Abstract
JunD is a versatile AP-1 transcription factor that can activate or repress a diverse collection of target genes. Precise control of junD expression and JunD protein–protein interactions modulate tumor angiogenesis, cellular differentiation, proliferation and apoptosis. Molecular and clinical knowledge of two decades has revealed that precise JunD activity is elaborated by interrelated layers of constitutive transcriptional control, complex post-transcriptional regulation and a collection of post-translational modifications and protein–protein interactions. The stakes are high, as inappropriate JunD activity contributes to neoplastic, metabolic and viral diseases. This article deconvolutes multiple layers of control that safeguard junD gene expression and functional activity. The activity of JunD in transcriptional activation and repression is integrated into a regulatory network by which JunD exerts a pivotal role in cellular growth control. Our discussion of the JunD regulatory network integrates important open issues and posits new therapeutic targets for the neoplastic, metabolic and viral diseases associated with JunD/AP-1 expression.
Keywords: JunD, AP-1, RNA helicase A and post-transcriptional control element, HTLV-1, cancer, heart failure
Introduction
The activating protein-1 (AP-1) transcription factor is a collection of dimeric complexes composed of members of three families of DNA-binding proteins: Jun (c-Jun, JunB, v-Jun, JunD), Fos (Fra-1 and Fra-2, c-Fos, FosB) and ATF/CREB (ATF1 through 4, ATF-6, β-ATF, ATFx) (Hai and Curran, 1991; Persengiev and Green, 2003; Milde-Langosch, 2005). The AP-1 component proteins are characterized structurally by their leucine-zipper dimerization motif and basic DNA-binding domain. They can either activate or repress transcription and this versatile functional activity is dependent on the specific components of the dimeric complex and the cellular environment (Eferl and Wagner, 2003; Hess et al., 2004). AP-1 figures prominently in transcriptional regulation of early response genes (reviewed by Jochum et al., 2001; Mechta-Grigoriou et al., 2001; Eferl and Wagner, 2003). A feature that characterizes the typical jun family members (junB and c-jun) is their dramatic transcriptional induction by cell growth factors. Their protein products (a) are regulated post-translationally by phosphorylation, (b) bind as heterodimers (some can also bind as homodimers) to the palindromic DNA sequence TGAC/GTCA, also known as the 12-O-tetradecanoyl-phorbol-13-acetate (TPA)-response element (TRE), (c) transmit Ras-mediated transformation signals and (d) participate in the control of apoptosis (Mechta-Grigoriou et al., 2001). Tight control of their gene expression fits hand-in-hand with programmed cell growth.
The junD gene is the most recent addition to the jun family and was identified in a screen of a mouse 3T3 cell cDNA library (Hirai et al., 1989). Hirai et al. (1989) demonstrated JunD binds to the TRE in vitro by DNA electrophoretic mobility-shift assays. JunD behaved similarly to previously identified Jun proteins to transactivate an AP-1-responsive promoter in conjunction with c-Fos. The domain structure of JunD matches other AP-1 component proteins. However, the expression pattern of junD diverges from the well-characterized growth factor-inducible pattern of the c-jun and junB early response genes (Hirai et al., 1989; Pfarr et al., 1994). Transcription of junD is constitutive in quiescent cells and is not induced by addition of serum. In addition, junD mRNA exhibits a divergent expression pattern across tissues. Transgenic mice studies demonstrated c-Jun and JunB are essential for embryonic development, whereas JunD is dispensable and junD−/− mice are viable (Thepot et al., 2000). Together, these features implicated a distinct gene regulation profile and possible function for JunD in relation to c-Jun and JunB (Pfarr et al., 1994). Experimental results over the past two decades have validated these predictions and provided appreciable molecular and clinical knowledge of JunD. This information has yet to be integrated into a cohesive model of the JunD-regulatory network.
This article reviews the significant body of molecular and clinical knowledge of (a) control of junD gene expression at the transcriptional and post-transcriptional levels and (b) post-translational modifications and alternative protein–protein interactions of JunD and their effect on the functional activity of JunD. We integrate JunD transcriptional activation and repression of a diverse collection of target genes into a regulatory network that is pivotal to cellular growth control.
Regulation of JUND gene expression
junD is not regulated by a typical immediate-early gene transcriptional mechanism
The mRNA template of typical AP-1 component proteins is undetectable in quiescent cells but robustly induced by serum stimulation (Herschman, 1991). By contrast, junD mRNA is detectable in quiescent cells, and neither serum stimulation nor TPA treatment significantly increase steady state expression (Hirai et al., 1989; Herschman, 1991). In contrast to typical AP-1 proteins, JunD protein is degraded within the first 30 min following serum stimulation of a quiescent cell population (Pfarr et al., 1994). Subsequently, JunD protein reemerges and steadily increases as cells progress to G1 (Pfarr et al., 1994). This opposite trend in comparison to other AP-1 family members implicates unique post-transcriptional and/or post-translational control mechanisms in the regulation of junD expression.
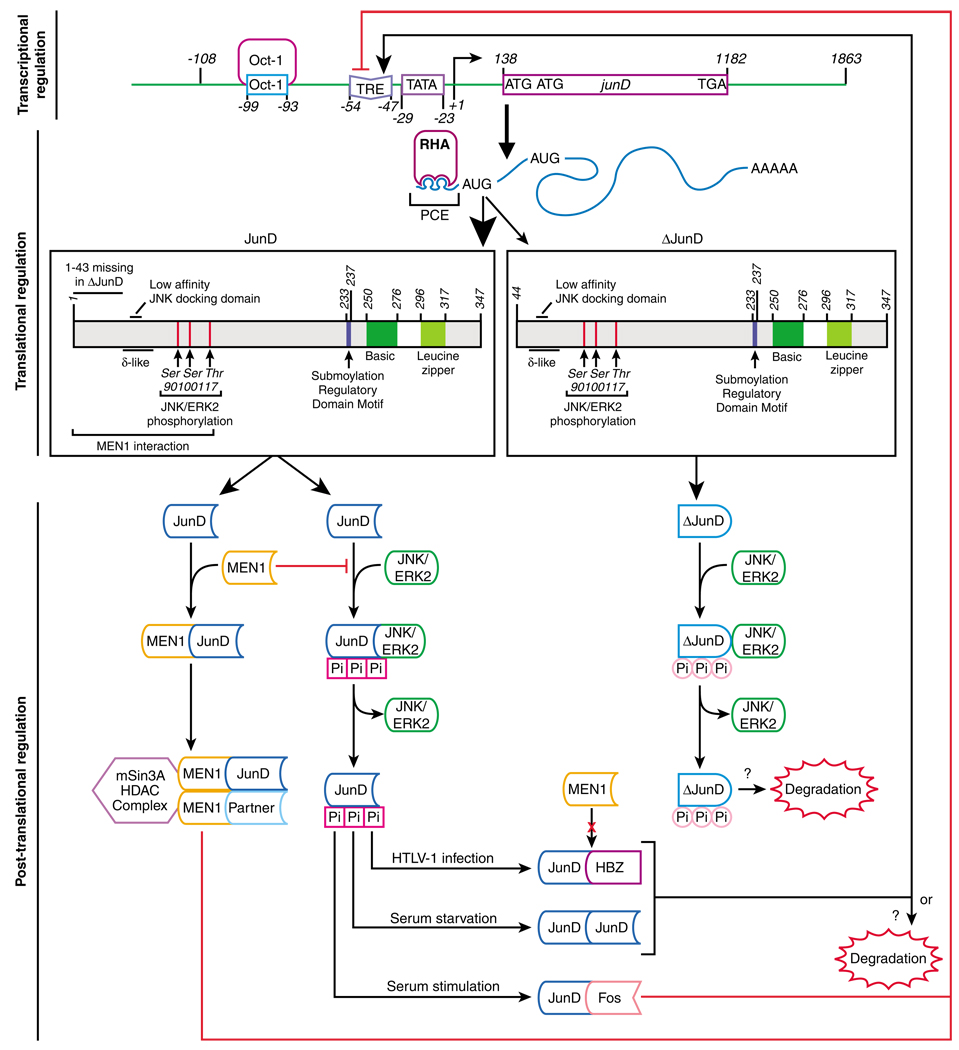
Given the drastic differences in the transcriptional regulation between junD and the other jun gene family members, the junD promoter/enhancer sequence may be expected to lack cis-elements that mediate the transcriptional induction characteristic of c-jun and other AP-1 genes. However, junD contains a conserved TRE in the junD enhancer region (Figure 1). Experiments in a heterologous reporter system determined that the TRE is TPA-inducible and recognized by AP-1 (de Groot et al., 1991). However, this TRE is not induced by AP-1 in the natural context of the junD promoter/enhancer. Instead, this transcription unit is rendered constitutive by an octamer-binding transcription factor 1 site adjacent to the TRE site (Figure 1). Two explanations for the lack of TPA induction of this TRE are that (1) octamer-binding transcription factor 1 binding sterically precludes AP-1 binding to the TRE site or (2) octamer-binding transcription factor 1 binding to the octamer site maximally activates the promoter (de Groot et al., 1991). However, conservation of the TRE in the junD promoter argues for a functional role. Two possible roles for the TRE include binding of JunD homodimers, which generates a positive autoregulatory loop during serum starvation (Figure 1; Berger and Shaul, 1991) and serum stimulation of c-Fos, which leads to downregulation of junD transcription by JunD-cFos heterodimeric AP-1 recruited to this TRE (Figure 1; Berger and Shaul, 1991, 1994).
