Abstract
More than twenty recombinant monoclonal antibodies are approved as therapeutics. Almost all of these are based on the whole IgG isotype format, but vary in the origin of the variable regions between mouse (chimeric), humanized mouse and fully human sequences; all of those with whole IgG format employ human constant region sequences. Currently, the opposing merits of the four IgG subclasses are considered with respect to the in vivo biological activities considered to be appropriate to the disease indication being treated. Human heavy chain genes also exhibit extensive structural polymorphism(s) and, being closely linked, are inherited as a haplotype. Polymorphisms (allotypes) within the IgG isotype were originally discovered and described using serological reagents derived from humans; demonstrating that allotypic variants can be immunogenic and provoke antibody responses as a result of allo-immunization. The serologically defined allotypes differ widely within and between population groups; therefore, a mAb of a given allotype will, inevitably, be delivered to a cohort of patients homozygous for the alternative allotype. This publication reviews the serologically defined human IgG allotypes and considers the potential for allotype differences to contribute to or potentiate immunogenicity.
Key words: human IgG, polymorphisms, IgG allotypes, antibody therapeutics, immunogenicity, anti-therapeutic antibody, IgG glycosylation
Introduction
Recombinant human protein (rP) and glycoprotein (rGP) therapeutics are established in the clinic. However, a variety of adverse reactions are reported that may differ between individual therapeutics, between the same therapeutic produced by different companies or different lots produced by the same company. Even when these parameters are controlled, there still remains the final “black box:” the patient and individual disease manifestations. A feature of all rP/rGP is a potential to be immunogenic i.e., cause the generation of anti-therapeutic antibodies (ATA). Such antibodies may neutralize the therapeutic, result in enhanced clearance or precipitate severe adverse reactions. To limit the generation of ATA, an rP/rGP should, ideally, have exactly the same structure as the natural product since departure from such structural fidelity constitutes “altered-self,” and the potential to be immunogenic.
The defined molecular structure of a natural protein/glycoprotein results from multiple intra-cellular processes that include co- and post-translational modifications (CTM; PTM). However, it is important to recognize that the structure assigned to a “natural” protein/glycoprotein is determined for molecules that have been resident in bodily fluid(s), prior to isolation and purification employing multiple protocols. In contrast, human rP/rGP therapeutics are produced in xenogeneic tissues such as Chinese hamster ovary (CHO cells) or mouse NS0 cells that may yield product without the necessary human type CTM and PTM, or add non-human CTM and PTM. Following secretion, the rP/rGP is exposed to the culture medium, to products of the producer cell line and subject to rigorous down-stream processing, formulation, storage and a defined delivery protocol. High productivity may compromise the cellular machinery for CTM and PTM with consequent poor product quality; it is essential therefore to apply qualitative as well as quantitative criteria at an early stage in clone selection. Despite these difficulties, both actual and potential, rapid progress has been made for both the productivity and stability of rP/rGP produced in mammalian cells. These issues have been explored in numerous review articles.1–4
Whilst the above parameters that might predispose antibodies to be immunogenic are widely recognized, and appropriate steps taken to monitor and control them, the consequences of administration to diverse human populations is less well developed. Human populations exhibit multiple genotypes and phenotypes, and one would not wish to add structural differences due to polymorphisms to the inherent propensity for rP/rGP products to exhibit structural heterogeneity. However, all currently licensed recombinant IgG antibody therapeutics have been developed as a single polymorphic (allotypic) form; therefore, administration to patients homozygous for the alternative allotype(s) presents added potential for immunogenicity. This review is intended to provide a summary of the serologically defined allotypes present within human IgG heavy and light chains, and the potential for recombinant monoclonal antibody therapeutics (mAbs) to be immunogenic when administered to diverse human populations.5
Immunogenicity of mAbs
It is no surprise that Orthoclone-OKT3 (muromonab), a licensed mouse IgG2a anti-CD3 antibody used in treatment of acute rejection in transplant patients, is immunogenic in humans, i.e., causes development of anti-therapeutic antibody (ATA) responses in patients.6–8 However, use of this drug continues as it is effective and the patient is only exposed to it for a short time. A significant reduction in immunogenicity and ATA was achieved with the development of chimeric antibodies, bearing mouse heavy and light chain variable domains (VH, VL) and human constant regions (CH, CL). Humanization of the mouse VH and VL, by “transplanting,” at the DNA level, the mouse or rat complementarity regions (CDR) into human VH/VL framework regions (FR) further reduced potential immunogenicity. In practice, such “humanized” domains usually resulted in reduced affinity or specificity such that some mouse or rat residues had to be re-introduced, with consequent increase in potential immunogenicity. With the advent of “fully” human antibodies, generated from phage display libraries or mice transgenic for human immunoglobulin genes, it was anticipated that immunogenicity and the development of ATA might be circumvented.6–8 In practice the first such antibody, adalimumab (Humira) resulted in ∼12% incidence of ATA in patients with rheumatoid arthritis. This was subsequently reduced to ∼2–3% for patients also receiving the non-steroid antiinflammatory drug methotrexate, a mild immunosuppressant.9
Initially, one may wonder why a fully human IgG mAb should be immunogenic; however, on reflection it may seem inevitable. The hallmark of an antibody is its specificity, not just for a particular target, but also for a unique structural feature on that target-the epitope. This is achieved within a secondary immune response that is characterised by somatic hypermutation and selection. Thus, each antibody is a structurally unique molecule with a unique epitope-binding site, the paratope. mAbs generated from phage display libraries or transgenic mice are unique to an individual, human or mouse, and may be perceived as “foreign” to a unique recipient. Given this inherent potential for immunogenicity, other structural disparities should be avoided. Allotypic variations constitute such a disparity between individuals and population groups.
