Abstract
Plant hormones have pivotal roles in almost every aspect of plant development. Over the past decades, physiological and genetic studies have revealed that hormone action in plants is determined by complex interactions between hormonal signalling pathways. Evidence is accumulating for the existence of crosstalk between the auxin and jasmonate (JA) signalling pathways. Recently, the JASMONATE ZIM-domain (JAZ) proteins have been identified as the long-sought repressors of JA signalling. Here, we show that expression of JAZ1/TIFY10A is not solely inducible by JA, but that it is also an early auxin-responsive gene. Furthermore, we could show that the auxin-inducible expression of JAZ1/TIFY10A is independent of the JA signalling pathway but is controlled by the auxin/indole-3-acetic acid-auxin response transcription factor signalling pathway. Our results provide evidence for the existence of at least two different input signals regarding JAZ1/TIFY10A expression and thus support the idea of an intimate molecular interplay between auxin and JA signalling.
Keywords: JAZ, MeJA, NLS, TIFY, ARF
Introduction
Plant hormones are implicated in every aspect of plant growth and development, as well as in the response to environmental cues. Both positive and negative regulators are involved in hormonal signal-transduction pathways to optimize the efficiency of the stimulus (Ulmasov et al, 1999; Ikeda et al, 2001; Sasaki et al, 2003). These regulators are often targeted for degradation by the ubiquitin–proteasome pathway, which is orchestrated by Skp1–Cullin–F-box (SCF)-type E3 ubiquitin ligases. The SCF transport inhibitor response 1 (SCFTIR1) complex has a central role in the signalling of the plant hormone auxin by targeting auxin/indole-3-acetic acid (Aux/IAA) proteins on auxin response (Dharmasiri et al, 2005; Kepinski & Leyser, 2005). Aux/IAAs are short-lived nuclear proteins that dimerize auxin-response transcription factors (ARFs). The proteasome-mediated degradation of Aux/IAAs results in the release of ARFs, which subsequently can interact with DNA motifs, designated auxin -responsive elements, to activate or repress auxin target genes (Ulmasov et al, 1999). Similar SCF-mediated regulation has been shown for gibberellic acid, ethylene and jasmonates—jasmonate (JA) and other oxylipin derivatives (for a review, see Yu et al, 2007)—of which the jasmonates' signalling pathway is most reminiscent of that of auxin. JASMONATE ZIM-domain (JAZ) proteins belonging to the larger family of the TIFY proteins (Vanholme et al, 2007) act as repressors of JA signalling (Chini et al, 2007; Thines et al, 2007). Perception of bioactive JAs by the F-box protein CORONATINE INSENSITIVE 1 (COI1) causes degradation of JAZ proteins through the ubiquitin–proteasome pathway, which, in turn, activates the expression of JA-responsive genes.
Besides the overall similarity between both pathways, physiological and genetic studies have suggested a complex and little understood crosstalk (Kazan & Manners, 2008). For example, treating Arabidopsis plants for 48 h with methyljasmonate (MeJA) resulted in a significant increase of free IAA levels (Dombrecht et al, 2007). Conversely, ARF6 and ARF8 have been shown to promote JA production in Arabidopsis flowers (Nagpal et al, 2005). In this report, we provide new insight into this close molecular crosstalk by showing that JAZ1/TIFY10A is an auxin-responsive gene. Furthermore, the auxin-inducible expression of JAZ1/TIFY10A is controlled by ARFs and does not depend on the JA signalling pathway.
