics and achieved 25–30% refolding within 36 hours similar to wild type molecule and took about 70 hours to achieve maximum refolding. Mut2 did not achieve significant percentage of refolding before 60 hours. The extent of functional Fab obtained in this experiment is an indication of the refolding potential of a molecule following complete denaturation. Mut1, mut3 and mut4 showed a high percentage of refolding of LC and Fd chains that were completely denatured and reduced in the presence of guanidine hydrochloride and DTE. Not only the LC and Fd chains refolded, they also associated to form the correct disulfide bonds and attain the right quarternary structure to give functional, monomeric Fab. The wild type chains also showed appreciable refolding. However, mut2 showed extremely slow refolding kinetics and only a small fraction of the total protein was able to refold and attain a functional conformation.
Figure 5.
In vitro renaturation of B6 LC and B6 Fd to form functional Fab. Inclusion bodies of B6 LC and B6 Fd obtained from one litre culture each were denatured and allowed to renature as described in Materials and Methods. The renaturation sample was tested for agglutination activity at different time points to estimate the extent of renaturation and B6 Fab formation.
The in vitro renaturation mix after 72 hours was dialysed and subjected to a 4-step column chromatography procedure as described in Methods to purify monomeric Fab in functional form. Fractions at each step were analysed for active Fab in agglutination assay and for purity of protein on non-reducing SDS-PAG.
The yield of Fab obtained was highest for mut3 followed by mut1 and mut4. mut2 showed degradation products with little full-length protein (data not shown). In comparison to wild type B6 Fab, which was obtained at a folding yield of 40 mg from 4 litre of renatured material, the yield of mutants was 1.5–2.0 fold more (Table 2). This improvement in yield is extremely significant in applications requiring large-scale production. Since mut2 showed degradation in a repeat preparation, it was omitted from further studies.
Table 2.
Agglutination activity and yield of B6 Fab obtained by in vitro renaturation of B6 LC and B6 Fd
| Protein | Agglutination activitya | Yield of in vitro refolded proteinb |
| Mut1 | 1024 | 65 mg (16%) |
| Mut3 | 512 | 88 mg (22%) |
| Mut4 | 512 | 60 mg (15%) |
| Wt | 256 | 40 mg (10%) |
The agglutination activity is reported for 30 µg of each protein.
The in vitro yield is reported for protein obtained from four litre renatured material containing 200 mg each of B6 LC and B6 Fd.
Biophysical characterisation of mutants.
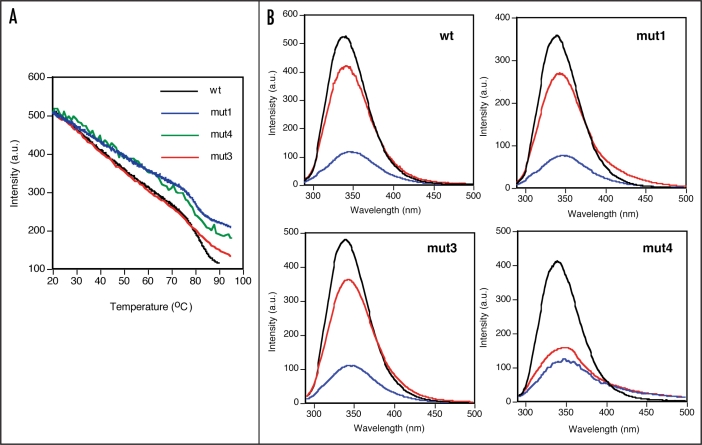
Thermal denaturation. Wild type and mutant Fab proteins were analysed for their intrinsic fluorescence emission spectra. All the proteins had similar spectra with emission maxima at 340 nm after excitation at 280 nm (Fig. 6B, black line).
Figure 6.
Comparison of thermal stability of wild type and mutant B6 Fab proteins. (A) Change in fluorescence intensity as a function of increase in temperature. (B) Fluorecence emission spectra of native (black line), thermally denatured (at 90°C, blue line) and renatured (cooled to 20°C, red line) B6 Fab wt and mutant proteins.
The Fab proteins were subjected to thermal denaturation and the changes in flourecence emission at 347 nm were recorded from 20 to 90°C. All the mutants were extremely stable as evident from the high thermal denaturation temperatures in the range of 75–85°C (Fig. 6A). While a two-phase transition was observed for the wt as well as mut1 and mut4, mut3 showed a broad transition with a small change in fluorescence intensity, suggestive of noncooperative denaturation behaviour. The transition temperature of mut3 was also higher as compared to wt and the other mutants. The change was more prominent for mut4 than for mut1, which showed thermal denaturation trends very similar to wt. The linear decrease in fluorescence intensity of all the clones upon thermal denaturation from 20–75°C is indicative of the gradual change in the tyrosine and tryptophan environment of the molecules.