Figure 1.
The regulation of JunD activity is operated by interrelated layers of transcriptional, post-transcriptional and post-translational control. The top panel summarizes general features of the human junD gene structure and its transcriptional regulation. Positions within the promoter region of the octamer-binding transcription factor 1, TPA response element (TRE) and TATA-binding sites are indicated with sequence numbering relative to the transcription start site (+1). The transcription of junD is constitutive and subject to an autoregulatory loop. The middle panel summarizes the structure of the primary transcription product and two translation products. JunD is more abundant than the N-terminal truncated ΔJunD, as indicated by the thicker arrow. The mRNA contains one exon and no introns. Two alternative translation initiation codons are utilized and these mRNA templates contain either a 138-nt (JunD) or a 681-nt (ΔJunD) 5′ UTR. The 5′ UTR forms stem loop structures that that act as a 5′ terminal PCE. PCE interaction with RHA derepresses mRNA translation and is necessary for junD translation. Domains of the JunD protein are indicated: basic and leucine zipper domains, MEN1 interaction region, the δ-like domain, JNK phosphorylation sites and the sumoylation regulatory domain motif. The N-terminus of JunD is truncated in ΔJunD, which profoundly alters the profile of protein–protein interactions. The lower panel summarizes protein–protein interactions that modulate the versatile functional activity of JunD. The N-terminal low-affinity docking site is important for efficient JunD phosphorylation by JNK, as indicated with red squares labeled Pi. ΔJunD, which lacks this domain is not efficiently phosphorylated by JNK, as indicated by the pink circles labeled Pi. Also, ΔJunD does not interact with MEN1. Interaction of JunD with MEN1 inhibits transcription of JunD target genes. MEN1 interaction with JunD has been proposed to be inhibited for JunD-HBZ heterodimers. MEN1 recruits an HDAC complex that renders the JunD dimer a repressor. Phosphorylation by JNK/ERK2 also is inhibited by MEN1. The identity of the dimerization partner of JunD is determined by the cell type and growth conditions. Little is known about the mechanism of JunD protein degradation. ERK, extracellular signal-regulated kinase; HDAC, histone deacetylase; JNK, Jun N-terminal kinase; MEN1, multiple endocrine neoplasia type-1; PCE, post-transcriptional control element; RHA, RNA helicase A; TPA, 12-O-tetradecanoyl-phorbol-13-acetate; TRE, TPA-response element.
junD is regulated by a unique post-transcriptional control mechanism
Given the constitutive activity of the junD promoter, the main changes in the abundance of JunD protein are regulated prominently downstream of transcription. Interrogation of junD has revealed complex features of the junD transcript that implicate a specialized post-transcriptional regulatory mechanism. First, the junD transcription product is intronless, which circumvents the process of intron removal from pre-mRNA in the nucleus. Intron removal has been shown to facilitate mRNA translation in the cytoplasm and presently is attributed to the activity of a multi-component exon junction complex that is deposited near exon–exon junctions (Braddock et al., 1994; Matsumoto et al., 1998; Wiegand et al., 2003; Nott et al., 2004; Tange et al., 2004; Gudikote et al., 2005). Second, the junD gene encodes a G/C-rich (86%) and relatively long 5′ untranslated region (UTR; Figure 1; Short and Pfarr, 2002). Third, initiation of translation at alternative AUG codons results in two biochemically distinct isoforms of JunD that orchestrate different protein–protein interactions (Okazaki et al., 1998; Short and Pfarr, 2002). The two isoforms of JunD are a full-length, 39-kDa protein, and a 34-kDa protein that lacks 43 amino acids at the N-terminus (ΔJunD in Figure 1; Short and Pfarr, 2002). Efficient cap-dependent translation initiation is favored by relatively short (less than 100 nt) and unstructured 5′ UTR and by single AUG translation initiation codon embedded in a robust consensus Kozak sequence (5′-GCC(A/G)CCAUGG-3′) (Kozak, 1984a, 1984b; Merrick and Hershey, 1996; Yilmaz et al., 2006). The longer and structured 5′ UTR and alternative initiation codons in junD are features discordant with efficient cap-dependent translation initiation.
Short and Pfarr (2002) investigated the possibility that the complex features of the junD 5′ UTR promote internal initiation by ribosome recruitment to an internal ribosome entry site. Cap-independent internal ribosome entry is utilized by some viruses and cellular RNAs that exhibit a long and structured 5′ UTR (reviewed by Baird et al., 2006). Bicistronic reporter assays in HeLa, COS-1 and CHO cells determined that the 5′ UTR of rat junD mRNA does not function as an internal ribosome entry site to promote internal initiation (Short and Pfarr, 2002). These results indicated that translation initiation of the junD mRNA is dependent on ribosome scanning. Efficient translation initiation of junD RNA was predicted to involve RNA–protein interactions that neutralize structural barriers within the 5′ UTR to derepress ribosome scanning and promote efficient translation (Short and Pfarr, 2002). A clue in deciphering junD translational control derived from parallels drawn from the retrovirus model system.
Interaction between RNA helicase A and the 5′ terminal PCE is necessary for efficient translation of junD
Similar to junD, the complex 5′ UTR of retrovirus mRNA templates poses structural barriers to robust cap-dependent translation (Yilmaz et al., 2006). Highly conserved structural cis-acting replication motifs conserved among all retrovirus 5′ UTRs impede ribosome scanning (Yilmaz et al., 2006). Similar to junD, internal ribosome entry site activity in the 5′ UTR has been ruled out for spleen necrosis virus, reticuloendotheliosis virus A and human T-cell leukemia virus type 1 (HTLV-1) (Bolinger et al., 2007). Instead, they contain a unique post-transcriptional control element (PCE) that promotes efficient cap-dependent translation (Butsch et al., 1999; Roberts and Boris-Lawrie, 2003; Hartman et al., 2006; Bolinger et al., 2007). PCE is an orientation-dependent (Butsch et al., 1999), ~150-nt structural element located adjacent to the RNA cap site (+ 1) comprised of two functionally redundant stem-loop structures (Roberts and Boris-Lawrie, 2000, 2003). Proteomic analysis identified that PCE interacts specifically with RNA helicase A (RHA) (Hartman et al., 2006). RHA is a ubiquitous nucleocytoplasmic shuttle protein that also is known as the DEIH (Asp–Glu–Ile–His) box polypeptide 9 and nuclear helicase II (Zhang and Grosse, 2004). The RHA/PCE interaction is necessary for efficient retrovirus translation (Hartman et al., 2006; Bolinger et al., 2007). Given the related characteristics of the junD 5′ UTR, de novo JunD protein synthesis was investigated. First, rat junD 5′ UTR exhibited PCE activity in reporter assays (Hartman et al., 2006). Second, the rate of de novo junD protein synthesis was drastically reduced upon downregulation of RHA in simian cells (Hartman et al., 2006). Third, subcellular fractionation experiments determined that RHA does not affect cytoplasmic accumulation of junD, but is necessary for polyribo-some association of junD. Co-immunoprecipitation studies determined that junD mRNA interacts with RHA in both the nucleus and cytoplasm, positing the hypothesis that RHA is recruited co-transcriptionally (Hartman et al., 2006). The current model is that RHA-junD PCE RNA interaction neutralizes structural barriers within the 5′ UTR to derepress ribosome scanning and promote efficient translation.