Results And Discussion
Identification of JAZ1/TIFY10A
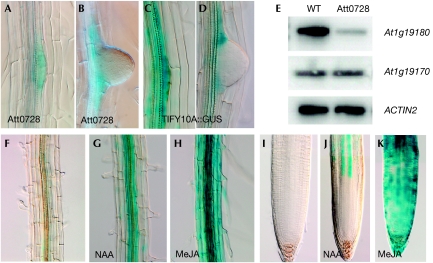
When studying the interaction between plants and plant-parasitic nematodes, we previously identified the Att0728 line by a promoter trap experiment (Barthels et al, 1997). Besides the reported β-glucuronidase (GUS) activity in the developing feeding sites of plant-parasitic nematodes (supplementary Fig S1 online; Barthels et al, 1997), GUS staining was also observed during lateral root initiation and at the base of lateral root primordia (Fig 1A,B). By using inverse PCR, we show here that the Att0728 line carries a transfer DNA (T-DNA) insertion in the promoter region of the At1g19180 gene 1.4 kb upstream from the predicted coding region (supplementary Fig S2 online). The Att0728 line showed a highly reduced At1g19180 expression level compared with that of the corresponding wild type (Fig 1E). Furthermore, a 1.4-kb At1g19180 promoter fragment fused to the GUS gene showed essentially the same GUS pattern as the Att0728 line; that is, during lateral root initiation and at the base of lateral roots (Fig 1C,D). We therefore concluded that At1g19180 was trapped and that its promoter controlled the expression of the GUS-containing T-DNA in the Att0728 line.
Figure 1.
Tissue-specific and hormone-inducible JAZ1/TIFY10A expression. (A–D) GUS expression in the Att0728 promoter trap (A,B) and TIFY10A∷GUS (C,D) lines during LR initiation (A,C) and at the base of the LR after emergence (B,D). (E) Reverse transcription–PCR gel blot showing the transcript level of At1g19180, At1g19170 and ACTIN2 in C24 WT and the homozygous Att0728 line. (F–K) TIFY10A∷GUS expression in mature root (F–H) and primary root meristem (I–K) on mock (F,I), auxin (10 μM NAA, 2 h; G,J) and jasmonate (25 μM MeJA, 2 h; H,K) treatment. GUS, β-glucuronidase; LR, lateral root; WT, wild type.
JAZ1/TIFY10A is localized in nuclear bodies
The At1g19180 gene product was initially catalogued as TIFY10A, a member of the plant-specific TIFY family (Vanholme et al, 2007). Shortly thereafter a subset of TIFY proteins was classified as JAZ proteins (Chini et al, 2007; Thines et al, 2007), leading to the double nomenclature JAZ1/TIFY10A. Previously, a nuclear localization has been shown for several TIFY proteins (Nishii et al, 2000; Chini et al, 2007; Thines et al, 2007; Yan et al, 2007), but a nuclear localization signal (NLS) had hitherto not been defined. In an effort to identify the essential region for nuclear import, we set up a deletion screen for JAZ1/TIFY10A in tobacco (Nicotiana tabacum) Bright Yellow 2 cells (Fig 2). The full-length construct as well as deletion constructs Δ1, which lack the amino-terminal domain, and Δ3, which has only the carboxy-terminal region, all showed a strong nuclear localization. However, with Δ3, green fluorescent protein (GFP) fluorescence was observed in the nucleolus, indicating that the NLS is still present but that essential nucleoplasma retention signals are missing, leading to the accumulation of the basic protein in the nucleolus (Meng et al, 2007). Deletion construct Δ2, which lacks the N-terminal and C-terminal regions, was localized both in the nucleus and in the cytoplasm, a pattern also observed when expressing free GFP. These results indicate that the C-terminal region is responsible for the nuclear localization of JAZ1/TIFY10A. This region contains a stretch of basic amino acids conserved among the JAZ proteins. Although NLSs are characterized by basic amino acids, this basic motif alone was not sufficient to function as an NLS. Neither Δ4 nor Δ5 directed GFP specifically to the nucleus (Fig 2). By using construct Δ6, we could show that five additional non-basic amino acids restored the NLS. This observation is consistent with earlier reports (Boulikas, 1994; Sekimoto et al, 2004) and reveals that the function of the basic core of an NLS is influenced by its flanking residues. Recently, by using both yeast two-hybrid and protein pull-down experiments the C-terminal region (designated as the Jas motif; Yan et al, 2007) has been shown to interact with COI1 in a JA–Ile-dependent manner (Katsir et al, 2008; Melotto et al, 2008). Together with our findings a dual role for the Jas motif can be put forward: one role in protein–protein interactions and one role in subcellular protein sorting.
Figure 2.