The samples heated up to 90°C were analysed for changes in the intrinsic fluorscence emission spectra. As shown in Figure 6B, all the samples showed a drop in fluorescence intensity and a shift in emission maxima from 340 nm to 347 nm, indicating that the proteins undergo substantial change in conformation with alterations in the tryptophan and tyrosine environment in the protein, resulting in the observed change in spectra. A scan of the samples cooled to 20°C to enable refolding of the molecule showed that mut3 like wt achieved more than 80% refolding as judged by the recovery of fluorescence intensity as well as the maxima back to 340 nm while mut1 achieved about 70% refolding. However, mut4 achieved only 40% refolding indicating that mut4 had lost its structure on thermal denaturation, which could not be regained on cooling. These results of thermal denaturation-renaturation are in complete agreement with the results obtained for chemical denaturation-renaturation experiment where also the refolding yield of mut3 and mut1 were higher than that of mut4.
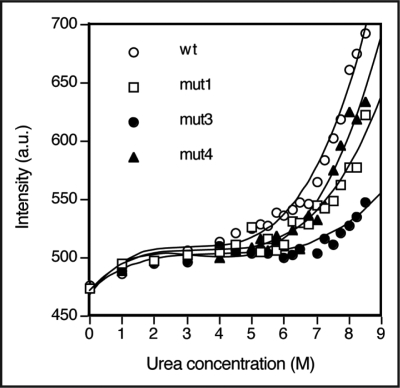
Urea denaturation. All the three mutants and wild type B6 showed a shift in the emission maxima from 340 nm without urea to 347 nm in 8.5 M urea at 25°C (data not shown). However, none of the mutants or wild type proteins showed a typical denaturation transition even in 8.5 M urea, indicating that the proteins were highly resistant to complete protein denaturation. This may be due to high stability of the disulfide bonded Fab molecule. Nevertheless, there was an increase in fluorescence intensity observed at ∼5–6 M urea for wt, mut4 and mut1 (Fig. 7). Mut3 was extremely stable and there was no appreciable change even upto 7 M urea. Fluorescence intensity changes upon denaturation were also small for mut3 compared to wt and other mutants. These results for mut3 are similar to the results obtained for the thermal denaturation experiments. Due to absence of clear post-transition zone, the thermodynamic parameters for protein denaturation could not be evaluated.
Figure 7.
Urea denaturation curves of wild type and mutant B6 Fab monitored for fluorescence intensity at 347 nm at 25°C.
Functional stability. To assess the stability of molecules on prolonged incubation at temperatures higher than recommended storage temperature (2–8°C), the wt and mutant proteins were incubated at 37°C for one month and were tested at defined time points for agglutination activity and for degradation products by immuno-blotting using anti-cmyc antibody, 9E10. All the proteins were stable for the period of testing i.e., one month at 37°C without any loss of agglutination activity and no degradation was observed (data not shown). This data substantiates the high stability of these proteins and their potential for widespread use in varied environments.
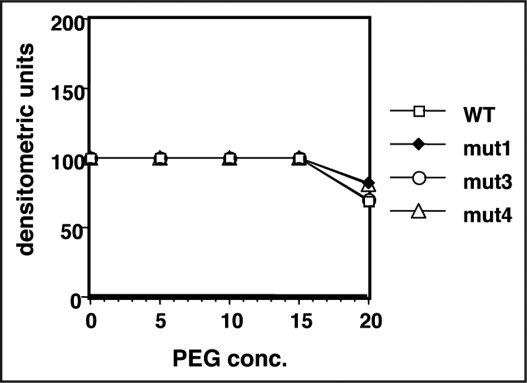
Solubility. To study the effect of mutations on the solubility of the folded proteins, we assessed their solubility in PEG 6000. PEG is known to decrease the solubility of proteins8 and is commonly used to induce protein crystallisation. Very similar concentrations of wild type and mutant proteins were obtained in supernatant after incubating proteins in various PEG concentrations and centrifuging the precipitated Fab protein. Densitometric scanning of SDS-PAG of supernatants showed that all the molecules had very similar solubility and there was a marginal drop in solubility at 20% PEG concentration (Fig. 8).
Figure 8.
Solubility of wild type and mutant B6 Fab. PEG was used to increase the protein concentration of the Fab samples above the solubility limit. The precipitated Fab was removed by centrifugation and Fab remaining in the supernatant was quantitated by SDS-PAGE and densitometric scanning.
Structural correlation of observed properties of mutants.