An important open issue is whether RHA translational enhancement on the junD PCE is constitutively active or inducible during the course of JunD biology. The steady increase in JunD protein level that starts 30 min after serum stimulation is proportional to the increase in the RHA protein level (C Bolinger, A Sharma and K Boris-Lawrie, unpublished data). By contrast, caspase 3 activity on RHA is a possible negative regulator of de novo JunD synthesis during induction of apoptosis (Takeda et al., 1999; Myohanen and Baylin, 2001; Abdelhaleem, 2003) and during serum starvation (A Sharma, C Bolinger and K Boris-Lawrie, unpublished data). The comprehensive role of RHA/JunD PCE activity in junD post-transcriptional control remains to be fully elucidated.
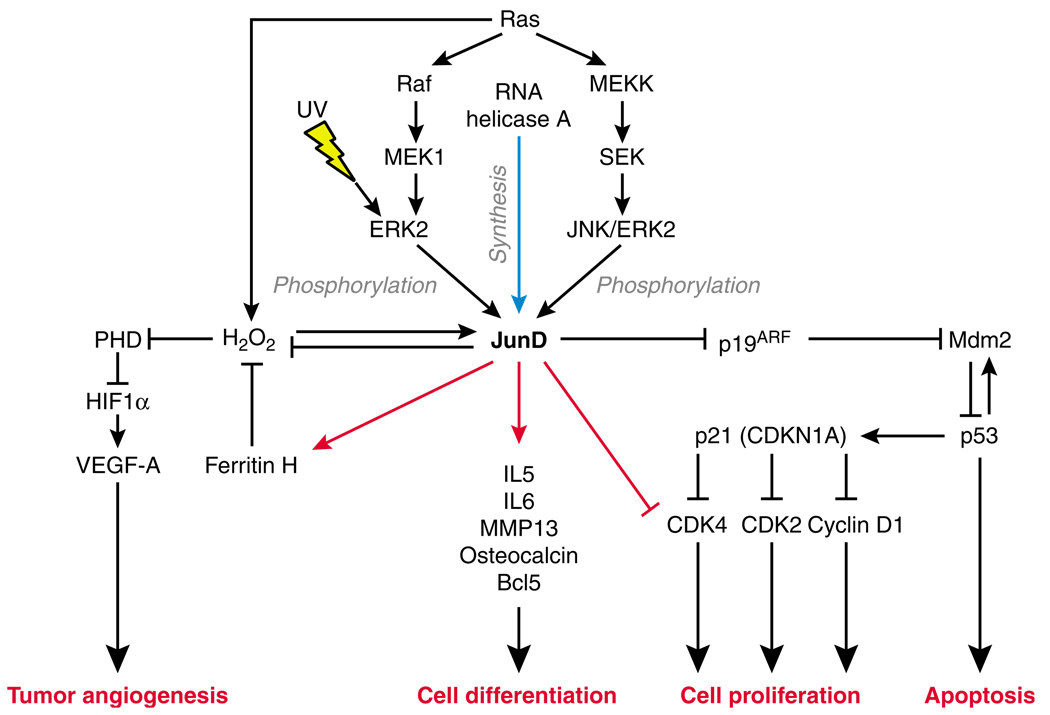
We speculate that induction of JunD translation governed by the RHA–PCE interaction represents a potent regulatory point for JunD protein production and would robustly amplify JunD transcriptional target genes (Figure 2). By influencing the expression of growth control genes such as p21 (CDKN1), which controls cell-cycle progression at G1 (Li et al., 1994), and p19ARF (CDKN2A), a regulator of the p53 pathway via MDM2 (Kamijo et al., 1998; Pomerantz et al., 1998; Zhang et al., 1998), RHA–PCE regulation of JunD is likely to have important consequences for control of cell growth and proliferation. The fact that JunD regulation of these important cellular processes has been conserved among mammalian systems and the conservation of junD PCE activity in primate and rodent supports the importance of RHA–PCE regulation in JunD biology.
Figure 2.
Proposed network operated by JunD activation and repression of a diverse collection of target genes modulates tumor angiogenesis, cellular differentiation, proliferation and apoptosis. Increased JunD protein synthesis occurs upon derepression of efficient junD mRNA translation by RNA helicase A (indicated by blue arrow). The Ras signal transduction cascade activates phosphorylation of JunD. Phosphorylated JunD exhibits versatile activity to selectively repress or activate transcription selected direct target genes (indicated by red lines). Protein–protein interactions, post-translational modification and/or other molecular interactions are indicated by black lines. RHA.
The microRNA pathway may contribute to post-transcriptional regulation of junD
MicroRNA regulation of human genes is estimated to be prominent (≥30% of human genes) (Willingham and Gingeras, 2006). The junD 681-nt 3′ UTR harbors several microRNA target sequences predicted by the Sanger Institute database miRBase (JM Hernandez and K Boris-Lawrie, unpublished data). The observation that RHA interacts with the RNA-induced signaling complex in human cells and acts as an small interfering RNA-loading factor (Robb and Rana, 2007) may provide another link between RHA and junD gene expression. We speculate that RHA is a gatekeeper that associates with PCE to facilitate efficient JunD protein synthesis, or associates with a complementary microRNA to enact translation suppression. We currently are testing these possibilities.
Post-translational modification and alternative protein–protein interactions modulate JunD
JunD diverges from c-Jun and JunB and is not subject to ubiquitin-mediated degradation
Post-translational modification and alternative viral and cellular protein–protein interactions contribute to the versatility of JunD in effecting change in cellular environments. The observed alteration in JunD protein stability during quiescence (Pfarr et al., 1994) has yet to be elucidated in detail. Interestingly, the initial decrease in steady-state JunD protein levels observed upon serum stimulation correlates with a small increase in molecular weight (Pfarr et al., 1994). One unresolved possibility is that JunD is post-translationally modified by factors that target JunD for proteolytic cleavage and/or degradation. Indeed, phosphorylation has been shown to precede ubiquitination of many transcription factors (reviewed by Harper and Elledge, 1999; Punga et al., 2006).
Ubiquitination and proteasome degradation prominently govern levels of c-Jun and JunB (Treier et al., 1994; Fuchs et al., 1997). Their ubiquitination is mediated through the δ-domain (Treier et al., 1994; Fuchs et al., 1997), which first was defined as 27 residues (amino acids 30–57 in c-Jun) present in the N-terminal region of c-Jun, JunB, and is deleted in v-Jun (Morgan et al., 1993; Fuchs et al., 1997). JunD is ubiquitinated inefficiently in spite of the presence of a δ-like domain comprising amino acids 43–69 (Musti et al., 1996). This difference could contribute to the longer half-life of JunD relative to c-Jun (360 and 90 min, respectively) (Treier et al., 1994; Musti et al., 1996). Treier et al. (1994) have shown that deletion of the first seven amino acids of the δ-domain severely reduces ubiquitination of c-Jun. In addition, ubiquitination of the Jun proteins is dependent on physical interaction between the Jun N-terminal kinase (JNK) and a binding region present in c-Jun and JunB (Fuchs et al., 1997). This region is absent in JunD and ΔJunD, which further compromises ubiquitination (Kallunki et al., 1996; Musti et al., 1996).
Pfarr et al. (1994) hypothesized that the stability of JunD is increased during serum starvation, which suggests that JunD protein degradation is subject to negative regulation under this condition. However, the degradation mechanism for JunD has yet to be elucidated as indicated by the question mark in Figure 1.