Subcellular localization of JAZ1/TIFY10A–GFP in tobacco BY2 cells. (A) Schematic representation of JAZ1/TIFY10A protein regions fused with GFP (amino acids are indicated in parentheses). (B) GFP signal in BY2 cells (inset, free GFP). (C) Close-up of nuclear GFP localizations. Arrows indicate nuclear bodies. Scale bars, 10 μm. BY, Bright Yellow; GFP, green fluorescent protein; Jas, jasmonate-associated; NTD, amino-terminal domain.
In planta expression of truncated versions of JAZ1/TIFY10A, JAZ3/TIFY6B and JAZ10/TIFY9 lacking the Jas motif showed JA insensitivity, phenocopying coi1 mutants (Chini et al, 2007; Thines et al, 2007; Yan et al, 2007). These results indicate a role for the N–terminus and/or tify domain in regulating the expression of JA-responsive genes. An interesting observation from our deletion screen was that the full-length JAZ1/TIFY10A protein and construct Δ1 were localized in the nucleus as discrete speckles, often referred to as nuclear bodies (Fig 2). As the speckled pattern was not observed with constructs lacking the tify domain, it is tempting to speculate that this domain is involved in this patterning, suggesting that it can be designated as a nucleoplasma retention signal. Recently, Chung & Howe (2009) showed that the tify motif mediates homomeric and heteromeric interactions between JAZ proteins, which is in agreement with the model of Meng et al (2007) that suggests that protein–protein interactions are necessary for the localization of proteins to the nucleoplasm.
Knocking down TIFY10A enhances sensitivity to MeJA
Recently, JAZ1/TIFY10A, together with its closest homologues, has been shown to act as a repressor of JA signalling, and constitutive expression of a truncated JAZ1 derivative creating dominant-negative mutants has been shown to promote JA insensitivity (Thines et al, 2007). The lack of knockout mutants for JAZ1/TIFY10A prevented the study of the endogenous full-length protein. We used the Att0728-tagged line to tackle this problem. In addition, artificial microRNA (amiRNA) lines were constructed (supplementary Fig S3 online). The JAZ1/TIFY10A knockdown lines were grown on several MeJA concentrations and primary root growth and lateral root density were analysed. These characteristics have been extensively used to screen for JA-insensitive mutants (Chini et al, 2007; Thines et al, 2007; Yan et al, 2007). Both Att0728 and amiRNATIFY10A plants showed a stronger response to MeJA compared with their corresponding controls: primary root growth was more inhibited (Fig 3A), whereas the number of lateral roots increased significantly (Fig 3B). In addition, the level of transcripts of three molecular JA markers was higher in JAZ1/TIFY10 knockdown lines (Fig 3C). These results indeed show that JAZ1/TIFY10A is one of the JAZ repressors involved in JA signalling.
Figure 3.
JAZ1/TIFY10A mutant phenotypes. (A) Root length of TIFY10 knockdown and knockout plants compared with their respective controls grown for 9 days on MeJA-containing medium. (B) Number of lateral roots per centimetre root length of Col-0 WT and amiRNATIFY10A seedlings grown for 9 days on NAA and MeJA-containing medium compared with mock–treated plants. (C) qRT-PCR analysis of the expression of AOS, LOX3 and MYC2 in Col-0 WT and amiRNATIFY10A plants. Averaged transcript levels are plotted relative to the WT control. Error bars indicate standard error; asterisks in (A,B) mark significant differences as compared with the respective controls (*P<0.05; **P<0.01). MeJA, methyljasmonate; NAA, naphthalene acetic acid; qRT, quantitative real time; WT, wild type.
The observation that full knockout lines of JAZ2/TIFY10B, JAZ5/TIFY11A, JAZ7/TIFY5B and JAZ9/TIFY7 did not alter JA signalling suggested functional redundancy among the JAZ proteins (Thines et al, 2007). To investigate whether JAZ1/TIFY10A would be functionally redundant with its closest homologue JAZ2/TIFY10B, we generated double mutants between Att0728 and a tify10b full knockout line (supplementary Fig S3 online). Surprisingly, no extra response could be seen when comparing Att0728 and tify10b × Att0728 (Fig 3A), indicating that TIFY10A and TIFY10B are not redundant and that further sensitivity to MeJA is probably countered by other JAZ proteins.