Human IgG Isotypes, Allotypes and Idiotypes
Human antibodies or immunoglobulins (Ig) were originally defined as gamma (γ) globulins because of their mobility to the gamma region on paper electrophoresis. Subsequently, nine isotypes were defined structurally and serologically, mostly with antisera raised in guinea pigs or rabbits. The nine isotypes (iso - same) are present in normal human serum and each isotype expresses a unique profile of effector functions.10–12 Each isotype was further defined by the unique amino acid sequence of the constant regions of their heavy chains and subsequently the sequence of the IGHC genes that encode these constant regions.13,14 Protein sequencing revealed the CH regions to be comprised of three or four repeating homologous sequences (domains). This was confirmed at the gene level by the demonstration that each domain is encoded within a separate exon, interspersed by introns. The heavy chains of the classes are designated as α (IgA), µ (IgM), δ (IgD), γ (IgG), ε (IgE), respectively. The γ heavy chains of the IgG comprise 3 domains, CH1, CH2 and CH3, with a “hinge” sequence intervening between the CH1 and CH2 domains. When referring to individual IgG subclasses, the heavy chains are enumerated as γ1, γ2, γ3 and γ4, respectively. Two types of light chain, kappa (κ) and lambda (λ), were also originally defined serologically, and subsequently by protein and gene sequences. Each H2L2 module expresses either two kappa or two lambda light chains to form H2κ2 or H2δ2 hetero-dimers.
Allotypy within human IgG was first described by Grubb who showed that certain human sera would agglutinate erythrocytes sensitized with human incomplete anti-Rh antibody.15–17 Extensive polymorphism (allotypy) within human IgG heavy and light chains was subsequently recognized by careful serological typing, using human reagents obtained from multiparous women, multiple transfused individuals and normal blood donors (∼5%). Thus, the discovery of this polymorphism demonstrates that exposure of an individual to IgG of a non-self allotype can induce an anti-allotype response. Gene sequencing studies have revealed extensive structural polymorphisms;13,14 however, this review will only consider serologically defined allotypes and their amino acid sequence correlates.
By definition allotypes are shared amongst individuals within population groups, although studies in rabbits showed that antibodies raised within an individual rabbit could be immunogenic to another rabbit of the same allotype. The unique epitopes recognized were termed idiotypes. The term idiotype is now in common use to indicate the uniqueness of a monoclonal antibody, although definition of the term has lead to controversial debate and esoteric arguments in the past. It might be instructive, therefore, to consider its origin in more detail. Extensive allo-immunizations and serological studies revealed and characterized serologically defined allotypy within rabbit heavy and light chains. By extension, Oudin hyper-immunized rabbits with Salmonella typhi and used the sera to generate immune complexes that were administered to other rabbits of the same allotype. The recipient rabbits produced a serologically unique response that was specific for the antibodies raised in the donor rabbit and did not cross-react with anti-Salmonella typhi antibodies raised in other rabbits of the same allotype.18 In a definitive publication, the terms isotype and allotype were extended to include idiotype and justified as follows:18 “The first part of the word (idio, from Greek, peculiar) is justified by the extreme peculiarity of the antigenic specificities in question. An idiotype is a peculiar kind of protein antigen defined by its idiotypic specificity.”
The important point is that idiotypy was defined, serologically, following allo-immunization protocols. At the same time Kunkel was investigating human monoclonal proteins isolated from the sera of patients with multiple myeloma. He immunized rabbits with these proteins, used cross-absorption experiments to show that each expressed unique antigenic determinants (epitopes) and coined the term “individual antigenic specificity” (IAS); he distinguished this phenomenon from idiotypy because the serologic reagents were generated by xeno-immunization, not allo-immunization.19 With the passage of time, both phenomena were referred to as idiotypy. In the current context, mAb treatment of patients may lead to both allo-immunization and/or xeno-immunization that results in antisera that may recognize isotypic, allotypic and idiotypic epitopes. Whilst it is generally held that the idiotype reflects the uniqueness of the combined VH and VL sequences, there is evidence that the idiotype can be influenced by the heavy chain constant region with which they are expressed.20,21
Allotypy and Haplotypes Within Human IgG Heavy and Light Chains
In 1976 the World Health Organization sponsored an expert committee meeting at which the nomenclature for human immunoglobulin allotypes was systematized and a numerical system was proposed to replace the alphabetical system (Table 1).22,23 Both systems may be encountered in the literature; particularly when reference is made to original publications in which the allotypes were defined. Allotypes of IgG proteins are defined by the expression of unique epitope(s) recognized by unique serologic reagent(s). Allotypes expressed on the constant region of IgG heavy chain are designated as Gm (Genetic marker) together with the subclass, e.g., G1m, and the allotype number (or letter), e.g. G1m1 [or G1m(a)], G3m5 [or G3m(b1)]. Polymorphisms within IgA and kappa light chains are designated A2m, e.g., A2m1, and Km, e.g., Km1 respectively. Serological polymorphisms have not been reported for lambda chains; however, there are multiple lambda chain isotypes and the number of IGLC (Cλ) genes can vary between individuals.24
Table 1.