JunD activity is positively regulated by mitogen activated protein kinase
Phosphorylation of JunD at the N-terminus by mitogen activated protein kinase JNK positively regulates trans-activation activity (Adler et al., 1994; Yazgan and Pfarr, 2002). As highlighted in Figure 1, JunD is phosphorylated by JNK at serines 90 and 100, which are conserved phosphorylation sites in c-Jun, and at threonine 117 (Yazgan and Pfarr, 2002). Downregulation of JNK by estrogen decreases JunD phosphorylation, which in turn decreases expression of the junD mRNA. These observations indicate that phosphorylation is necessary for JunD transcriptional trans-activation of the junD promoter and that JNK stimulates the autoregulatory loop that modulates junD expression (Srivastava et al., 1999). Although the 43 N-terminal residues of JunD do not bind JNK (and do not promote ubiquitination, as discussed above), residues 49–59 of JunD provide a low-affinity docking domain for JNK that is necessary for phosphorylation (Figure 1, efficient phosphorylation is designated by the red squares labeled Pi) (Kallunki et al., 1996; Fuchs et al., 1997; Yazgan and Pfarr, 2002). Although these phosphorylation sites are retained in the ΔJunD isoform, JNK binding and phosphorylation are less efficient in vitro compared with JunD (Figure 1, inefficient phosphorylation is designated by the pink circles labeled Pi) (Yazgan and Pfarr, 2002). Yazgan and Pfarr (2002) speculated that ΔJunD is a less efficient phosphorylation substrate because of conformational changes attributable to the N-terminal truncation of ΔJunD. Results of co-transfection experiments support the conclusion that the less efficient phosphorylation of ΔJunD results in weaker transcriptional activity (Yazgan and Pfarr, 2002).
JunD also is regulated by the extracellular signal-regulated kinase (ERK)–mitogen-activated protein kinase pathway, also known as the MEK–ERK kinase cascade. The evidence is that MEK1–ERK1/2 specific inhibitors PD98059 and UO126 reduce the phosphorylated form of JunD, and that constitutively active forms of the ERK2 enhance JunD phosphorylation (presumably at the same positions as JNK) (Gallo et al., 2002). Figure 2 summarizes the outcome of Ras signaling of ERK2 and JNK/ERK2 on JunD. Phosphorylated JunD exhibits versatile activity in repressing transcription of target genes involved in cell proliferation, apoptosis and tumor angiogenesis, and activating target genes involved in cell differentiation. The role of JunD in differentiation of osteoblast and keratinocyte lineages is described in more detail below.
MEN1 represses JunD activity by two mechanisms
Multiple endocrine neoplasia type-1 (MEN1) is a broadly expressed tumor suppressor and mutations in this gene contribute to tumorigenesis in a diverse range of tissues (reviewed by Agarwal et al., 2003; Dreijerink et al., 2006). The interaction of JunD with MEN1 is mediated by N-terminal residues 1–120 (Figure 1; Agarwal et al., 1999). Both of N- and C-terminal regions of MEN1 are needed for efficient JunD binding (Gobl et al., 1999). MEN1 negatively effects ERK- and JNK-dependent phosphorylation of JunD, without affecting the activation of either kinase (Gallo et al., 2002). The observation that a deletion mutant of MEN1 interferes with ERK2-dependent phosphorylation of JunD, but does not suppress phosphorylation by JNK, indicates that MEN1 affects each of these pathways by a distinct mechanism (Gallo et al., 2002).
As highlighted in Figure 1, MEN1 interaction represses JunD transcriptional activity by inhibition of JunD phosphorylation by JNK and ERK2 (Agarwal et al., 1999; Gobl et al., 1999, 2002; Naito et al., 2005), and also by recruitment of the mSin3A–histone deacetylase (HDAC) complex (Kim et al., 2003). This effect of MEN1 on JunD function involves changes in chromatin structure in the promoter regions of JunD-MEN1 transcriptional targets. MEN1-mediated repression is sensitive to the HDAC inhibitor trichostatin A (Gobl et al., 1999; Kim et al., 2003). One possible mechanism is that the recruitment of the HDAC complex is dependent on sumoylation of JunD that occurs upon formation of the JunD–MEN1 complex; a similar mechanism has been described for ELK1 (Yang and Sharrocks, 2004). A regulatory domain motif that mediates sumoylation, which first was identified in four members of the C/EBP family (C/EBPα, C/EBPβ, C/EBPδ and C/EBPε), is present in JunD (Figure 1; Kim et al., 2002). The JunD-regulatory domain motif corresponds to the sequence LKDEP (amino acids 233–237), which is a putative sumoylation site. The regulatory domain motif is well conserved in c-Jun and JunB, and was determined to have a negative effect on transcription (Kim et al., 2002).
Downregulation of MEN1 in osteoblasts increases JunD transcriptional activity via the junD transcription autoregulatory loop, which in turn increases steady-state JunD protein level (Naito et al., 2005). Since MEN1 interaction is mediated by the JunD N-terminus, ΔJunD does not bind MEN1 and is not susceptible to MEN1 repression (Yazgan and Pfarr, 2002). The expectation that ΔJunD is constitutively active could be offset by reduced transcriptional activity that is attributed to inefficient phosphorylation (Yazgan and Pfarr, 2001, 2002).
MEN1 repression of JunD activity in T cells reduces transcription of the Nur77 locus by recruitment of mSin3A–HDAC (Kim et al., 2003, 2005). During T-cell receptor-mediated thymocyte apoptosis, T-cell receptor activation can derepress Nur77 transcription via the protein kinase C pathway (Kim et al., 2005). Protein kinase C mediates phosphorylation of serine 100 residue of JunD by activating JNK and ERK2, which in turn may mediate the recruitment of p300 to cooperatively activate transcription of Nur77 (Kim et al., 2005). The recruitment of p300 derepresses JunD activity, presumably by reversing HDAC-mediated silencing of the Nur77 promoter (Kim et al., 2005). Repression of MEN1 also allows ERK2-mediated JunD phosphorylation, thus providing a pleiotropic effect (Gallo et al., 2002). The phosphorylation of JunD is dependent on repression of MEN1, but not on recruitment of p300.
JunD activity is altered in heterodimers with human retrovirus leucine zipper protein
Decade-long persistent infection with HTLV-1 can progress to an aggressive, chemotherapy-refractive infectious adult T-cell leukemia (Matsuoka and Jeang, 2007). JunD is postulated to contribute to this process by forming active heterodimers with the HTLV-1 basic leucine zipper factor (HBZ) (Thebault et al., 2004). HBZ also forms heterodimers with c-Jun and JunB (Larocca et al., 1989; Basbous et al., 2003; Cavanagh et al., 2006). HBZ represses c-Jun activity by impairing DNA binding and decreasing c-Jun protein stability, which culminates in reduced expression of c-Jun target genes (Matsumoto et al., 2005). By contrast, HBZ heterodimerizes with either JunD (Figure 1) or JunB and trans-activates expression of AP-1 target genes (Figure 1; Thebault et al., 2004). Studies in a rabbit model system have demonstrated HBZ plays an important role in HTLV-1 infectivity and persistence (Arnold et al., 2006).
HBZ contains an as yet-to-be understood modulatory domain that upregulates JunD transcriptional activity (Hivin et al., 2006). Hivin et al. (2006) have shown that the modulatory activity is not attributable to changes in HBZ–JunD interaction or DNA binding, and they speculated this domain inhibits recruitment of MEN1 or another negative regulator of JunD (Figure 1). A possible scenario is that the HBZ interaction with JunD induces a conformational change that eliminates efficient interaction with MEN1.
JunD activity may figure prominently in the model for HTLV-1 persistence. JunD/HBZ heterodimers are postulated to induce downregulation of cellular proliferation, lymphocyte activation and viral transcription to favor viral latency and persistence and coordinately slow outgrowth of transformed cells. Specifically, HBZ is postulated to enhance JunD activity on target genes (Kuhlmann et al., 2007) that inhibit cell growth (Thebault et al., 2004) and coordinately downregulate c-Jun activity on cellular growth genes and the HTLV-1 genes (Thebault et al., 2004; Matsumoto et al., 2005). However, if the HBZ–JunD interaction interferes with MEN1-mediated JunD transcriptional repression of cellular genes, dysregulation of cellular signaling pathways could occur and drive the cell fate toward neoplastic transformation.
An important caveat of the majority of published studies is the reliance on overexpression of HBZ, rather than physiologic expression from provirus. In authentic infection, HBZ expression is expected to be significantly lower than in standard overexpression systems (Arnold et al., 2006; Li and Green, 2007). This critical difference will undoubtedly affect studies of the physiological significance of HBZ in dysregulation of cellular gene expression. Another important consideration toward understanding HBZ activity is the elucidation of the stoichiometry of HBZ-JunD heterodimers in relation to other AP-1 complexes in infected cells.