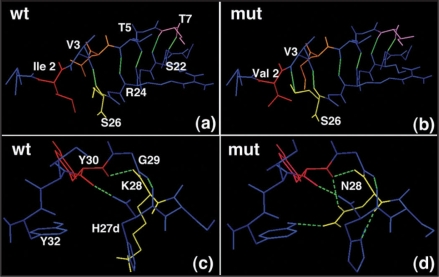
The importance of FR1 in folding and stability of antibody fragments is well recognised9 and it has been found that changing FR1 sequences post-PCR to consensus sequence for the corresponding subgroup results in dramatic improvement of refolding yields.10 B6 scFv has three residues in FR1 of VL at positions L2, L4 and L7 that were changed to the consensus residues. Isoleucine at L2 position in wild type B6 was probably a result of PCR amplification with degenerate primers. Valine is a highly conserved residue in subgroup II of kappa variable domain at position L2. When Val L2 is present, the carbonyl oxygen of Val L3 makes a hydrogen bond with hydrogen of Ser L26 and the side chain hydoxyl of Ser L26 makes a hydrogen bond with Val L3 hydrogen (Fig. 9B). Presence of the bulky side chain of Isoleucine at L2 position causes the Ser L26 side chain to point in different direction from the L3 residue, thereby preventing the formation of the hydrogen bond (Fig. 9A). This change in hydrogen bonding network in turn destabilises the core of the domain. Replacement of Ile L2 with Val L2 in the mutants allows the reformation of the consensus hydrogen bonding pattern for VL domain and could play a role in the increased stability of the mutant fragments. A very important stabilising mutation selected from the library was Asn residue in place of Lys at position L28. Unlike the CDR1 residue Tyr L30 which was found invariant and therefore played an essential role in antigen binding, Lys L28 was found to be replaced by Asn in more than 80% of the selected clones. Detailed analysis of the mutants shows that this substitution has a strong effect on the hydrogen bonding pattern of the domain and stabilisation of the CDR1 loop for favourable interactions with the antigen. While the Lys residue only participates in formation of one hydrogen bond through its carbonyl oxygen (Fig. 9C), the Asn residue is present in an altered spatial configuration that allows it to form four hydrogen bonds with the neighbouring residues, two of which are formed with the crucial Tyr L30. Moreover, the long side chain of Lys causes His L27d imidazole ring to be present away from the main chain preventing the imidazole nitrogen from participating in hydrogen bonding. The substitution with Asn changes the conformation of the CDR allowing imidazole nitrogen of His L27d to form hydrogen bond with Asn L28 hydrogen (Fig. 9D). The extensive changes caused by Asn substitution on the hydrogen bonding pattern of the VL domain could result in the improved stability of the mutants.
Figure 9.
Hydrogen bonding pattern in wild type and mutant B6 Fab. (A and B) Pattern of bonding between L3 Val and L26 Ser. (C and D) interaction of L28 Lys (wt) or Asn (mut) with neighbouring residues. For explanation, see the text.
Discussion
Our antibody evolution approach has resulted in the isolation of improved Fab molecules having superior binding, increased folding yield, and greater thermodynamic stability with potential commercial applications. Our results showed that both the CDRs and framework were determinants of the binding and folding characteristics of an antibody, and fine-tuning of the antibody sequence could dramatically improve its physicochemical properties. The improved sequences resulted in superior antibody fragments in both scFv and Fab format. Thus, it is possible to turn a natural antibody into one with favourable properties by changing only a few residues.
There are other approaches to identify residues that improve activity, thermodynamic stability and folding yield of scFv. Detailed analysis of the three dimensional structure of the antibody followed by mutagenesis of the identified problematic residues has been used in specific examples.11 A second approach employs random mutagenesis of antibody sequences followed by selection for a desired property.12 The third approach involves changing the length and/or sequence of CDRs, generally CDR3 of the heavy chain followed by selection for better properties.13,14 We have employed an approach that uses the sequence and structure information from naturally occurring antibodies as a template to improve newly isolated antibodies.
Alignment of an antibody sequence with germline sequences helps in identifying residues in the sequence, which are not normally present in antibodies at those positions. These residues could be PCR artifacts incorporated in the antibody sequence during the process of cloning antibody genes from hybridoma/PBMCs. Replacement of these residues with residues normally present at those positions in the antibodies of the specific subgroup will therefore help in improving the folding and binding properties of the antibody. Simultaneously analysis of the variable domain model in conjunction with the structures of its closest homologues will help in identifying residues that are inappropriate and hinder correct folding and attainment of a suitable conformation. These residues can then be mutated to more suitable residues to isolate a superior antibody molecule.
Our data clearly shows that this two-prong approach combining sequence knowledge with structural information can improve existing antibody molecules. Targeting selected residues is advantageous because it narrows down the number of combinations to be tried, thereby reducing the required library sizes to achievable numbers unlike the random mutagenesis approach where the theoretical library sizes are much higher than the practical sizes achieved. Large library sizes limit the diversity of sequences at hand and the best binders may never be available for selection. A small library size ensures that all the variants theoretically calculated will be available for selection, so that the best molecule in that pool can be obtained.
To explore the effect of all possible combinations of mutant sequences on activity, folding and stability of B6, we employed the method of PCR-based mutagenesis using degenerate nucleotides wherein each selected position could accommodate a mutant residue or retain the wild type residue. This strategy led to creation of a diverse, unbiased library containing ∼106 sequence combinations. Most studies describing antibody improvement employing different methods analyse effects of substitution of individual residues first and later combine the selections to check for further improvement. Our approach of library construction allows both single substitutions and multiple substitutions in various combinations to be screened simultaneously for selection of the best variants.
