Abstract
Post-translational histone modifications have essential roles in controlling nuclear processes; however, the specific mechanisms regulating these modifications and their combinatorial activities remain elusive. Cyclin-dependent kinase 9 (CDK9) regulates gene expression by phosphorylating transcriptional regulatory proteins, including the RNA polymerase II carboxy-terminal domain. Here, we show that CDK9 activity is essential for maintaining global and gene-associated levels of histone H2B monoubiquitination (H2Bub1). Furthermore, CDK9 activity and H2Bub1 help to maintain correct replication-dependent histone messenger RNA (mRNA) 3′-end processing. CDK9 knockdown consistently resulted in inefficient recognition of the correct mRNA 3′-end cleavage site and led to increased read-through of RNA polymerase II to an alternative downstream polyadenylation signal. Thus, CDK9 acts to integrate phosphorylation during transcription with chromatin modifications to control co-transcriptional histone mRNA processing.
Keywords: cyclin-dependent kinase, histone, mRNA processing, monoubiquitination, RNA polymerase II C-terminal domain
Introduction
Covalent histone modifications regulate the chromatin environment and are essential for correct transcriptional regulation (Berger, 2007). Histone H3 Lys 4 trimethylation (H3K4me3) is associated with active gene promoters, whereas histone H2B monoubiquitination (H2Bub1) is found in the transcribed regions of active genes (Berger, 2007; Weake & Workman, 2008). Genome-wide analyses of histone modifications have shown that the regulation of gene expression is more complex than initially thought, and revealed extensive cross talk between chromatin modifications and the transcriptional machinery (Egloff & Murphy, 2008; Suganuma & Workman, 2008).
H2Bub1 is carried out by the E3 ubiquitin ligase complex RNF (ring finger protein) 20/40 in humans (Kim et al, 2005; Zhu et al, 2005). RNF20 has tumour suppressor activity as loss of its expression results in a tumorigenic and more invasive phenotype (Shema et al, 2008). Surprisingly, although H2Bub1 is present on most active genes in humans, only a subset of the genes actually requires H2Bub1 for expression (Minsky et al, 2008; Shema et al, 2008). Thus, further nuclear functions might require H2Bub1 for their activity.
The RNA polymerase II (RNAPII) carboxyl-terminal domain (CTD) contains a conserved heptapeptide repeat that occurs 52 times in humans (Egloff & Murphy, 2008). Ser 5 of the repeat is phosphorylated at the promoter, and is associated with messenger RNA (mRNA) capping and the onset of transcription elongation. Ser 2 (P-Ser 2) is phosphorylated by cyclin-dependent kinase 9 (CDK9) in the transcribed region and is required for post-initiation transcription events such as elongation, splicing and mRNA processing. CDK9 further stimulates elongation by phosphorylating negative elongation factor-E (NELF-E) and suppressor of Ty homologue 5 (SUPT5H). Similar to P-Ser 2, H2Bub1 is enriched in the transcribed region of actively transcribed genes (Minsky et al, 2008). Although P-Ser 2 and H2Bub1 are disconnected in yeast, a link in humans has not been investigated (Xiao et al, 2005; Wood et al, 2007).
Unlike most mRNAs, replication-dependent histone transcripts end in a conserved stem–loop recognized by the stem–loop binding protein (SLBP) and cleaved by the U7 small nuclear ribonucleoprotein (snRNP; Marzluff et al, 2008). Loss of correct histone mRNA processing by the depletion of SLBP or U7 increases the formation of polyadenylated histone transcripts (Sullivan et al, 2001; Wagner et al, 2007). Replication-dependent histone genes have been identified as targets of both H2Bub1 (RNF20) and the CDK9 target NELF-E (Narita et al, 2007; Shema et al, 2008). Although CDK9 activity has been linked to the transcription and processing of polyadenylated mRNAs, its role in histone mRNA 3′-end processing remains unknown. Furthermore, the contribution of specific histone modifications to mRNA processing is also unknown. Here, we report that CDK9 regulates H2Bub1 through a CTD-dependent mechanism involving the RNA polymerase II associated factor (PAF)–RNF20/40 complex. Furthermore, CDK9 and H2Bub1 are required for correct replication-dependent histone mRNA 3′-end processing, and together reveal a new role for CTD phosphorylation and chromatin modifications in controlling this process.