JAZ1/TIFY10A interconnects JA and auxin signalling
A detailed analysis of the TIFY10A promoter showed the presence of several AREs (supplementary Fig S2 online). Owing to the presence of these elements, transcription can be activated within minutes on application of auxin (Katagiri et al, 1989; Singh et al, 1990; Ulmasov et al, 1995). Furthermore, data from AtGenExpress (Goda et al, 2008), in which the transcriptional response of seedlings to seven phytohormones was recorded, showed an induced JAZ1/TIFY10A expression on a 30-min auxin treatment and suggested that JAZ1/TIFY10A is the only early auxin-inducible JAZ gene (supplementary Fig S4 online). Auxin-inducible expression of JAZ1 was also observed by Vanneste et al (2005; supplementary Fig S5 online), but not by Nemhauser et al (2004) and Peng et al (2009). As auxin induction of JAZ1 was not consistent in various microarray experiments, expression of the JAZ1/TIFY10A gene was tested for its response to changes in auxin level (by using 10 μM naphthalene acetic acid (NAA)) in a time-course experiment using quantitative real-time polymerase chain reaction (qRT–PCR). This analysis indeed showed a very early and transient upregulation of the JAZ1/TIFY10A transcript (Fig 4A). To investigate whether lower, more physiologically relevant auxin concentrations could regulate JAZ1 expression, we carried out an auxin dose–response study. Upregulation of JAZ1 in seedlings treated for 30 min with NAA could be observed from a concentration of 1 μM onwards, similar to the behaviour of Aux/IAA genes (Fig 4B). The auxin-induced promoter activity was confirmed using the TIFY10A∷GUS lines. In contrast to non-treated TIFY10A∷GUS lines, strong GUS staining could be observed in the vascular cylinder (Fig 1F,G,I,J; supplementary Fig S6 online). Interestingly, the auxin response differed from the MeJA response. By using the latter hormone, upregulation of JAZ1/TIFY10A could be observed in almost all root cells (Fig 1H,K).
Figure 4.
JAZ1/TIFY10A expression is induced by auxin. (A) Time-course experiment of JAZ1/TIFY10A expression in auxin-treated seedlings (10 μM NAA). (B) JAZ1/TIFY10A, Aux/IAA1, Aux/IAA5 and Aux/IAA19 expression in WT seedlings treated for 30 min with increasing auxin concentrations. (C) Expression of JAZ1/TIFY10A in WT, arf6, arf8 and arf6arf8 backgrounds on mock, NAA (10 μM 30 min), MeJA (25 μM 30 min) and NAA+MeJA treatments (*P<0.01; **P<0.001). (D) JAZ1/TIFY10A expression in mock and auxin-treated WT, HS∷axr3 and HS∷shy2 backgrounds. MeJA, methyljasmonate; NAA, naphthalene acetic acid; WT, wild type.
Given the fast upregulation of the JAZ1/TIFY10A transcript on auxin treatment, we considered that the Aux/IAA–ARF signalling pathway might regulate JAZ1/TIFY10A expression. Previously, ARF6 and ARF8 were reported to promote JA production in flowers (Nagpal et al, 2005). Therefore, we checked the expression of JAZ1/TIFY10A in arf6 and arf8 single mutants as well as in the arf6arf8 sesquimutant (arf6/arf6 arf8/ARF8). In both arf6 and arf8, the auxin-induced expression was highly reduced and, interestingly, no JAZ1/TIFY10A auxin response could be observed in the sesquimutant (Fig 4C), suggesting that ARF6 and ARF8 both mediate the auxin-induced expression of JAZ1/TIFY10A. In none of the arf mutant backgrounds was the MeJA-induced expression of JAZ1/TIFY10A affected (Fig 4C). In addition, expression of JAZ1/TIFY10A was analysed in the aux/iaa mutants HS∷axr3/iaa17 and HS∷shy2/iaa3 on auxin treatment. In both mutant backgrounds, the auxin-induced JAZ1/TIFY10A expression was abolished on heat-shock treatment (Fig 4D).