We used phage display as a platform for selection of improved B6 variants from this library. In phage display, folding characteristics of the molecule, resistance to aggregation and thermodynamic stability are important criteria for display, since only correctly folded, functionally active molecules displayed on phage surface can bind to antigen and get selected in panning. Therefore, a higher copy number per phage is an indication of better folding and stability of a molecule. Our selection design is based on a selection for functionality i.e., binding of the phage displayed molecule to the antigen on cell surface. Sequestering of binding sites on the RBC surface by the addition of wild type soluble antibody served as a good selection pressure in this study and allowed the recovery of phage-displayed scFv that had several fold higher binding than wild type B6 displayed on phages.
Another key result obtained from this study was that individual mutations selected in the scFv format, when shuffled with each other and subjected to selection in a different structural background as Fab, led to isolation of further improved antibody molecules. These evolved molecules selected by phage display exemplify their improved characteristics both during in vivo expression and functionality assays, as well as during in vitro denaturation-renaturation studies. All the mutants showed higher display density on phages, and also gave higher yields of functional soluble protein in periplasmic expression as compared to wild type.
Denaturation studies are capable of yielding information about the native state in terms of its intrinsic stability, cooperativity in folding and nature of forces responsible for maintaining the structure of the protein. In denaturation studies, the mutants exhibited improved characteristics. Compared to wt B6, the mut1, mut3 and mut4 have more extensive hydrogen bonding network that imparts greater thermodynamic stability to folding intermediates and the completely folded molecule. Also, replacement of bulky side-chain residues by shorter side-chain residues improves the spatial orientation of functional groups increasing favourable interactions and resulting in tighter VH/VL packing.
Increased activity, improved stability and higher yields are essential properties for antibodies to be used in diverse applications. Most of the work on in vitro maturation of antibody fragments has been done using scFv antibody fragments that bind small antigens and haptens. This work is perhaps the first complete study describing evolution of Fab fragment of an antibody that binds to a cell surface molecule, selection using whole cells and the systematic biochemical and biophysical characterisation of the evolved molecules. Using a germline family consensus-based and structure-based predictive engineering method, we have isolated a series of mutants with improved characteristics that might be useful as a reagent in a sensitive and cost-effective HIV diagnostic assay.
Materials
B6 is an anti-human RBC murine monoclonal antibody, which was isolated by conventional hybridoma technology in our laboratory. The DNA encoding LC and Fd of B6 were cloned using PCR based methods (our unpublished data). Fab fragment of B6 (B6 Fab) is used as a reagent for haemagglutination based detection of HIV.6 AGM13 is a derivative of M13K07 made in the lab.
E. coli strain TG1 (supE Δ(hsdM-mcrB)5(rk−mk−McrB−)thi Δ(lacproAB) [F'traD36, LacIqΔ(lacZ)M15]) was used as host for titering culture supernatants and amplification of phages. Restriction Enzymes and Modifying Enzymes were procured from Roche and chemicals from USB, Roche and SIGMA. All chromatography columns and material Q-Sepharose fast flow, SP-Sepharose fast flow, Sephacryl S-200 (high resolution), Sephadex G-25 (medium) were purchased from GE Amersham. Toyo-Butyl gel (M) was obtained from Tosohaas, Montgomeryville, PA, USA.
Anti-cmyc MAb, 9E10 was produced using hybridoma obtained from ATCC, Manassas, Va. HRP-conjugated secondary antibodies were obtained from Jackson ImmunoResearch Laboratories, West Grove, PA.
Methods
Identification of candidate residues for mutagenesis.
The sequence of the variable domains of B6 was aligned with the murine germline sequences database and analysis was performed using DNAPLOT version 2.0.1 using V BASE version 1.0. B6 uses a VL segment of the VKII subgroup and VH segment of VHI family. The VH and VL residues were numbered according to the Kabat numbering scheme. Based on the alignment of the wild type B6 sequence with the germline database, residues that were either completely absent or were present at very low frequency at the specific positions in the antibody sequence were identified. The Chothia canonical classes of the CDRs were also identified using a set of key residue templates detailed by Chothia et al.15,16 as well as those used by Oxford Molecular's MbM software.17 Based on this analysis, the residues normally allowed at each position in antibody sequence were defined and the mismatched residues in B6 were identified.
A homology model of the Fv fragment of B6 was built in O, based on the structures of the anti-polysialic acid antibody (PDB entry 1plg), the anti-single stranded DNA antibody (PDB entry 1i8m), the anti-influenza virus neuraminidase antibody NC10 (PDB entry 1 nmc) and the anti-deoxyinosine-5′-monophosphate antibody JEL 103 (PDB entry 1mrf).
Based on the above comparison of the B6 VH and VL, residues which were highly conserved both in structural homologs and in closely related database sequences were delineated for mutagenesis (Suppl. Table S1) and the residues to be incorporated in those positions were identified (Table 1).
Library design and construction.