Results And Discussion
Transcription is not sufficient for maintaining H2Bub1
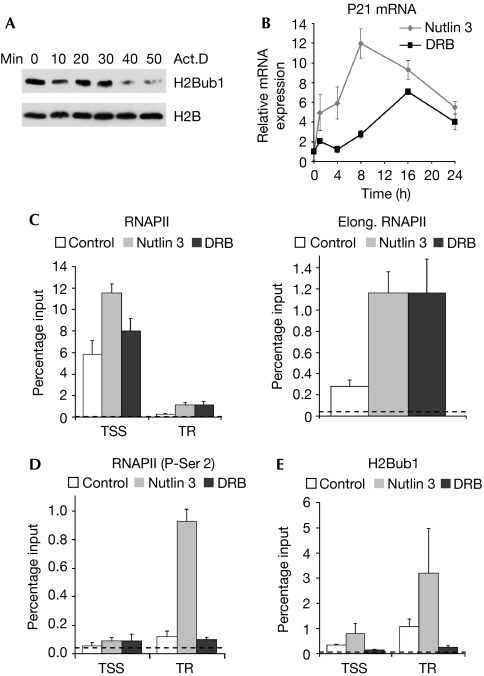
As H2Bub1 is associated with the transcribed regions of active genes, we tested whether transcription is essential for maintaining global levels of H2Bub1. Consistent with previous studies (Davie & Murphy, 1990; Minsky et al, 2008), actinomycin D rapidly decreases global levels of H2Bub1, confirming that active transcription is necessary for maintaining H2Bub1 and showing the rapid dynamics of H2B deubiquitination (Fig 1A). To test whether active transcription is sufficient for H2Bub1 accumulation, we exploited the p53 response gene p21, the transcription of which is uniquely induced by 5,6-dichloro-β-D-ribofuranosyl benzimidazole (DRB) despite a loss of P-Ser 2 (Gomes et al, 2006). p21 mRNA levels significantly increased following treatment with either DRB or the specific mouse double minute 2 (MDM2) inhibitor nutlin 3 (Fig 1B), and correlated with an increase in elongation of RNAPII on the p21 gene (Fig 1C), as detected by chromatin immunoprecipitation (ChIP). As reported previously, the levels of RNAPII at the transcriptional start site (TSS) were only modestly affected by either treatment, whereas both P-Ser 2 (Gomes et al, 2006) and H2Bub1 (Minsky et al, 2008) specifically increased in the transcribed region (Fig 1D,E) after nutlin 3 treatment. In contrast to total RNAPII, P-Ser 2 and H2Bub1 levels did not increase in the transcribed region after DRB treatment (Fig 1D,E).
Figure 1.
H2Bub1 depends on CDK9 activity rather than transcriptional activation per se. (A) H1299 cells were treated with actinomycin D for the indicated times to inhibit transcription, and protein lysates were analysed by Western blot for H2Bub1 and H2B protein levels. (B) Nutlin 3 and DRB both increase levels of p21 mRNA. U-2 OS cells were treated with 8 μM nutlin 3 or 50 μM DRB for the indicated times before RT–PCR analyses. Transcript levels were normalized to mitochondrial 16S ribosomal RNA and expressed relative to the control treatment; mean values+s.d., n=3. (C–E) P-Ser 2, rather than elongation by RNAPII, is essential for maintaining gene-associated H2Bub1. U-2 OS cells were treated with nutlin 3 or DRB for 8 h and analysed by ChIP using antibodies against (C) total RNAPII, (D) P-Ser 2 or (E) H2Bub1. qChIP values were normalized to their respective DNA inputs and expressed as DNA recovery (percentage input); mean values+s.d., n=3. The experimental background is shown as a dotted line. Act.D, actinomycin D; CDK9, cyclin-dependent kinase 9; ChIP, chromatin immunoprecipitation; DRB, 5,6-dichloro-β-D ribofuranosyl benzimidazole; H2Bub1, histone H2B monoubiquitination; mRNA, messenger RNA; qChIP, quantitative chromatin immunoprecipitation; RNAPII, RNA polymerase II; RT–PCR, reverse transcription PCR; TR, transcribed region; TSS, transcriptional start site.