Human immunoglobulin allotypes
| Isotype/type | Heavy chains | Light chains | |||
| IgG1 | IgG2 | IgG3 | IgA | κ | |
| Allotypes | G1m | G2m | G3m | A2m | Km |
| 1(a) | 23(n) | 21(g1) | 1 | 1 | |
| 2(x) | 28(g5) | 2 | 2 | ||
| 3(f) | 11(b0) | 3 | |||
| 17(z) | 5(b1) | ||||
| 13 (b3) | |||||
| 14 (b4) | |||||
| 10 (b5) | |||||
| 15(s) | |||||
| 16(t) | |||||
| 6(c3) | |||||
| 24(c5) | |||||
| 26(u) | |||||
| 27 (v) | |||||
NB: Alphabetical notation given within brackets.
All the allotypes presented in Table 1 are expressed on immunoglobulin constant regions. Since the genes encoding the constant region of the heavy chains (IGHC) are closely linked within the IGH gene locus they are inherited together as a haplotype with a low frequency of crossovers. However, crossover events have occurred during evolution resulting in present populations expressing characteristic haplotypes, hence the usefulness of the allotype system in population studies, Table 2.25,26 Thus, in northern Europe (The Netherlands) the haplotype Gm5*;3;23 that corresponds to the allotypes G3m5,10,11,13,14,26,27;G 1m3;G2m23 is present with a frequency of 0.45 and virtually all individuals are homozygous for the A2m1 allele. However, amongst Nigerian Africans the Gm5*;17;1 haplotype occurs with a frequency of 0.678, the Gm5*;3;23 haplotype is not encountered and the A2m2 allele is present with a frequency of 0.826. Prior to the development of DNA fingerprinting techniques the distribution of Gm haplotypes between individuals and population groups was employed in population studies, paternity testing and forensic science27–31 and provided a bridge between immunological and molecular analysis of IG.32
Table 2.
Prevalent Gm haplotypes in different populations
| Gm haplotypes | Gm haplotypes | Caucasoids | Negroids | Mongoloids |
| G3m;G1m;G2m | G3m;G1m;G2m | |||
| 5*;3;23 | b*;f;n | + | − | − |
| 5*;3;.. | b*;f;n- | + | − | − |
| 21*;17,1;.. | g*;za;n- | + | − | + |
| 21*;17,1,2;.. | g*;za;n- | + | − | + |
| 5*;17,1;.. | b*;za;n- | − | + | − |
| 6,24*;17,1;.. | c3c5*;za;n- | − | + | − |
| 6*;17,1;.. | c3*;za;n- | − | + | − |
| 15*;17,1;.. | s*;za;n- | − | + | − |
| 15,16*;17,1;.. | st*;za;n- | − | − | + |
| 5*;3,1;23 | b*;fa;n | − | − | + |
| 5*;3,1;.. | b*;fa;n- | − | − | + |
Allotype Expression and Structural Correlates With Amino Acid Residues (Eu/Imgt Numbering)
IgG1.
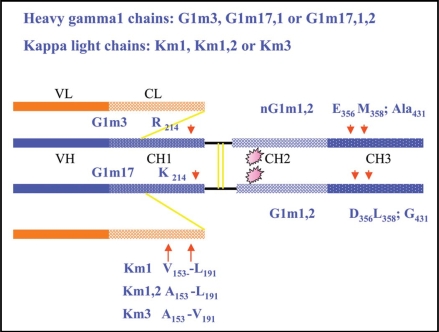
The heavy chains of IgG1 proteins may express G1m3, G1m17,1 or G1m17,1,2 allotypes or G1m(f), G1m(z,a), G1m(z,a,x), respectively.25 The structural correlations with amino acid residues are illustrated in Fig. 1; thus the constant region of G1m17,1, and G1m17,1,2 heavy chains differ from that of G1m3 heavy chains by three and four amino acids, respectively.25 The presence of CH1 arginine (R) at position 214 (IMGT R120; IMGT® www.imgt.org) correlates with G1m3 and that of CH1 lysine (IMGT K120) with G1m17. The presence of CH3 aspartate 356 (IMGT D12) and leucine 358 (IMGT L14) correlates with G1m1. In contrast, the presence of CH3 glutamate and methionine at 356 and 358 (IMGT E12, M14), as observed in G1m3 chains, does not constitute a “true” allotype because these amino acid residues are present in other IgG subclasses, and are expected not to be immunogenic to the individual. Amino acid residue that are present at allotypic positions within one IgG subclass (here IgG1), but are also present in other IgG isotypes are referred to as “isoallotype” (here “nG1m1” isoallotype), when they can be detected in vitro by antibody reagents. The presence of CH3 glycine at residue 431 (IMGT G110) correlates with expression of the G1m2 allotype. An alanine residue is present at that position in other heavy γ1 chains and also in other IgG subclasses; however, CH3 alanine 431 is not an isoallotype as no antibody reagent has been characterized. It is of interest to note that rituximab is G1m17 only (instead of G1m17,1) as the G1m1 allotype was removed by engineering the gene to replace the D-E-L (IMGT D12-E-L14) sequence by E-E-M (IMGT E12-E-M14); “to minimise the potential to be immunogenic.”33
Figure 1.
Correlations between IgG G1m and Km allotypes and amino acids
IgG2.