JunD is a central molecule in an intricate regulatory network
The ultimate function of JunD is to regulate transcription of target genes that help the cell cope with environmental signals perceived from the environment. As summarized in Table 1, JunD can act as an activator or a repressor of transcription of diverse cell type-specific genes involved in oxidative stress, cell proliferation and differentiation. As a consequence, JunD is associated with a broad array of clinical scenarios including neoplasia, bone, cardiac and epithelial cell biology. The wide spectrum of JunD disease associations is not surprising given its broad expression pattern (Hirai et al., 1989). Accordingly, the role that JunD plays in a given cell depends on the identity of its partner in the AP-1 dimer, the presence or absence of JunD post-transcriptional and post-translational regulators, and the presence of other transcription factors that combinatorially regulate JunD transcriptional target genes.
Table 1.
Features of selected JunD transcriptional targets
| Gene | Cell system | JunD activity | Pathway | References |
|---|---|---|---|---|
| Ferritin H | Oxidative stress in human hepatoma cells | Activator | Antioxidant response during oxidative stress | Tsuji (2005) |
| CDK4 | Polyamine depletion in human intestinal epithelial cells | Repressor | Cell proliferation | Xiao et al. (2007) |
| IL-5 | Human primary T cells | Activator | Cell differentiation | Schwenger et al. (2002) |
| IL-6 | Human androgen independent prostate cancer cells | Activator | Cell differentiation | Zerbini et al. (2003) |
| Bcl6 | Mouse germinal center B cells | Activator | Cell differentiation | Arguni et al. (2006) |
| Osteocalcin | Mouse osteoblasts | Activator | Cell differentiation | Naito et al. (2005); Akhouayri and St Arnaud (2007) |
| MMP13 | Human chondrocytes and fibroblasts | Activator | Cell differentiation | Uria et al. (1998); Ijiri et al. (2005) |
| C4.4A | Rat tumor cell lines | Activator | Cell proliferation | Fries et al. (2007) |
Abbreviations: CDK, cyclin-dependent kinase; IL, interleukin; MMP, matrix metalloprotease.
As highlighted in Table 2, downregulation of JunD affects normal progression of cell maturation and differentiation and contributes to uncontrolled cell proliferation. Accordingly, JunD generally is considered to be a negative regulator of cell proliferation. For instance, JunD exhibits a cell-dependent role in apoptosis and prevents cell death in adult mouse heart cells (Hilfiker-Kleiner et al., 2005) and in UV/H2O2-stressed mouse embryonic fibroblasts (Zhou et al., 2007), while enhancing UV-induced increases in caspase-3 activity and apoptosis in human myeloblastic leukemia ML-1 cells (Li et al., 2002b). Given its pivotal role in balancing cell proliferation, differentiation and apoptosis, and the association of JunD dysregulation with several neoplasms and metabolic diseases, JunD is an attractive target molecule for therapeutic intervention.
Table 2.
JunD modulates a broad array of cell types and clinical scenarios
| Cellular process | Treatment | Effect on JunD expression | Outcome | References |
|---|---|---|---|---|
| Human ovarian cell differentiation | LHRH and FSH/LH | JunD protein levels increase | JunD is upregulated during cell maturation to arrest the cells at G0 | Gunthert et al. (2002) |
| Human epithelial ovarian cancer | Disease | JunD mRNA levels decrease | Downregulation of junD may contribute to the malignant phenotype | Neyns et al. (1996) |
| Adult mouse hearts under chronic moderate pressure overload | Moderate thoracic aortic constriction | JunD protein levels decrease | JunD protects from cardiac hypertrophy, apoptosis, and angiogenesis under pressure overload | Hilfiker-Kleiner et al. (2005) |
| Failing human myocardium | Hearts obtained at the time of transplantation | junD mRNA levels decrease by factor of four | junD downregulation in myopathic heart | Pollack et al. (1997) |
| Chicken chondrocyte differentiation | Parathyroid hormone | JunD protein levels increase | JunD suppresses chondrocyte maturation | Kameda et al. (1997) |
| Mouse T-cell differentiation | Ubi-junDm junD−/− transgenic cells | Not applicable | JunD suppresses lymphocyte proliferation and has a negative effect on Th cell differentiation | Meixner et al. (2004) |
| Human myeloblastic leukemia | UV radiation and TPA | junD mRNA levels increase | UV upregulates junD in ML1 cells via PKC-coupled Erk-signaling pathway and plays a role in UV-induced cell death | Li et al. (2002b) |
| Splenic marginal zone lymphoma (SMZL) | Analysis of genome-wide gene expression | junD mRNA levels increase | junD as well as other AP-1 genes may to be autoregulated by a MAP kinase-independent mechanism in SMZL | Troen et al. (2004) |
| Human intestinal epithelial cells | Polyamine depletion | junD mRNA is stabilized | Polyamines downregulates junD mRNA post-transcriptionally | Li et al. (2002a) |
| Mouse embryonic fibroblasts | V-C or H2O2 | JunD protein level and transcriptional activity increase | JunD plays an antagonistic role to UV-induced and H2O2-induced apoptosis | Zhou et al. (2007) |
Abbreviations: FSH, follicle-stimulating hormone; LH, luteinizing hormone; LHRH, luteinizing hormone-releasing hormone; TPA, TPA, 12-O-tetradecanoyl-phorbol-13-acetate.
Abnormal levels of steady state junD mRNA and JunD protein have been reported in many types of cancer cells and in response to hormones and other factors (Table 2; Neyns et al., 1996; Pollack et al., 1997; Li et al., 2002a, b; Troen et al., 2004). Some studies solely measured the abundance of JunD protein. Given the prominent influence that post-transcriptional and post-translational regulation can exert in balancing JunD abundance and activity, an important consideration is to dissect the relative roles of transcriptional and post-transcriptional regulation in examples of JunD dysregulation. In at least one case, a variation in the stability of the JunD mRNA was identified as the reason for the phenotype (Table 2; Li et al., 2002a).
JunD negatively regulates Ras-mediated transformation
Ras regulates cell proliferation, apoptosis, tumor angiogenesis and accumulation of reactive oxygen species (reviewed by Finkel, 2006). Contrary to c-Jun, JunD acts as a negative regulator of Ras-mediated transformation by downregulating cell growth in response to Ras signal transduction (reviewed by Jochum et al., 2001; Mechta-Grigoriou et al., 2001; Eferl and Wagner, 2003, and summarized in Figure 2). The initial clue that JunD is a negative regulator of Ras was that overexpression of JunD caused a reduction in Ras-induced tumor growth (Pfarr et al., 1994). Consistently, immortalized JunD−/− mouse cell lines exhibit increased proliferation. However primary JunD−/− mouse embryonic fibroblasts exhibit upregulation of p19ARF, which causes early senescence, and p53-dependent apoptosis upon stress (Weitzman et al., 2000). These observations indicate that JunD protects against Ras-induced apoptosis and neutralizes Ras transformation. Contrary to c-Jun, JunD protects cells from oxidative stress by contributing to the downregulation of angiogenic transcription factors like hypoxia-inducible factor α (Gerald et al., 2004). A possible scenario is JunD trans-activation of ferritin (Figure 2), which may be attributable to JunD recruitment at two copies of a bidirectional AP-1 site. This site is located within an antioxidant/electrophile response element 4.5 kb upstream of the human ferritin H transcription start site (Figure 2; Table 1; Tsuji, 2005).
JunD as a positive regulator of cellular maturation
JunD plays a role in cellular maturation of several cell types. JunD positively regulates the expression of osteoblast-specific proteins, type I collagen, osteocalcin and alkaline phosphatase (Table 1; Naito et al., 2005; Akhouayri and St Arnaud, 2007). JunD is coexpressed at high levels with Fra-2 in fully differentiated osteoblasts (McCabe et al., 1996). Loss of function of the negative JunD regulator, MEN1, correlates with increased JunD expression in osteoblasts and upregulation of osteoblast markers (Hendy et al., 2005; Naito et al., 2005). A clue that JunD acts after the commitment of the cells to the osteoblast lineage is the observation that JunD regulates critical osteoblast genes, although JunD−/− mice have not been reported to demonstrate severe skeletal deformities characteristic of other osteoblast lineage genes. Since JunD regulates critical osteoblast genes, an important open issue is the potential for a bone phenotype in JunD-deficient mice under basal versus stress conditions.
JunD plays a role in additional cell types that undergo continuous renewal, differentiation and maturation: keratinocytes, spermatocytes and hematopoeitic cells. Keratinocytes express JunD in all layers of the dermis (Mehic et al., 2005). Jun proteins play a positive role in keratinocyte differentiation in part through positive effects on epidermal growth factor receptor expression (reviewed by Zenz and Wagner, 2006). JunD −/− mice do not exhibit an obvious skin phenotype, although this needs to be evaluated further. The potent function of c-Jun and the redundancy of other AP-1 complex members could contribute to the apparent dispensability of JunD in skin renewal.