To exclude the possibility that the auxin response of JAZ1/TIFY10A is because of a stimulation of MeJA signalling, we generated transgenic plants producing a JAZ1/TIFY10A–GFP fusion protein driven by the 35S promoter and checked its stability in the presence of NAA and MeJA. As expected, the GFP signal rapidly disappeared when transferred to MeJA-containing medium; however, when seedlings were treated with NAA, no degradation of JAZ1/TIFY10A–GFP could be observed (Fig 5A). These results indicate that auxin does not influence the stability of JAZ1/TIFY10A and suggest that auxin and JA act differently with regard to the transcriptional upregulation of JAZ1/TIFY10A. Next, we checked the expression of JAZ1/TIFY10A in the MeJA signalling mutants coi1-16 and myc2 on treatment with NAA and MeJA. In the coi1-16 background, the MeJA response was highly reduced compared with wild type, whereas the auxin-dependent induction of JAZ1/TIFY10A was unaffected (Fig 5B). Similar results were obtained using the myc2 mutant: a reduced MeJA induction and unaffected auxin-induced expression. These findings strongly suggest that auxin-dictated JAZ1/TIFY10A expression is independent of the MeJA signalling pathway. To investigate whether the auxin-inducible JAZ1/TIFY10A expression is important physiologically, we analysed lateral root outgrowth in response to NAA. In two independent experiments, the number of emerged lateral roots of the amiRNATIFY10A seedlings was found to be slightly but significantly increased relative to wild type in the presence of 0.1 μM NAA (Fig 3B). JAZ1/TIFY10A knockdown mutants thus show an increased lateral root formation on treatment with MeJA as with auxin. JAZ1/TIFY10 is known to bind to and inhibit the activity of MYC2, which, interestingly, was reported as a positive regulator of lateral root formation (Yadav et al, 2005). Together with the observed lateral root-specific JAZ1/TIFY10A expression pattern, these results reveal that a modulation of JAZ1/TIFY10A expression levels might be used by both MeJA and auxin to control lateral root formation.
Figure 5.
Auxin-induced expression of JAZ1/TIFY10A is independent of MeJA signalling. (A) Visualization of JAZ1/TIFY10A–GFP on MeJA (25 μM) and NAA (10 μM) treatment. (B) Expression of JAZ1/TIFY10A in Col-0 WT, coi1-16 and myc2 backgrounds on mock, NAA (10 μM), MeJA (25 μM) and NAA+MeJA treatments (*P<0.01; **P<0.001). MeJA, methyljasmonate; NAA, naphthalene acetic acid; WT, wild type.
The results presented here suggest that the auxin and JA signalling pathways are interconnected through JAZ1/TIFY10A (supplementary Fig S7 online). Furthermore, our results can be used to explain some of the observations of Chung et al (2008), in which the expression of three JAZ genes was determined in cycloheximide-treated coi1 mutants. The expression of JAZ7 and JAZ10 could not be induced by cycloheximide in coi1, showing that SCFCOI also contributes to JAZ turnover in the absence of exogenous MeJA. By contrast, the transcript level of JAZ1/TIFY10A was upregulated by cycloheximide (Chung et al, 2008). This observation can now be explained by the action of ARF proteins, which are derepressed in the presence of cycloheximide and further supports the fact that auxin-induced expression of JAZ1/TIFY10A is independent of JA signalling and seemingly specific to JAZ1/TIFY10A.
Methods
Hormone treatments, RNA extraction and qRT–PCR analysis. For the NAA time-course experiment, 10-day-old seedlings were transferred to mock MS medium or MS medium supplemented with 10 μM NAA (Sigma-Aldrich, St Louis, MO, USA) for 0, 15, 30, 60, 120, 240 and 360 min. For the NAA dose response, plants were incubated for 30 min on medium supplemented with 0, 0.01, 0.1, 1 or 10 μM NAA. For all other hormone treatments, 10-day-old plants were transferred to a medium containing 25 μM MeJA and 10 μM NAA, or 10 μM NAA and 25 μM MeJA, all for 30 min. For heat-shock experiments, 10-day-old plants were incubated for 2 h at 37°C. Afterwards, plants were incubated for 30 min at 21°C. Plants were then incubated for 30 min on auxin-containing (NAA 10 μM) or mock medium. After all treatments, plants were ground and total RNA was isolated with TriZol (Invitrogen, Merelbeke, Belgium) according to the manufacturer's instructions. Poly(dT) complementary DNA was prepared from 2 μg total RNA with Superscript III reverse transcriptase (Invitrogen) and quantified on a LightCycler 480 apparatus (Roche Diagnostics, Mannheim, Germany) with the SYBR Green I Master kit (Roche Diagnostics) according to the manufacturer's instructions. Target quantifications were carried out with specific primer pairs designed with the Beacon Designer 4.0 (Premier Biosoft International, Palo Alto, CA, USA). All individual reactions were done in triplicate. Data were analysed with qBase (Hellemans et al, 2007). Expression levels were normalized to those of EEF1α4 and CDKA, which showed no clear systematic changes in Ct value. The primers used to quantify gene expression levels are listed in supplementary Table S1 online. A t-test was carried out manually using the results obtained from qBase.