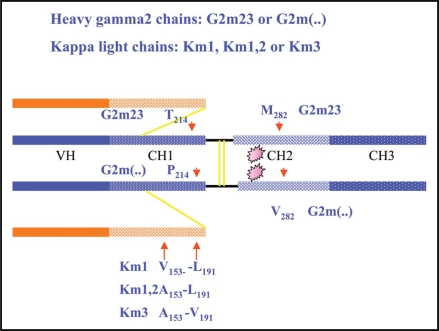
The heavy chains of IgG2 proteins may express the G2m23 or G2m(n) allotype that is correlated with the presence of CH2 methionine at residue 282 (IMGT M45.1), Fig. 2. The G2m23 chains are also characterized by the presence of CH1 threonine 189 (IMGT T92).34 The G2m(..) or G2m(n-) chains and gamma chains of other IgG subclasses have CH1 proline 189 (IMGT 92) and CH2 valine 282 (IMGT V45.1); however, these amino acid residues do not seem immunogenic and cannot be defined as isoallotypes as no mAb reagent has been characterized.
Figure 2.
Correlations between IgG G2m and Km allotypes and amino acids
IgG3.
It is evident from Table 1 that serologically defined allotypy within the IgG3 subclass is very complex. Assigning sequence correlates to IgG3 proteins expressing a given cluster of allotypes had to await dedicated analysis of multiple IgG3 protein sequences. The result of such analysis is summarized in Table 3; however, there is another layer of complexity that underlies these assignments. Thus, analysis of 19 DNA sequences of the G3m5* or G3m(b*) haplotype yielded 11 different allelic sequences; similarly analysis of 10 DNA sequences of the G3m21* or G3m(g1*) haplotype yielded 4 distinct sequences.35 It is not known whether amino acid changes due to these polymorphisms are immunogenic and could, potentially, be detected by unique serological reagents. An additional polymorphism results from differing numbers of hinge region exons. The hinge may be encoded by 2–5 exons; the first encoding a common hinge region with 1–4 repeats of a second exon.36 Thus, the hinge region protein sequence can vary from 27 to 83 amino acid residues, and can influence structural characteristics.
Table 3.
Amino acids involved in the expression of G3m allotypes
| CH2 | CH3 | |||||||||
| IMGT unique numbering: | 82 | 83 | 39 | 44 | 84 | 88 | 98 | 101 | 115 | 116 |
| EU index positions: | 291 | 292 | 379 | 384 | 397 | 409 | 419 | 422 | 435 | 436 |
| γ1 | Pro | Arg | Val | Asn | Val | Lys | Gln | Val | His | Tyr |
| γ2 | - | - | - | - | Met | - | - | - | - | - |
| γ4 | - | - | - | - | - | Arg | Glu | - | - | - |
| G3m5,10,11,13,14 | - | - | - | Ser | Met | - | - | lle | Arg | Phe |
| G3m5,6,10,11,14 | - | - | - | Ser | Met | - | Glu | lle | Arg | Phe |
| G3m5,6,11,24 | - | - | - | Ser | - | Arg | Glu | - | Arg | Phe |
| G3m10,11,13,15 | - | - | Met | Ser | - | - | - | lle | - | - |
| G3m10,11,13,15,16 | - | Trp | Met | Ser | - | - | - | lle | - | - |
| G3m21,28 | Leu | - | - | - | Met | - | - | lle | Arg | - |
| ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | ||||
| G3m allotypes: | 21 | 16 | 15 | 6 | 24 | 28 | 5 | |||
IgG4.
No allotypes have been defined for IgG4. The serologically defined IgG4 polymorphism being the isoallotypes nG4m(a) and nG4m(b). The nG4m(a) epitope that correlates with CH2 leucine 309 (IMGT L92) is also expressed on IgG1 and IgG3 heavy constant regions, whereas the nG4m(b) epitope that correlates with CH2 valine 309 (IMGT V92) is also expressed on the IgG2 heavy chain constant region.
Kappa and lambda light chains.
The IgG.κ/IgG.λ expression ratio is approximately 60/40. The human genome has one kappa constant (IGKC) gene but variable number of lambda constant (IGLC) genes; therefore, we need to consider two different forms of polymorphism, each of which may have consequences for immunogenicity of mAbs. There are three kappa chain allotypes designated Km1, Km2 and Km3 that define three Km alleles: Km1, that correlates with valine153 (IMGT V45.1) and leucine 191 (IMGT L101), Km1,2 that correlates with alanine 153 (IMGT A45.1 and leucine 191 and Km3 that correlates with alanine 153 and valine 191 (IMGT V101).37
Serologically defined allotypes of the constant region of lambda chains have not been reported; however, the number of IGLC genes varies between 7 and 11 depending on individual haplotypes.24,38–40 These isotypes can also be distinguished serologically. In the 7-gene haplotype IGLC1, IGLC2, IGLC3 and IGLC7 are functional, IGLC4 and IGLC5 are pseudogenes and IGLC6 is either functional in a rare haplotype39 or, more frequently, a pseudogene encoding a truncated sequence. The IGLC isotypes can be recognized serologically through the expression of the Mcg, Kern and Oz serological markers;41–43 thus protein products of the IGLC1 gene are Mcg+ Ke+ Oz−, IGLC2 (and IGCL3*04) Mcg− Ke− Oz−, IGLC3 Mcg− Ke− Oz+, IGLC6 and IGLC7 (and IGLC2*04) Mcg− Ke+ Oz−. The Mcg+ isotype correlates with asparagine and threonine at residues 112 and 114 (IMGT N1, T3); whereas, in contrast all Mcg- proteins have alanine and serine at these positions (IMGT A1, S3). Comparison of the C lambda sequences of two Mcg-/Oz- Bence Jones proteins (protein MOR and protein MCP encoded by the IGLC7 gene) has shown that position 193 (IMGT 82) initially claimed to be characteristic of the Mcg marker is not involved.43 Proteins having glycine at position 152 (IMGT G45) express the Kern (Ke) epitope whilst Ke- proteins have serine at this position. Proteins expressing the Oz+ epitope have a lysine at position 190 (IMGT 100), whilst Oz- proteins have an arginine at this position.43
Allotypy of Licensed Chimeric mAb Therapeutics.