JunD is the only AP-1 family member that is expressed at high levels in post-meiotic spermatocytes; the other AP-1 family members are expressed throughout early spermatocyte development (Alcivar et al., 1991). Male junD − /− mice are sterile and exhibit loss of expression of late markers of spermatogenesis, such as calspermin, BMP8 and RT7. However, the open issues remain as to whether or not these markers are direct transcriptional targets of JunD (Thepot et al., 2000). JunD-transgenic mice have decreased numbers of lymphocytes, indicating that JunD may play a negative role in lymphocyte maturation (Meixner et al., 2004).
As highlighted in Table 1, other cell developmental pathways for which JunD is important are maturation of ovarian cells and chondrocytes (Gunthert et al., 2002). Additional studies of the transcriptional regulation by JunD of late markers of cell type maturation will be important to establish the exact role of JunD in this context. The possible role of RHA/PCE translational control among the cell developmental pathways remains an open issue.
Perspectives
These transcriptional, post-transcriptional and post-translational control mechanisms represent interrelated conduits to ensure tight control of JunD functional activity (Figure 1). The outcome is JunD activation and repression of transcription of a diverse collection of target genes, which in turn operate a regulatory network that exerts a pivotal role in cellular growth control (Figure 2). The JunD-regulatory pathways that safeguard precise JunD activity are, in addition to junD gene sequence, potential loci for genetic mutations that dysregulate junD biology. Examples are MEN1 and other proteins that interaction with JunD. These pathways also offer therapeutic targets to treat junD dysregulation. For instance, a JunD transdominant mutation that prevents functional interaction with HTLV-1 HBZ may stall neoplastic transformation by HTLV-1. Continued investigation of the role of JunD in controlling proper growth of cells that undergo continuous renewal, differentiation and maturation (for example, keratinocytes, spermatocytes and hematopoeitic) is expected to identify selective therapeutic targeting of cancers and metabolic diseases.
Acknowledgements
We thank Ms Nicole Placek for input on early versions of the manuscript, Mr Tim Vojt for assistance on figure design and the reviewers for important suggestions. This work was supported by grants from the National Institutes of Health: RO1CA108882; P01CA16058 and P30CA100730.
References
- Abdelhaleem M. The actinomycin D-induced apoptosis in BCR-ABL-positive K562 cells is associated with cytoplasmic translocation and cleavage of RNA helicase A. Anticancer Res. 2003;23:485–490. [PubMed] [Google Scholar]
- Adler V, Unlap T, Kraft AS. A peptide encoding the c-Jun delta domain inhibits the activity of a c-jun amino-terminal protein kinase. J Biol Chem. 1994;269:11186–11191. [PubMed] [Google Scholar]
- Agarwal SK, Guru SC, Heppner C, Erdos MR, Collins RM, Park SY, et al. Menin interacts with the AP1 transcription factor JunD and represses JunD-activated transcription. Cell. 1999;96:143–152. doi: 10.1016/s0092-8674(00)80967-8. [DOI] [PubMed] [Google Scholar]
- Agarwal SK, Novotny EA, Crabtree JS, Weitzman JB, Yaniv M, Jr, Burns AL, et al. Transcription factor JunD, deprived of menin, switches from growth suppressor to growth promoter. Proc Natl Acad Sci USA. 2003;100:10770–10775. doi: 10.1073/pnas.1834524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhouayri O, St Arnaud R. Differential mechanisms of transcriptional regulation of the mouse osteocalcin gene by Jun family members. Calcif Tissue Int. 2007;80:123–131. doi: 10.1007/s00223-006-0102-7. [DOI] [PubMed] [Google Scholar]
- Alcivar AA, Hake LE, Kwon YK, Hecht NB. junD mRNA expression differs from c-jun and junB mRNA expression during male germinal cell differentiation. Mol Reprod Dev. 1991;30:187–193. doi: 10.1002/mrd.1080300304. [DOI] [PubMed] [Google Scholar]
- Arguni E, Arima M, Tsuruoka N, Sakamoto A, Hatano M, Tokuhisa T. JunD/AP-1 and STAT3 are the major enhancer molecules for high Bcl6 expression in germinal center B cells. Int Immunol. 2006;18:1079–1089. doi: 10.1093/intimm/dxl041. [DOI] [PubMed] [Google Scholar]
- Arnold J, Yamamoto B, Li M, Phipps AJ, Younis I, Lairmore MD, et al. Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1. Blood. 2006;107:3976–3982. doi: 10.1182/blood-2005-11-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basbous J, Arpin C, Gaudray G, Piechaczyk M, Devaux C, Mesnard JM. The HBZ factor of human T-cell leukemia virus type I dimerizes with transcription factors JunB and c-Jun and modulates their transcriptional activity. J Biol Chem. 2003;278:43620–43627. doi: 10.1074/jbc.M307275200. [DOI] [PubMed] [Google Scholar]
- Berger I, Shaul Y. Structure and function of human jun-D. Oncogene. 1991;6:561–566. [PubMed] [Google Scholar]
- Berger I, Shaul Y. The human junD gene is positively and selectively autoregulated. DNA Cell Biol. 1994;13:249–255. doi: 10.1089/dna.1994.13.249. [DOI] [PubMed] [Google Scholar]
- Bolinger C, Yilmaz A, Hartman TR, Kovacic MB, Fernandez S, Ye J, et al. RNA helicase A interacts with divergent lymphotropic retroviruses and promotes translation of human T-cell leukemia virus type 1. Nucleic Acids Res. 2007;35:2629–2642. doi: 10.1093/nar/gkm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braddock M, Muckenthaler M, White MR, Thorburn AM, Sommer-ville J, Kingsman AJ, et al. Intron-less RNA injected into the nucleus of Xenopus oocytes accesses a regulated translation control pathway. Nucleic Acids Res. 1994;22:5255–5264. doi: 10.1093/nar/22.24.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butsch M, Hull S, Wang Y, Roberts TM, Boris-Lawrie K. The 5′ RNA terminus of spleen necrosis virus contains a novel posttranscriptional control element that facilitates human immunodeficiency virus Rev/RRE-independent Gag production. J Virol. 1999;73:4847–4855. doi: 10.1128/jvi.73.6.4847-4855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh MH, Landry S, Audet B, Arpin-Andre C, Hivin P, Pare ME, et al. HTLV-I antisense transcripts initiating in the 3′ LTR are alternatively spliced and polyadenylated. Retrovirology. 2006;3:15. doi: 10.1186/1742-4690-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot RP, Karperien M, Pals C, Kruijer W. Characterization of the mouse junD promoter—high basal level activity due to an octamer motif. EMBO J. 1991;10:2523–2532. doi: 10.1002/j.1460-2075.1991.tb07792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreijerink KM, Hoppener JW, Timmers HM, Lips CJ. Mechanisms of disease: multiple endocrine neoplasia type 1—relation to chromatin modifications and transcription regulation. Nat Clin Pract Endocrinol Metab. 2006;2:562–570. doi: 10.1038/ncpendmet0292. [DOI] [PubMed] [Google Scholar]
- Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- Finkel T. Intracellular redox regulation by the family of small GTPases. Antioxid Redox Signal. 2006;8:1857–1863. doi: 10.1089/ars.2006.8.1857. [DOI] [PubMed] [Google Scholar]
- Fries F, Nazarenko I, Hess J, Claas A, Angel P, Zoller M. CEBPbeta, JunD and c-Jun contribute to the transcriptional activation of the metastasis-associated C4.4A gene. Int J Cancer. 2007;120:2135–2147. doi: 10.1002/ijc.22447. [DOI] [PubMed] [Google Scholar]
- Fuchs SY, Xie B, Adler V, Fried VA, Davis RJ, Ronai Z. c-Jun NH2-terminal kinases target the ubiquitination of their associated transcription factors. J Biol Chem. 1997;272:32163–32168. doi: 10.1074/jbc.272.51.32163. [DOI] [PubMed] [Google Scholar]