B6 scFv cloned in VL-linker-VH orientation in phagemid vector, named pVCB6 scFv10533042 was used as template for library construction. To introduce mutations, degenerate oligonucleotides, which encoded for the wild type or mutant residue at specific positions were used in PCR. Since the mutations were in different regions of the scFv, the entire scFv sequence was amplified in 5 fragments. Using pVCB6 scFv10533042 as template, five PCR reactions were carried out with mutagenesis primers using Expand High Fidelity Polymerase (Roche). The amplified fragments (140 bp, 100 bp, 350 bp, 140 bp and 130 bp) were purified on agarose gel and used as template in subsequent PCR in multiple steps to amplify a 750 bp product which encoded the complete scFv sequence.
This final spliced product was digested with Nhe I and Mlu I and cloned into similarly digested pVCB6 scFv10633042. This vector contains B6 scFv coding sequence which has a stop codon in CDR3 region of B6 scFv and therefore does not express full-length B6 scFv-gIIIp fusion protein. The ligation mix was electroporated into TG1 cells and regenerated in 25 ml SOC medium for 1 hour at 37°C. Aliquot of the regenerated mix was plated on LB agar plates containing 100 µg/ml ampicillin (amp) and 1% (w/v) glucose (LBAmpglu) and incubated at 37°C overnight to obtain colonies of transformants and calculate library size. The bulk of regeneration mix was diluted into 50 ml of prewarmed 2X YT broth containing 20 µg/ml ampicillin and 1% (w/v) glucose (2XYTAmpglu) and grown at 37°C for 1 hr. Ampicillin was added to a final concentration of 100 µg/ml and the culture was grown for another two hours at 37°C to reach A600 of 0.8. The culture was then infected with helper phage AGM13 at MOI of 20 and phages were produced overnight at 32°C by diluting ten times in 2X YT containing 100 µg/ml ampicillin and 50 µg/ml kanamycin. The culture supernatant was titrated as colony forming units on LBAmpglu plates to determine the phagemid titre.
Panning.
For panning experiments, intact human RBCs were used as antigen. In the first round of panning, 1 × 109 RBCs were mixed with 1 × 1010 phages (in culture supernatant) in a total volume of 2.0 ml in PBS and incubated at 37°C for 30 minutes with end-to-end mixing. The sample was then centrifuged at RT for 5 minutes at 2,000 xg to pellet RBCs. The supernatant was aspirated and the pellet was washed five times with 10 ml PBS containing 0.2% BSA (PBS-BSA) followed by five times with 10 ml PBS. The washed pellet was suspended in 500 µl PBS and transferred to a microfuge tube. 50 µl Trypsin (1 mg/ml in PBS) was added and the sample was incubated at 37°C for 20 minutes followed by centrifugation at 4°C, 13,000 xg for 10 minutes. The supernatant (Pan I elute) was stored in another tube and used for determining titre of eluted phages as ampr transductants.
To amplify phages for the next round of panning, TG1 cells were infected at MOI of 0.1 with phages eluted above by incubating cells with phages at 37°C for 90 minutes. The infected cells were then pelleted to remove trypsin and the pellet was resuspended in original volume in 2XYTAmpglu. The culture was grown to A600 of 0.3 and infected with helper phage AGM13 at MOI of 20 and phages were produced overnight at 32°C. The culture supernatant was used to determine phage titre.
For the second round of panning, 5 × 108 RBCs were mixed with 1 × 109 phages (amplified from Pan I eluate) and 1 µg B6 Fab protein in a total volume of 2 ml and panning was carried out as described above for the 1st round. The eluate of 2nd panning (Pan II eluate) was used to determine titre of phages in eluate and was amplified for the third round of panning.
For the third round of panning, 5 × 108 RBCs were mixed with 1 × 109 phages (amplified Pan II eluate) and 10 µg B6 Fab protein and panning was carried out as described above. The Pan III eluate was also titrated to determine phage titre.
Analysis of individual clones.
Colonies were grown in 150 µl LBAmpglu in 96-well plate at 37°C for 3 hrs. This culture was then used for sequencing and phage production. For sequencing, 5 µl culture was diluted 1:20 in water and 5 µl of this was used as template in 10 µl PCR using primers M13R and U257 and High Fidelity Mix (Roche). 3 µl of the PCR product was analysed by agarose gel electrophoresis, and the remaining volume was treated with Exo-SAP (USB) to degrade unincorporated oligonucleotides and used for sequencing with primers PelBss and U251. For phage production 20 µl culture was transferred into another 96-well plate containing 20 µl helper phage AGM13 at MOI 20 and incubated at 37°C for 90 minutes followed by addition of 160 µl of 2 × YT containing 100 µg/ml ampicillin and 50 µg/ml kanamycin. The culture was grown overnight at 32°C to produce phages. The plate was centrifuged at 3,000 xg to pellet cells and the phage containing supernatant was obtained.
Agglutination assay.