It can be appreciated from Fig. 1 that there are six amino acid residues differences between IgG1 proteins that are G1m3;Km1 and G1m17,1,2;Km3, respectively, and this could constitute a significant antigenic “barrier.” Biopharmaceutical companies producing and marketing mAbs rarely disclose the allotype of their product or give any rationale for their choice. This reflects earlier research imperatives that concentrated on the development of genetic engineering techniques for the generation of chimeric and humanized antibodies, and companies obtained a IGHC gene from an available source. The allotype may be predicted from the sequence; however, access to sequence data is not always easy and different sources can yield conflicting data. Currently, IgG1 mAb therapeutics of both predominant allotypes are licensed and in the market place.
Using a panel of mouse anti-human IgG reagents,44,45 we serologically tested a panel of licensed mAbs to determine the allotype distribution; the results are summarized in Table 4. It will be seen from Table 2 that some express the G1m3 allotype commonly expressed within caucasoid populations and others the G1m17,1 allotype commonly expressed among, caucasoid, negroid and mongoloid populations.26 Our serological typing yielded apparently anomalous results for trastuzumab (Herceptin) and omalizumab (Xolair). These proteins appeared, serologically, to have heavy chains that were G1m17 instead of G1m17,1. This was in agreement for trastuzumab with the heavy chain having been engineered to introduce the iso-allotpe nG1m1 correlated to CH3 E356 - E - M358 (IMGT E12, M14).33 This approach has been extended to generate an IgG1 protein with a “null” allotype sequence by the additional replacement of CH1 arginine 214 (G1m3) or lysine 214 (G1m17), respectively, with threonine; threonine 214 IMGT T120), present in IgG2, has not been shown to confer immunogenicity.46
Table 4.
Allotypes of licensed antibody therapeutics
| Generic name | Specificity | Construct | Allotype |
| Rituximab | CD20 | mhIgG1κ | G1m17,1; Km3 |
| Daclizumab | CD25 | hIgG1κ | G1m17,1; Km3 |
| Trastuzumab | p185Her-2 | hIgG1κ | G1m17; Km3 |
| Infliximab | TNFα | mhIgG1κ | G1m17; Km3 |
| Basiliximab | CD25 | mhIgG1κ | G1m3; Km3 |
| Palivizumab | pF-RSV | hIgG1κ | G1m3; Km3 |
| Alemtuzumab | CD52 | hIgG1κ | G1m17,1; Km3 |
| Adalimumab | TNFα | humIgG1κ | G1m17,1; Km3 |
| Omalizumab | IgE-Fc | hIgG1κ | G1m17; Km3 |
| Cetuximab | erbB1 | mhIgG1κ | G1m3; Km3 |
mh, mouse/human chimeric; h, humanised; hum, “fully” human
Incidence of Anti-Allotype Responses
A search of the literature has not yielded any definitive reports in which anti-ATA responses have been analysed for the presence of anti-allotype antibodies. Numerous articles refer to the possibility of such responses, and others claim that all anti-ATA probed were accounted for by specificities for the variable regions of the ATA; however, the assays were only controlled by lack of reactivity with mAbs of an irrelevant specificity. Since these reagents were mostly produced “in-house,” they are likely to have been generated using the same IGHC gene, and would not detect anti-allotype antibodies. The assays were not controlled by testing for lack of reactivity with IgG proteins of the alternate allotype
The possible influence of an allotype discrepancy on ATA responses has been assessed for patients with Crohn's disease receiving infliximab. Of 118 patients receiving infliximab, that carries the G1m17,1 allotypes, 60 (48 %) were homozygous for the alternate G1m3; however, the proportion of patients positive for ATA was the same as for patients who were homozygous for G1m17,1 or heterozygous G1m17,1/G1m3.45 Therefore, exposure across an allotype difference did not predispose to the development of ATA antibodies in this study.
Allotypes and mAb Against Infectious Disease
Immunoglobulin levels may also be associated with allotype, and several studies have established that individuals homozygous for G2m23 have higher IgG2 subclass levels than those homozygous for G2m(..), heterozygotes having intermediate levels. Since protective immune responses to certain bacterial species are restricted to the IgG2 isotype, these findings can and do have implications for disease susceptibility. Such an effect has been reported for post-immunization levels of antibody to Haemophilus influenzae and Meningococcus C polysaccharides with individuals lacking the G2m23 and Km1 allotypes having an increased risk of infection.47–49 These finding suggest that G2m23 mAbs may be optimal as therapeutic and/or prophylactics to combat bacterial infections; however, the FcγRIIa status of the patient must also be considered.50 There is emerging evidence of an interaction between Gm and HLA allotypes on disease susceptibility.52–55
IgG Polymorphism Within Non-Human Primates
It is recognized that immunogenicity in non-human primates does not, necessarily, anticipate immunogenicity in humans; however, such studies are required, prior to regulatory approval. In addition to sequence differences between human and non-human primate IgG isotypes there are differences due to polymorphisms. There are also polymorphism differences within non-human primate species and between those sourced from different countries. When conducting necessary studies in non-human primates it is relevant, therefore, to establish their source and the extent to which the colony used results from inbreeding and the consequent polymorphism diversity, or lack of it.