- Gallo A, Cuozzo C, Esposito I, Maggiolini M, Bonofiglio D, Vivacqua A, et al. Menin uncouples Elk-1, JunD and c-Jun phosphorylation from MAP kinase activation. Oncogene. 2002;21:6434–6445. doi: 10.1038/sj.onc.1205822. [DOI] [PubMed] [Google Scholar]
- Gerald D, Berra E, Frapart YM, Chan DA, Giaccia AJ, Mansuy D, et al. JunD reduces tumor angiogenesis by protecting cells from oxidative stress. Cell. 2004;118:781–794. doi: 10.1016/j.cell.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Gobl AE, Berg M, Lopez-Egido JR, Oberg K, Skogseid B, Westin G. Menin represses JunD-activated transcription by a histone deacetylase-dependent mechanism. Biochim Biophys Acta. 1999;1447:51–56. doi: 10.1016/s0167-4781(99)00132-3. [DOI] [PubMed] [Google Scholar]
- Gudikote JP, Imam JS, Garcia RF, Wilkinson MF. RNA splicing promotes translation and RNA surveillance. Nat Struct Mol Biol. 2005;12:801–809. doi: 10.1038/nsmb980. [DOI] [PubMed] [Google Scholar]
- Gunthert AR, Grundker C, Hollmann K, Emons G. Luteinizing hormone-releasing hormone induces JunD-DNA binding and extends cell cycle in human ovarian cancer cells. Biochem Biophys Res Commun. 2002;294:11–15. doi: 10.1016/S0006-291X(02)00427-8. [DOI] [PubMed] [Google Scholar]
- Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. Skipping into the E2F1-destruction pathway. Nat Cell Biol. 1999;1:E5–E7. doi: 10.1038/8952. [DOI] [PubMed] [Google Scholar]
- Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat Struct Mol Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- Hendy GN, Kaji H, Sowa H, Lebrun JJ, Canaff L. Menin and TGF-beta superfamily member signaling via the Smad pathway in pituitary, parathyroid and osteoblast. Horm Metab Res. 2005;37:375–379. doi: 10.1055/s-2005-870152. [DOI] [PubMed] [Google Scholar]
- Herschman HR. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- Hilfiker-Kleiner D, Hilfiker A, Kaminski K, Schaefer A, Park JK, Michel K, et al. Lack of JunD promotes pressure overload-induced apoptosis, hypertrophic growth, and angiogenesis in the heart. Circulation. 2005;112:1470–1477. doi: 10.1161/CIRCULATIONAHA.104.518472. [DOI] [PubMed] [Google Scholar]
- Hirai SI, Ryseck RP, Mechta F, Bravo R, Yaniv M. Characterization of junD: a new member of the jun proto-oncogene family. EMBO J. 1989;8:1433–1439. doi: 10.1002/j.1460-2075.1989.tb03525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hivin P, Arpin-Andre C, Clerc I, Barbeau B, Mesnard JM. A modified version of a Fos-associated cluster in HBZ affects Jun transcriptional potency. Nucleic Acids Res. 2006;34:2761–2772. doi: 10.1093/nar/gkl375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijiri K, Zerbini LF, Peng H, Correa RG, Lu B, Walsh N, et al. A novel role for GADD45beta as a mediator of MMP-13 gene expression during chondrocyte terminal differentiation. J Biol Chem. 2005;280:38544–38555. doi: 10.1074/jbc.M504202200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochum W, Passegue E, Wagner EF. AP-1 in mouse development and tumorigenesis. Oncogene. 2001;20:2401–2412. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- Kallunki T, Deng T, Hibi M, Karin M. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- Kameda T, Watanabe H, Iba H. C-Jun and JunD suppress maturation of chondrocytes. Cell Growth Differ. 1997;8:495–503. [PubMed] [Google Scholar]
- Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Lee JE, Cho EJ, Liu JO, Youn HD. Menin, a tumor suppressor, represses JunD-mediated transcriptional activity by association with an mSin3A-histone deacetylase complex. Cancer Res. 2003;63:6135–6139. [PubMed] [Google Scholar]
- Kim H, Lee JE, Kim BY, Cho EJ, Kim ST, Youn HD. Menin represses JunD transcriptional activity in protein kinase C theta-mediated Nur77 expression. Exp Mol Med. 2005;37:466–475. doi: 10.1038/emm.2005.57. [DOI] [PubMed] [Google Scholar]
- Kim J, Cantwell CA, Johnson PF, Pfarr CM, Williams SC. Transcriptional activity of CCAAT/enhancer-binding proteins is controlled by a conserved inhibitory domain that is a target for sumoylation. J Biol Chem. 2002;277:38037–38044. doi: 10.1074/jbc.M207235200. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984a;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984b;12:3873–3893. doi: 10.1093/nar/12.9.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann AS, Villaudy J, Gazzolo L, Castellazzi M, Mesnard JM, Duc DM. HTLV-1 HBZ cooperates with JunD to enhance transcription of the human telomerase reverse transcriptase gene (hTERT) Retrovirology. 2007;4:92. doi: 10.1186/1742-4690-4-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocca D, Chao LA, Seto MH, Brunck TK. Human T-cell leukemia virus minus strand transcription in infected T-cells. Biochem Biophys Res Commun. 1989;163:1006–1013. doi: 10.1016/0006-291x(89)92322-x. [DOI] [PubMed] [Google Scholar]
- Li L, Liu L, Rao JN, Esmaili A, Strauch ED, Bass BL, et al. JunD stabilization results in inhibition of normal intestinal epithelial cell growth through P21 after polyamine depletion. Gastroenterology. 2002a;123:764–779. doi: 10.1053/gast.2002.35386. [DOI] [PubMed] [Google Scholar]
- Li M, Green PL. Detection and quantitation of HTLV-1 and HTLV-2 mRNA species by real-time RT-PCR. J Virol Methods. 2007;142:159–168. doi: 10.1016/j.jviromet.2007.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Dai W, Lu L. Ultraviolet-induced junD activation and apoptosis in myeloblastic leukemia ML-1 cells. J Biol Chem. 2002b;277:32668–32676. doi: 10.1074/jbc.M203519200. [DOI] [PubMed] [Google Scholar]
- Li Y, Jenkins CW, Nichols MA, Xiong Y. Cell cycle expression and p53 regulation of the cyclin-dependent kinase inhibitor p21. Oncogene. 1994;9:2261–2268. [PubMed] [Google Scholar]
- Matsumoto J, Ohshima T, Isono O, Shimotohno K. HTLV-1 HBZ suppresses AP-1 activity by impairing both the DNA-binding ability and the stability of c-Jun protein. Oncogene. 2005;24:1001–1010. doi: 10.1038/sj.onc.1208297. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Wassarman KM, Wolffe AP. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 1998;17:2107–2121. doi: 10.1093/emboj/17.7.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7:270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- McCabe LR, Banerjee C, Kundu R, Harrison RJ, Dobner PR, Stein JL, et al. Developmental expression and activities of specific fos and jun proteins are functionally related to osteoblast maturation: role of Fra-2 and Jun D during differentiation. Endocrinology. 1996;137:4398–4408. doi: 10.1210/endo.137.10.8828501. [DOI] [PubMed] [Google Scholar]
- Mechta-Grigoriou F, Gerald D, Yaniv M. The mammalian Jun proteins: redundancy and specificity. Oncogene. 2001;20:2378–2389. doi: 10.1038/sj.onc.1204381. [DOI] [PubMed] [Google Scholar]
- Mehic D, Bakiri L, Ghannadan M, Wagner EF, Tschachler E. Fos and jun proteins are specifically expressed during differentiation of human keratinocytes. J Invest Dermatol. 2005;124:212–220. doi: 10.1111/j.0022-202X.2004.23558.x. [DOI] [PubMed] [Google Scholar]
- Meixner A, Karreth F, Kenner L, Wagner EF. JunD regulates lymphocyte proliferation and T helper cell cytokine expression. EMBO J. 2004;23:1325–1335. doi: 10.1038/sj.emboj.7600133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC, Hershey JWB. In: Translational Control. Hershey JWB, Mathews DH, Sonenberg N, editors. Cold Spring Harbor Laboratory Press; Plainview: 1996. pp. 31–69. [Google Scholar]
- Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. Eur J Cancer. 2005;41:2449–2461. doi: 10.1016/j.ejca.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Morgan IM, Asano M, Havarstein LS, Ishikawa H, Hiiragi T, Ito Y, et al. Amino acid substitutions modulate the effect of Jun on transformation, transcriptional activation and DNA replication. Oncogene. 1993;8:1135–1140. [PubMed] [Google Scholar]