Sixty microliters of culture supernatant containing phages or periplasm or column fraction or an appropriate dilution (including sample before column and flow through obtained during loading) was taken in a 96-well round-bottom microtitre plate (Wells 1–12). Two-fold serial dilutions were made in PBS-BSA (PBS containing 0.2% BSA) (Wells A-H). To each well, 30 µl of 2% human RBC suspension (washed RBC pellet reconstituted v/v to 2% in PBS-BSA) was added. After mixing, the plate was incubated at 37°C for 1 hour. The plate was centrifuged at 800 xg at RT for 5 minutes. The supernatant was discarded by inversion and the pellet was suspended in remaining buffer by vortexing. 60 µl of anti-cmyc Mab or anti-gVIIIp Mab (1:1,000 dilution of ascitic fluid in PBS-BSA) was added per well. After mixing, the plate was incubated at 37°C for 1 hour. Agglutination was read visually. The maximum dilution of sample that gave agglutination (end point) was recorded.
Construction of Fab library and panning.
DNA of 10 scFv clones selected from scFv library were used as template in PCR to amplify the VL and VH variable domain sequences. Equal amounts of all the VL and VH amplified products were pooled to make VL and VH pool and purified on agarose gel. DNA encoding the constant domains CL and CH1 were amplified and spliced with VL pool and VH pool respectively to obtain LC and Fd. The LC and Fd fragments were further spliced to form Fab encoding bicistronic fragment. The amplified fragment was digested with Nhe I and Mlu I and cloned into similarly digested pVCB6 scFv10633042 vector.
The ligated mix was electroporated into TG1 cells and was plated on LBAmpglu plates. Eight-thousand colonies from two 90 mm plates were scraped in 4 ml 2XYT medium. 400 µl of the scraped library was diluted 1:100 into 2XYTAmpglu, grown to A600 ∼ 0.3 at 37°C, infected with AGM13 at MOI 20 and then grown overnight at 32°C to obtain culture supernatant containing Fab-displaying phages.
The Fab library was subjected to panning on human RBCs using protocol similar to that used for selection from scFv library. For the 1st round of panning, 1 × 109 RBCs were mixed with 1 × 1010 Fab library phages and panning was done. The Pan I eluate was titrated and amplified for the next round of panning as described for the scFv library. For the second round of panning, 5 × 108 RBCs were mixed with 1 × 1010 phages and 1 µg of B6 Fab protein and panning was carried out.
Competitive binding assay.
B6 Fab protein was diluted 2-fold serially in microtitre U-bottom plate in PBS-BSA to have a final volume of 15 µl/well. 15 µl culture supernatant containing B6 Fab phages was added to each well and mixed with protein. 30 µl 2% RBCs were added to the phage-protein mix, and incubated at 37°C for 1 hour. The plate was centrifuged at 800 xg at RT for 5 minute. The supernatant was discarded and the pellet was suspended in remaining buffer by vortexing. Sixty microlitre of anti-gVIIIp antibody (1:1,000 dilution) was added per well. After mixing, the plate was incubated at 37°C for 1 hr. Agglutination was read visually and the minimum concentration of B6 Fab protein till which no agglutination was observed was recorded.
Cloning of B6 Fab in secretion vector and periplasmic expression.
DNA of selected B6 Fab mutants was used as template in PCR to amplify Fab encoding sequence. The amplification product was digested with Nhe I and Mlu I and cloned into Nhe I-Mlu I digested low copy number T7-based expression vector pVNLB6Fab13416, carrying wild-type B6 Fab encoding segment preceded by PelB signal sequence. The recombinants obtained were then used to transform BL21 (λDE3) cells and plated on M9 medium containing 100 µg/ml ampicillin (M9Amp) and grown for 16 hrs at 30°C. Approximately 400 colonies were scraped into 150 ml M9Amp medium and grown at 30°C to A600 ∼ 0.6. IPTG was then added to a final conc. of 0.1 mM and incubation was continued for 90 minutes at 30°C. The culture was then processed for periplasm preparation by osmotic shock procedure.18
To check the in vivo folding yield of various B6 Fab mutants, B6 Fab protein was purified from 1.2 ml periplasmic fraction on Ni-NTA spin columns as per manufacturers instructions (Qiagen GmbH). The eluate was checked for presence of Fab by non-reducing SDS-PAGE. The gels were blotted and immunostained using HRP conjugate-Goat-anti mouse (Fab)2 fragment specific antibody and anti-cmyc antibody 9E10 followed by HRP-conjugated goat antimouse IgG (Fc) specific antibody. To analyse the presence of functional Fab, the eluate was also used in agglutination assay.
Cloning of B6 LC and B6 Fd in cytoplasmic expression vector.
DNA encoding B6 LC and B6 Fd was amplified from mutant phage clone DNA and cloned as Nhe I-Mlu I insert in pVCCD41140.7 pVCB6Fd1140 contains DNA sequence encoding Fd region of B6, as Nhe I-Mlu I fragment followed by DNA encoding a 10 amino acid tag, cmyc. pVCB6LC1140 is similar to pVCB6 Fd1140 but contains DNA sequence encoding light chain of B6 (B6 LC) as Nhe I-Mlu I fragment with a stop codon preceding Mlu I site.