Root growth analysis. To measure root growth, seedlings were grown vertically on the media mentioned. Nine days after germination, the number of lateral roots was counted using a binocular microscope and the plates were scanned on a colour copier CLC-iR C3200 (Canon, Tokyo, Japan). Root growth was measured using the software programme image j (http://rsb.info.nih.gov/ij/). The data were statistically analysed in SPSS (Chicago, IL, USA; version 16.0) using the Levene test for the analysis of homogeneity of variance and a one-way ANOVA (using Tukey's test with a significance level P<0.05 or P<0.01) for comparison of means.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by grants from Fonds Wetenschappelijk Onderzoek—Vlaanderen (G.0031.08) and Ghent University (Bijzonder Onderzoeksfonds postdoctoral fellowship to B.V.). L.P. is a Research Fellow of the Fonds Wetenschappelijk Onderzoek Vlaanderen.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barthels N et al. (1997) Regulatory sequences of Arabidopsis drive reporter gene expression in nematode feeding structures. Plant Cell 9: 2119–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T (1994) Putative nuclear localization signals (NLS) in protein transcription factors. J Cell Biochem 55: 32–58 [DOI] [PubMed] [Google Scholar]
- Chini A et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chung HS, Howe GA (2009) A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Koo AJ, Gao X, Jayanty S, Thines B, Jones AD, Howe GA (2008) Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol 146: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dombrecht B et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H et al. (2008) The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Lam E, Chua N-H (1989) Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature 340: 727–730 [DOI] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM (2008) Jasmonate signaling: toward an integrated view. Plant Physiol 146: 1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Meng L, Zhu Q, Tsai RY (2007) Nucleolar trafficking of nucleostemin family proteins: common versus protein-specific mechanisms. Mol Cell Biol 27: 8670–8682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M et al. (2008) A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J 55: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P et al. (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishii A, Takemura M, Fujita H, Shikata M, Yokota A, Kohchi T (2000) Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during the reproductive phase in Arabidopsis thaliana. Biosci Biotechnol Biochem 64: 1402–1409 [DOI] [PubMed] [Google Scholar]
- Peng ZY et al. (2009) Arabidopsis hormone database: a comprehensive genetic and phenotypic information database for plant hormone research in Arabidopsis. Nucleic Acids Res 37: D975–D982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A et al. (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Sekimoto T, Fukumoto M, Yoneda Y (2004) 14-3-3 suppresses the nuclear localization of threonine 157-phosphorylated p27(Kip1). EMBO J 23: 1934–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Dennis ES, Ellis JG, Llewellyn DJ, Tokuhisa JG, Wahleithner JA, Peacock WJ (1990) OCSBF-1, a maize ocs enhancer binding factor: isolation and expression during development. Plant Cell 2: 891–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu GH, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCFCO11 complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ (1995) Composite structure of auxin response elements. Plant Cell 7: 1611–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1999) Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA 96: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G (2007) The tify family previously known as ZIM. Trends Plant Sci 12: 239–244 [DOI] [PubMed] [Google Scholar]
- Vanneste S et al. (2005) Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17: 3035–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S (2005) A basic helix–loop–helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17: 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chetelat A, Reymond P, Pagni M, Dubugnon L, Farmer EE (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Wu J, Xu N, Peng M (2007) Roles of F-box proteins in plant hormone responses. Acta Biochim Biophys Sin (Shanghai) 39: 915–922 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information