- Musti AM, Treier M, Peverali FA, Bohmann D. Differential regulation of c-Jun and JunD by ubiquitin-dependent protein degradation. Biol Chem. 1996;377:619–624. doi: 10.1515/bchm3.1996.377.10.619. [DOI] [PubMed] [Google Scholar]
- Myohanen S, Baylin SB. Sequence-specific DNA binding activity of RNA helicase A to the p16INK4a promoter. J Biol Chem. 2001;276:1634–1642. doi: 10.1074/jbc.M004481200. [DOI] [PubMed] [Google Scholar]
- Naito J, Kaji H, Sowa H, Hendy GN, Sugimoto T, Chihara K. Menin suppresses osteoblast differentiation by antagonizing the AP-1 factor, JunD. J Biol Chem. 2005;280:4785–4791. doi: 10.1074/jbc.M408143200. [DOI] [PubMed] [Google Scholar]
- Neyns B, Katesuwanasing, Vermeij J, Bourgain C, Vandamme B, Amfo K, et al. Expression of the jun family of genes in human ovarian cancer and normal ovarian surface epithelium. Oncogene. 1996;12:1247–1257. [PubMed] [Google Scholar]
- Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki S, Ito T, Ui M, Watanabe T, Yoshimatsu K, Iba H. Two proteins translated by alternative usage of initiation codons in mRNA encoding a JunD transcriptional regulator. Biochem Biophys Res Commun. 1998;250:347–353. doi: 10.1006/bbrc.1998.9331. [DOI] [PubMed] [Google Scholar]
- Persengiev SP, Green MR. The role of ATF/CREB family members in cell growth, survival and apoptosis. Apoptosis. 2003;8:225–228. doi: 10.1023/a:1023633704132. [DOI] [PubMed] [Google Scholar]
- Pfarr CM, Mechta F, Spyrou G, Lallemand D, Carillo S, Yaniv M. Mouse JunD negatively regulates fibroblast growth and antagonizes transformation by ras. Cell. 1994;76:747–760. doi: 10.1016/0092-8674(94)90513-4. [DOI] [PubMed] [Google Scholar]
- Pollack PS, Pasquarello LM, Budjak R, Fernandez E, Soprano KJ, Redfern BG, et al. Differential expression of c-jun and junD in end-stage human cardiomyopathy. J Cell Biochem. 1997;65:245–253. doi: 10.1002/(sici)1097-4644(199705)65:2<245::aid-jcb9>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2′s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- Punga T, Bengoechea-Alonso MT, Ericsson J. Phosphorylation and ubiquitination of the transcription factor sterol regulatory element-binding protein-1 in response to DNA binding. J Biol Chem. 2006;281:25278–25286. doi: 10.1074/jbc.M604983200. [DOI] [PubMed] [Google Scholar]
- Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Roberts TM, Boris-Lawrie K. The 5′ RNA terminus of spleen necrosis virus stimulates translation of nonviral mRNA. J Virol. 2000;74:8111–8118. doi: 10.1128/jvi.74.17.8111-8118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TM, Boris-Lawrie K. Primary sequence and secondary structure motifs in spleen necrosis virus RU5 confer translational utilization of unspliced human immunodeficiency virus type 1 reporter RNA. J Virol. 2003;77:11973–11984. doi: 10.1128/JVI.77.22.11973-11984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenger GT, Kok CC, Arthaningtyas E, Thomas MA, Sanderson CJ, Mordvinov VA. Specific activation of human inter-leukin-5 depends on de novo synthesis of an AP-1 complex. J Biol Chem. 2002;277:47022–47027. doi: 10.1074/jbc.M207414200. [DOI] [PubMed] [Google Scholar]
- Short JD, Pfarr CM. Translational regulation of the JunD messenger RNA. J Biol Chem. 2002;277:32697–32705. doi: 10.1074/jbc.M204553200. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Weitzmann MN, Cenci S, Ross FP, Adler S, Pacifici R. Estrogen decreases TNF gene expression by blocking JNK activity and the resulting production of c-Jun and JunD. J Clin Invest. 1999;104:503–513. doi: 10.1172/JCI7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y, Caudell P, Grady G, Wang G, Suwa A, Sharp GC, et al. Human RNA helicase A is a lupus autoantigen that is cleaved during apoptosis. J Immunol. 1999;163:6269–6274. [PubMed] [Google Scholar]
- Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Thebault S, Basbous J, Hivin P, Devaux C, Mesnard JM. HBZ interacts with JunD and stimulates its transcriptional activity. FEBS Lett. 2004;562:165–170. doi: 10.1016/S0014-5793(04)00225-X. [DOI] [PubMed] [Google Scholar]
- Thepot D, Weitzman JB, Barra J, Segretain D, Stinnakre MG, Babinet C, et al. Targeted disruption of the murine junD gene results in multiple defects in male reproductive function. Development. 2000;127:143–153. doi: 10.1242/dev.127.1.143. [DOI] [PubMed] [Google Scholar]
- Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- Troen G, Nygaard V, Jenssen TK, Ikonomou IM, Tierens A, Matutes E, et al. Constitutive expression of the AP-1 transcription factors c-jun, junD, junB, and c-fos and the marginal zone B-cell transcription factor Notch2 in splenic marginal zone lymphoma. J Mol Diagn. 2004;6:297–307. doi: 10.1016/S1525-1578(10)60525-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y. JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress. Oncogene. 2005;24:7567–7578. doi: 10.1038/sj.onc.1208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uria JA, Jimenez MG, Balbin M, Freije JM, Lopez-Otin C. Differential effects of transforming growth factor-beta on the expression of collagenase-1 and collagenase-3 in human fibroblasts. J Biol Chem. 1998;273:9769–9777. doi: 10.1074/jbc.273.16.9769. [DOI] [PubMed] [Google Scholar]
- Weitzman JB, Fiette L, Matsuo K, Yaniv M. JunD protects cells from p53-dependent senescence and apoptosis. Mol Cell. 2000;6:1109–1119. doi: 10.1016/s1097-2765(00)00109-x. [DOI] [PubMed] [Google Scholar]
- Wiegand HL, Lu S, Cullen BR. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc Natl Acad Sci USA. 2003;100:11327–11332. doi: 10.1073/pnas.1934877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT, Gingeras TR. TUF love for ‘junk’ DNA. Cell. 2006;125:1215–1220. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Xiao L, Rao JN, Zou T, Liu L, Marasa BS, Chen J, et al. Induced JunD in intestinal epithelial cells represses CDK4 transcription through its proximal promoter region following polyamine depletion. Biochem J. 2007;403:573–581. doi: 10.1042/BJ20061436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Mol Cell. 2004;13:611–617. doi: 10.1016/s1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- Yazgan O, Pfarr CM. Differential binding of the Menin tumor suppressor protein to JunD isoforms. Cancer Res. 2001;61:916–920. [PubMed] [Google Scholar]
- Yazgan O, Pfarr CM. Regulation of two JunD isoforms by Jun N-terminal kinases. J Biol Chem. 2002;277:29710–29718. doi: 10.1074/jbc.M204552200. [DOI] [PubMed] [Google Scholar]
- Yilmaz A, Bolinger C, Boris-Lawrie K. Retrovirus translation initiation: Issues and hypotheses derived from study of HIV-1. Curr HIV Res. 2006;4:131–139. doi: 10.2174/157016206776055039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenz R, Wagner EF. Jun signalling in the epidermis: from developmental defects to psoriasis and skin tumors. Int J Biochem Cell Biol. 2006;38:1043–1049. doi: 10.1016/j.biocel.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Zerbini LF, Wang Y, Cho JY, Libermann TA. Constitutive activation of nuclear factor kappaB p50/p65 and Fra-1 and JunD is essential for deregulated interleukin 6 expression in prostate cancer. Cancer Res. 2003;63:2206–2215. [PubMed] [Google Scholar]
- Zhang S, Grosse F. Multiple functions of nuclear DNA helicase II (RNA helicase A) in nucleic acid metabolism. Acta Biochim Biophys Sin (Shanghai) 2004;36:177–183. doi: 10.1093/abbs/36.3.177. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- Zhou H, Gao J, Lu ZY, Lu L, Dai W, Xu M. Role of c-Fos/JunD in protecting stress-induced cell death. Cell Prolif. 2007;40:431–444. doi: 10.1111/j.1365-2184.2007.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]