Expression and isolation of inclusion bodies, assembly of Fab molecules and purification of monomeric Fab.
Expression was carried out for BL21 (λDE3) cells transformed with pVCB6Fd1140 and pVCB6LC1140 and Inclusion Bodies isolated from one litre culture each of B6 Fd and B6 LC were solubilised and renatured in 4 litre mix to assemble Fab at 10°C for 70 hours using protocol as described earlier.7 The ongoing renaturation was checked in agglutination assay as described above. The renatured material was processed as described previously7 to purify monomeric Fab. The 1st step in purification was anion-exchange chromatography using SP-Sepharose FF. All the 5 proteins showed similar binding and elution profile on SP-Sepharose. The fractions containing functional protein were pooled and subjected to Hydrophobic chromatography on Toyobutyl. Well-folded Fab molecules do not bind to the hydrophobic resin and come in the flow through collected during loading. However, misfolded molecules, which aggregate during refolding bind to the resin and are eluted at zero salt. Both mut1 and mut3 preparation showed presence of very little aggregates on butyl column. However, mut2 and mut4 showed a high percentage of aggregates (data not shown). The flow through of butyl column containing monomeric Fab was treated with iodoacetamide to block free sulfhydryl groups and desalted. The sample was then subjected to anion-exchange chromatography on Q-Sepharose FF. The protein obtained after Q-Sepharose column chromatography was loaded on a 480 ml Sephacryl S-200 column. The column was developed with PBS at a flow rate of 1 ml/min. The elution was monitored at 280 nm and 4 ml fractions were collected. All the proteins gave a symmetrical peak, which eluted at the position corresponding to monomeric Fab. The fractions were analysed by SDS-PAGE under non-reducing conditions and by agglutination assay. Fractions containing active, monomeric Fab fusion protein were pooled.
Thermodynamic stability.
Each purified Fab protein was dissolved in 1 ml PBS, pH 7.2 to a final concentration of 0.5 µM and a temperature scan was carried out from 20°C to 90°C with a scan rate of 1°C per minute and fluorescence emission at 347 nm was recorded with excitation at 280 nm with both excitation and emission slits of 1 nm in Cary Eclipse (Varian) spectroflourimeter. The intrinsic fluorescence spectra were also recorded from 290 nm to 500 nm with an excitation wavelength of 280 nm for each Fab protein incubated at 0.5 µM concentration (1) at 20°C, (2) taken to 95°C with change of 1°C/min and spectra taken at 95°C (3) sample cooled back to 20°C and spectra recorded at 20°C.
Equilibrium urea denaturation.
Purified Fab proteins were incubated at 0.5 µM concentration in increasing concentration of urea (2 M–8.5 M in 0.5 M steps) in PBS, pH 7.2 in a total volume of 1 ml at 25°C for 12 hours. Fluorescence emission spectra were then recorded for samples at 25°C from 290 nm to 450 nm with exitation wavelength of 280 nm. The changes in the emission maxima at 347 nm obtained from these scans were used to determine the urea concentration at which the protein unfolded.
Solubility measurements.
Twenty micrograms of purified Fab protein at a concentration of 1 mg/ml was incubated with increasing concentration of PEG6000 (Fluka) in a total volume of 40 µl in PBS for 1 hour at RT. The sample was then centrifuged at 15,000 xg for 30 minutes to pellet any precipitated Fab protein and the supernatant was analysed by SDS-PAGE. The gels were stained with Coomassie Brilliant Blue R-250 and scanned densitometrically to estimate the amount of soluble Fab as a function of PEG concentration.19
Functional stability.
Fab proteins were incubated at a conc., of 100 µg/ml in PBS at 37°C and RT and agglutination assay and immunoblot was performed at specific time points to monitor the activity of B6 Fab.
Supplementary Material
Acknowledgements
The Department of Science and Technology, Government of India, financially supported the work.