Conclusion
Anti-therapeutic antibody responses are commonly encountered, particularly in chronic diseases when patients are dosed on a continuing, although possibly irregular, basis. There are many parameters that can contribute to immunogenicity, resident both in the quality of the therapeutic, the mode of administration, the genetic constitution of the recipient etc. It is known that ATA may compromise the patient because they can be neutralizing, result in enhanced clearance or precipitate adverse reactions. Informed patient management requires quantitation of circulating levels of the therapeutic, and detection of any ATA response. When an ATA is detected it is advised that the isotype profile of the ATA be determined; we suggest that preliminary epitope specificity might also be informative and that it should include distinction between variable region structure and constant region allotypy. Should anti-allotype responses be encountered, the development of two or more allotypic variants would be indicated.
Footnotes
Previously published online as a mAbs E-publication: www.landesbioscience.com/journals/mabs/article/9122
References
- 1.Walsh G, Jefferis R. Post-translational modifications in the context of therapeutic proteins. Nat Biotechnol. 2006;24:1241–1252. doi: 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- 2.Jenkins N, Murphy L, Tyther R. Post-translational modifications of recombinant proteins: significance for biopharmaceuticals. Mol Biotechnol. 2008;39:113–118. doi: 10.1007/s12033-008-9049-4. [DOI] [PubMed] [Google Scholar]
- 3.Jenkins N. Modification of therapeutic proteins: challenges and prospects. Cytotechnology. 2007;53:121–125. doi: 10.1007/s10616-007-9075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grillberger L, Kreil TR, Nasr S, Reiter M. Emerging trends in plasma-free manufacturing of recombinant protein therapeutics expressed in mammalian cells. Biotechnol J. 2009;4:186–201. doi: 10.1002/biot.200800241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jefferis R. Immunoglobulins: Allotypes. In: Roitt IM, Delves PJ, editors. Encyclopaedia of Immunology. 2nd edition. Florida: Saunders; 1998. [Google Scholar]
- 6.Chirino AJ, Ary ML, Marshall SA. Minimizing the immunogenicity of protein therapeutics. Drug Discov Today. 2004;9:82–90. doi: 10.1016/S1359-6446(03)02953-2. [DOI] [PubMed] [Google Scholar]
- 7.Reichert JM, Valge-Archer VE. Development trends for monoclonal antibody cancer therapeutics. Nat Rev Drug Discov. 2007;6:349–356. doi: 10.1038/nrd2241. [DOI] [PubMed] [Google Scholar]
- 8.Moutel S, Perez F. Antibodies—Europe. Engineering the next generation of antibodies. Biotechnol J. 2008;3:298–300. doi: 10.1002/biot.200800011. [DOI] [PubMed] [Google Scholar]
- 9.Radstake TR, Svenson M, Eijsbouts AM, van den Hoogen FH, Enevold C, van Riel PL, et al. Formation of antibodies against infliximab and adalimumab strongly correlates with functional drug levels and clinical responses in rheumatoid arthritis. Ann Rheum Dis. 2008 Nov 19; doi: 10.1136/ard.2008.092833. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 10.Woof JM, Burton DR. Human antibody-Fc receptor interactions illuminated by crystal structures. Nat Rev Immunol. 2004;4:89–99. doi: 10.1038/nri1266. [DOI] [PubMed] [Google Scholar]
- 11.Nezlin R, Ghetie V. Interactions of immunoglobulins outside the antigen-combining site. Adv Immunol. 2004;82:155–215. doi: 10.1016/S0065-2776(04)82004-2. [DOI] [PubMed] [Google Scholar]
- 12.Jefferis R. Antibody therapeutics: isotype and glycoform selection. Expert Opin Biol Ther. 2007;7:1401–1413. doi: 10.1517/14712598.7.9.1401. [DOI] [PubMed] [Google Scholar]
- 13.Lefranc G, Lefranc M-P. The Immunoglobulin Facts Book. London: Academic Press; 2001. pp. 1–458. [Google Scholar]
- 14.Guidicelli V, Chaume D, Lefranc M-P. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucl Acids Res. 2005:256–261. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grubb R. Agglutination of erythrocytes coated with incomplete anti-Rh by certain rheumatoid arthritic sera and some other sera; the existence of human serum groups. Acta Pathol Microbiol Scand. 1956;39:95–197. [PubMed] [Google Scholar]
- 16.Grubb R. Advances in human immunoglobulin allotypes. Exp Clin Immunogenet. 1995;12:191–197. doi: 10.1159/000424871. [DOI] [PubMed] [Google Scholar]
- 17.Grubb R. Allotypes: perspectives and future directions. Exp Clin Immunogenet. 1995;12:217–221. doi: 10.1159/000424874. [DOI] [PubMed] [Google Scholar]
- 18.Oudin J, Michel M. Idiotypy of rabbit antibodies. I. Comparison of idiotypy of antibodies against Salmonella typhi with that of antibodies against other bacteria in the same rabbits, or of antibodies against Salmonella typhi in various rabbits. J Exp Med. 1969;130:595–617. doi: 10.1084/jem.130.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natvig JB, Kunkel HG. Human immunoglobulins: classes, subclasses, genetic variants, and idiotypes. Adv Immunol. 1973;16:1–59. doi: 10.1016/s0065-2776(08)60295-3. [DOI] [PubMed] [Google Scholar]