Abbreviations
- bp
base pair
- BSA
bovine serum albumin
- CDR
complementarity determining region
- CH1
first constant domain of the antibody heavy chain
- Fab fragment
antigen-binding fragment consisting of LC and Fd
- Fd
antibody heavy chain fragment consisting of VH and CH1
- FR
framework region of antibody variable domains
- Fv
antibody fragment consisting of VL and VH in non-covalent assembly
- HIV
human immunodeficiency virus
- HRP
horse radish peroxidase
- IPTG
isopropyl-β-D-thiogalactoside
- kb
kilobase
- LC
light chain of antibody
- MAb
monoclonal antibody
- mut
mutant
- NTA
nitrilotriacetic acid
- PAG
polyacrylamide gel
- PBS
phosphate buffered saline (20 mM phosphate buffer, pH 7.2 containing 145 mM NaCl)
- PEG
polyethylene glycol
- RBCs
human red blood cells
- RT
room temperature
- scFv
single-chain antibody fragment consisting of VL and VH connected by a peptide linker
- VL
variable domain of the antibody light chain
- VH
variable domain of the antibody heavy chain
- wt
wild type
Note
Supplementary materials can be found at: www.landesbioscience.com/supplement/GuptaMABS1-3-Sup.pdf
Footnotes
Previously published online as a mAbs E-publication: http://www.landesbioscience.com/journals/mabs/article/8231
References
- 1.Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, et al. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol. 1996;14:309–314. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- 2.Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu Rev Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 3.Hoogenboom HR, de Bruine AP, Hufton SE, Hoet RM, Arends JW, Roovers RC. Antibody phage display technology and its applications. Immunotechnology. 1998;4:1–20. doi: 10.1016/s1380-2933(98)00007-4. [DOI] [PubMed] [Google Scholar]
- 4.Griffiths AD, Duncan AR. Strategies for selection of antibodies by phage display. Curr Opin Biotechnol. 1998;9:102–108. doi: 10.1016/s0958-1669(98)80092-x. [DOI] [PubMed] [Google Scholar]
- 5.Nagy ZA, Hubner B, Lohning C, Rauchenberger R, Reiffert S, Thomassen-Wolf E, et al. Fully human, HLA-DR-specific monoclonal antibodies efficiently induce programmed death of malignant lymphoid cells. Nature medicine. 2002;8:801–807. doi: 10.1038/nm736. [DOI] [PubMed] [Google Scholar]
- 6.Gupta A, Chaudhary VK. Whole-blood agglutination assay for on-site detection of human immunodeficiency virus infection. J Clin Microbiol. 2003;41:2814–2821. doi: 10.1128/JCM.41.7.2814-2821.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta A, Gupta S, Chaudhary VK. Recombinant fusion proteins for haemagglutination-based rapid detection of antibodies to HIV in whole blood. J Immunol Methods. 2001;256:121–140. doi: 10.1016/s0022-1759(01)00435-5. [DOI] [PubMed] [Google Scholar]
- 8.Bhat R, Timasheff SN. Steric exclusion is the principal source of the preferential hydration of proteins in the presence of polyethylene glycols. Protein Sci. 1992;1:1133–1143. doi: 10.1002/pro.5560010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benhar I, Pastan I. Identification of residues that stabilize the single-chain Fv of monoclonal antibodies B3. J Biol Chem. 1995;270:23373–23380. doi: 10.1074/jbc.270.40.23373. [DOI] [PubMed] [Google Scholar]
- 10.McCartney JE, Tai MS, Hudziak RM, Adams GP, Weiner LM, Jin D, et al. Engineering disulfide-linked single-chain Fv dimers [(sFv')2] with improved solution and targeting properties: anti-digoxin 26–10 (sFv')2 and anti-c-erbB-2 741F8 (sFv')2 made by protein folding and bonded through C-terminal cysteinyl peptides. Protein Eng. 1995;8:301–314. doi: 10.1093/protein/8.3.301. [DOI] [PubMed] [Google Scholar]
- 11.Jung S, Honegger A, Pluckthun A. Selection for improved protein stability by phage display. J Mol Biol. 1999;294:163–180. doi: 10.1006/jmbi.1999.3196. [DOI] [PubMed] [Google Scholar]
- 12.Daugherty PS, Chen G, Iverson BL, Georgiou G. Quantitative analysis of the effect of the mutation frequency on the affinity maturation of single chain Fv antibodies. Proc Natl Acad Sci USA. 2000;97:2029–2034. doi: 10.1073/pnas.030527597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbas CF, 3rd, Hu D, Dunlop N, Sawyer L, Cababa D, Hendry RM, et al. In vitro evolution of a neutralizing human antibody to human immunodeficiency virus type 1 to enhance affinity and broaden strain cross-reactivity. Proc Natl Acad Sci USA. 1994;91:3809–3813. doi: 10.1073/pnas.91.9.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thom G, Cockroft AC, Buchanan AG, Candotti CJ, Cohen ES, Lowne D, et al. Probing a protein-protein interaction by in vitro evolution. Proc Natl Acad Sci USA. 2006;103:7619–7624. doi: 10.1073/pnas.0602341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chothia C, Lesk AM. Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol. 1987;196:901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- 16.Chothia C, Lesk AM, Tramontano A, Levitt M, Smith-Gill SJ, Air G, et al. Conformations of immunoglobulin hypervariable regions. Nature. 1989;342:877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- 17.Martin AC, Thornton JM. Structural families in loops of homologous proteins: automatic classification, modelling and application to antibodies. J Mol Biol. 1996;263:800–815. doi: 10.1006/jmbi.1996.0617. [DOI] [PubMed] [Google Scholar]
- 18.Nossal NG, Heppel LA. The release of enzymes by osmotic shock from Escherichia coli in exponential phase. J Biol Chem. 1966;241:3055–3062. [PubMed] [Google Scholar]
- 19.Nieba L, Honegger A, Krebber C, Pluckthun A. Disrupting the hydrophobic patches at the antibody variable/constant domain interface: improved in vivo folding and physical characterization of an engineered scFv fragment. Protein Eng. 1997;10:435–444. doi: 10.1093/protein/10.4.435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.