- 20.Torres M, Casadevall A. The immunoglobulin constant region contributes to affinity and specificity. Trends Immunol. 2008;29:91–97. doi: 10.1016/j.it.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Dam TK, Torres M, Brewer CF, Casadevall A. Isothermal titration calorimetry reveals differential binding thermodynamics of variable region-identical antibodies differing in constant region for a univalent ligand. J Biol Chem. 2008;283:31366–31370. doi: 10.1074/jbc.M806473200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Recommendations for the Nomenclature of Human Immunoglobins International Union of Immunological Sciences. Eur J Biochem. 1974;45:5–6. [Google Scholar]
- 23.WHO, author. Review of the notation for allotypic and related markers of human immunoglobulins. Eur J Immunol. 1976;6:59. [PubMed] [Google Scholar]
- 24.Ghanem N, Dariavach P, Bensmana M, Chibani J, Lefranc G, Lefranc MP. Polymorphism of immunoglobulin lambda constant region genes in populations from France, Lebanon, and Tunisia. Exp Clin Immunogenet. 1988;5:186–195. [PubMed] [Google Scholar]
- 25.Lefranc G, Lefranc M-P. IMGT Repertoire (IG and TR) IMGT®, the international ImMunoGeneTics information system®; Gm allotype and Gm haplotypes> Allotypes. http://www.imgt./textes/IMGTrepertoire/Proteins/allotypes/human/IGH/IGHC/Hu_IGHCallotypes1.html. [Google Scholar]
- 26.Lefranc G, Lefranc MP. IMGT Repertoire (IG and TR) MGT®, the international ImMunoGeneTics information system®; Prevelent Gm haplotypes of the human IGHG3,IGHG1 and IGHG2 alleles in different populations. Allotypes. http://www.imgt.org/textes/IMGTrepertoire/Proteins/allotypes/human/IGH/IGHC/Hu_IGHCallotypes2.html. [Google Scholar]
- 27.Lefranc G, Loiselet J, Rivat L, Ropartz C. Gm, Km and Isf allotypes in the Lebanese population. Acta Anthropogenetica. 1976;1:34–45. [Google Scholar]
- 28.Dugoujon JM, Hazout S, Loirat F, Mourrieras B, Crouau-Roy B, Sanchez-Mazas A. GM haplotype diversity of 82 populations over the world suggests a centrifugal model of human migrations. Am J Phys Anthropol. 2004;125:175–192. doi: 10.1002/ajpa.10405. [DOI] [PubMed] [Google Scholar]
- 29.Calderón R, Lodeiro R, Varela TA, Fariña J, Ambrosio B, Guitard E, et al. GM and KM immunoglobulin allotypes in the Galician population: new insights into the peopling of the Iberian Peninsula. BMC Genet. 2007;8:37. doi: 10.1186/1471-2156-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kloosterman A. Application of immunoglobulin allotyping in forensic stain analysis: reliability and sensitivity of Gm and Km typing. Exp Clin Immunogenet. 1989;6:123–132. [PubMed] [Google Scholar]
- 31.Schanfield MS. Application of immunoglobulin heavy chain (GM, AM) and light chain (KM) allotypes to cases of disputed paternity. Exp Clin Immunogenet. 1989;6:112–122. [PubMed] [Google Scholar]
- 32.Lefranc G, Chaabani H, Van Loghem E, Lefranc MP, De Lange G, Helal AN. Simultaneous absence of the human IgG1, IgG2, IgG4 and IgA1 subclasses: immunological and immunogenetical considerations. Eur J Immunol. 1983;13:240–244. doi: 10.1002/eji.1830130312. [DOI] [PubMed] [Google Scholar]
- 33.Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, et al. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci, USA. 1992;89:4285–4289. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hougs L, Svejgaard A, Barrington T. The first constant-domain (CH1) exon of human IGHG2 is polymorphic and in strong linkage disequilibrium with the CH2 exon polymorphism encoding the G2m(n+) allotype in Caucasians. Immunogenetics. 2001;52:242–248. doi: 10.1007/s002510000278. [DOI] [PubMed] [Google Scholar]
- 35.Dard P, Lefranc MP, Osipova L, Sanchez-Mazas A. DNA sequence variability of IGHG3 alleles associated to the main G3m haplotypes in human populations. Eur J Hum Genet. 2001;9:765–772. doi: 10.1038/sj.ejhg.5200700. [DOI] [PubMed] [Google Scholar]
- 36.Dard P, Huck S, Frippiat JP, Lefranc G, Langaney A, Lefranc MP, et al. The IGHG3 gene shows a structural polymorphism characterized by different hinge lengths: sequence of a new 2-exon hinge gene. Hum Genet. 1997;99:138–411. doi: 10.1007/s004390050328. [DOI] [PubMed] [Google Scholar]
- 37.Lefranc G, Lefranc M-P. IMGT Repertoire (Ig and TR) MGT®, the international ImMunoGenTics information system®; Km allotypes. http://imgt.cines.fr/textes/IMGTrepertoire/Proteins/allotypes/human/IGK/IGKC/Hu_IGKCallotypes.html#t1. [Google Scholar]
- 38.Hieter PA, Hollis GF, Korsmeyer SJ, Waldmann TA, Leder P. Clustered arrangement of immunoglobulin lambda constant region genes in man. Nature. 1981;294:536–540. doi: 10.1038/294536a0. [DOI] [PubMed] [Google Scholar]
- 39.Dariavach P, Lefranc G, Lefranc MP. Human immunoglobulin C lambda 6 gene encodes the Kern+Oz-lambda chain and C lambda 4 and C lambda 5 are pseudogenes. Proc Natl Acad Sci USA. 1987;84:9074–9078. doi: 10.1073/pnas.84.24.9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niewold TA, Murphy CL, Weiss DT, Solomon A. Characterization of a light chain product of the human JC lambda 7 gene complex. J Immunol. 1996;157:4474–4477. [PubMed] [Google Scholar]
- 41.Jefferis R, Reimer CB, Skvaril F, de Lange G, Ling NR, Lowe J, et al. Evaluation of monoclonal antibodies having specificity for human IgG sub-classes: results of an IUIS/WHO collaborative study. Immunol Lett. 1985;10:223–252. doi: 10.1016/0165-2478(85)90082-3. [DOI] [PubMed] [Google Scholar]
- 42.Walker MR, Solomon A, Weiss DT, Deutsch HF, Jefferis R. Immunogenic and antigenic epitopes of Ig. XXV. Monoclonal antibodies that differentiate the Mcg+/Mcg− and Oz+/Oz− C region isotypes of human lambda L chains. J Immunol. 1988;140:1600–1604. [PubMed] [Google Scholar]
- 43.Lefranc G, Lefranc M-P. IMGT Repertoire (IG and TR) IMGT®, the international ImMunoGeneTics information system®; Isotypes: Human IGLC. http://imgt.cines.fr/textes/IMGTrepertoire/Proteins/isotypes/human/IGL/IGLC/Hu_IGLCisotypes.html. [Google Scholar]
- 44.Jefferis R, Reimer CB, Skvaril F, de Lange GG, Goodall DM, Bentley TL, et al. Evaluation of monoclonal antibodies having specificity for human IgG subclasses: results of the 2nd IUIS/WHO collaborative study. Immunol Lett. 1992;31:143–168. doi: 10.1016/0165-2478(92)90141-a. [DOI] [PubMed] [Google Scholar]
- 45.Magdelaine-Beuzelin C, Vermeire S, Goodall M, Baert F, Noman M, Assche GV, et al. IgG1 heavy chain-coding gene polymorphism (G1m allotypes) and development of antibodies-to-infliximab. Pharmacogenet Genomics. 2009;19:383–387. doi: 10.1097/FPC.0b013e32832a06bf. [DOI] [PubMed] [Google Scholar]
- 46.Gorman SD, Clark MR. Humanisation of monoclonal antibodies for therapy. Sem Immunol. 1990;2:457–466. [PubMed] [Google Scholar]
- 47.Ambrosino DM, Schiffman G, Gotschlich EC, Schur PH, Rosenberg GA, DeLange GG, et al. Correlation between G2m(n) immunoglobulin allotype and human antibody response and susceptibility to polysaccharide encapsulated bacteria. J Clin Invest. 1985;75:1935–1942. doi: 10.1172/JCI111909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Granoff DM, Suarez BK, Pandey JP, Shackelford PG. Genes associated with the G2m(23) immunoglobulin allotype regulate the IgG subclass responses to Haemophilus influenzae type b polysaccharide vaccine. J Infect Dis. 1988;157:1142–1149. doi: 10.1093/infdis/157.6.1142. [DOI] [PubMed] [Google Scholar]
- 49.Sarvas H, Rautonen N, Mäkelä O. Allotype-associated differences in concentrations of human IgG subclasses. J Clin Immunol. 1991;11:39–45. doi: 10.1007/BF00918793. [DOI] [PubMed] [Google Scholar]
- 50.Oxelius VA. Immunoglobulin constant heavy G subclass chain genes in asthma and allergy. Immunol Res. 2008;40:179–191. doi: 10.1007/s12026-007-0007-1. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez ME, van der Pol WL, Sanders LA, van de Winkel JG. Crucial role of FcgammaRIIa (CD32) in assessment of functional anti-Streptococcus pneumoniae antibody activity in human sera. J Infect Dis. 1999;179:423–433. doi: 10.1086/314603. [DOI] [PubMed] [Google Scholar]
- 52.Dugoujon JM, Cambon-Thomsen A. Immunoglobulin allotypes (GM and KM) and their interactions with HLA antigens in autoimmune diseases: a review. Autoimmunity. 1995;22:245–260. doi: 10.3109/08916939508995322. [DOI] [PubMed] [Google Scholar]
- 53.Propert D. Immunoglobulin allotypes and RFLPs in disease association. Exp Clin Immunogenet. 1995;12:198–205. doi: 10.1159/000424872. [DOI] [PubMed] [Google Scholar]
- 54.Pandey JP. Immunoglobulin GM and KM allotypes and vaccine immunity. Vaccine. 2000;19:613–617. doi: 10.1016/s0264-410x(00)00255-3. [DOI] [PubMed] [Google Scholar]
- 55.Pandey JP, Nietert PJ, Klaamas K, Kurtenkov O. A genetic variant of immunoglobulin gamma2 is strongly associated with immunity to mucin 1 in patients with breast cancer. Cancer Immunol Immunother. 2009 Apr 14; doi: 10.1007/s00262-009-0709-4. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rogers KA, Scinicariello F, Attanasio R. IgG Fc receptor III homologues in non-human primate species: genec characterization and ligand interactions. J Immunol. 2006;177:3848–3856. doi: 10.4049/jimmunol.177.6.3848. [DOI] [PubMed] [Google Scholar]
- 57.Scinicariello F, Engleman CN, Jayashankar L, McClure HM, Attanasio R. Rhesus macaque antibody molecules: sequences and heterogeneity of alpha and gamma constant regions. Immunology. 2004;111:66–74. doi: 10.1111/j.1365-2567.2003.01767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]