CDK9 activity maintains global H2Bub1 levels
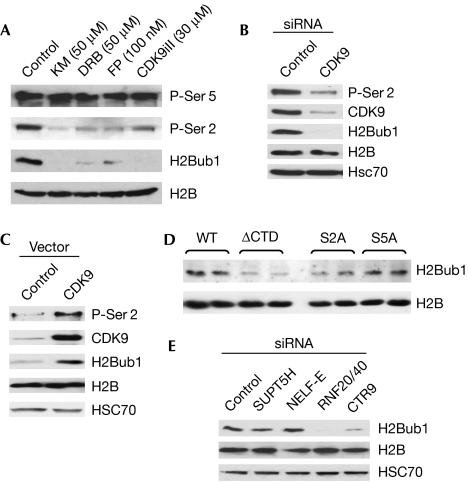
As H2Bub1 and P-Ser 2 are present primarily in the transcribed regions of active genes, we hypothesized that CDK9 is essential for H2Bub1. Therefore, we treated cells with four chemical CDK9 inhibitors and analysed total levels of H2Bub1. Each significantly decreased both H2Bub1 and P-Ser 2 (Fig 2A). These effects were dose-dependent (supplementary Fig S1A online; data not shown) and specific, as the levels of H2B and P-Ser 5 were unaltered (Fig 2A); RNF20/40 protein levels were also unaffected (supplementary Fig S1B online).
Figure 2.
CDK9 activity is essential for maintaining global levels of H2Bub1. (A) H1299 cells were treated with the specific CDK9 inhibitors KM, DRB, FP or CDK9 inhibitor II (CDK9iII) at the indicated concentrations for 4 h and analysed by Western blot analysis. (B,C) H1299 cells were transfected with (B) control or CDK9 siRNAs or (C) expression plasmids and analysed by Western blot with the indicated antibodies. (D) The CTD and Ser 2 are necessary for maintaining H2Bub1 levels. HEK293 cells were transfected with α-amanitin-resistant RNAPII large subunit expression constructs containing wild-type (WT), deletion or mutant CTD. (E) Regulation of H2Bub1 by SUPT5H, NELF-E, RNF20/40 and PAF1. H1299 cells were transfected and analysed by Western blot using the indicated siRNAs and antibodies. H2B and HSC 70 are shown as loading controls. CDK9, cyclin-dependent kinase 9; CTD, carboxyl-terminal domain; DRB, 5,6-dichloro-β-D-ribofuranosyl benzimidazole; FP, flavopiridol; H2Bub1, histone H2B monoubiquitination; HEK, human embryonic kidney; HSC70, heat shock cognate protein 70; KM, KM05283; NELF-E, negative elongation factor E; PAF, polymerase II associated factor; RNAPII, RNA polymerase II; RNF, ring finger protein; siRNA, small interfering RNA; SUPT5H, suppressor of Ty 5 homologue.
To verify further the role of CDK9, we transfected cells with specific small interfering RNAs (siRNAs) against CDK9. These decreased the level of both P-Ser 2 and H2Bub1 (Fig 2B; supplementary Fig S1C online). This effect was rescued by re-expressing the mouse CDK9 (supplementary Fig S1D online), and the overexpression of CDK9 alone increased the level of both P-Ser2 and H2Bub1 (Fig 2C).
To determine whether CTD phosphorylation is required for H2Bub1, we transfected cells with α-amanitin-resistant RNAPII constructs with wild-type CTD, no CTD or CTD with specific mutations at Ser 2 or Ser 5. Both an intact CTD and Ser 2, but not Ser 5, were essential for H2Bub1 (Fig 2D; supplementary Fig S1E online). Knockdown of SUPT5H or NELF-E had little effect on global levels of H2Bub1 (Fig 2E). Thus, CDK9 affects H2Bub1 primarily in a CTD-dependent manner. Consistent with a previous report (Zhu et al, 2005), we also observed a decrease in H2Bub1 after knockdown of the PAF component CTR9 (Fig 2E).
RNF40 is essential for maintaining global H2Bub1 levels
Conflicting reports have left the role of RNF40 in H2Bub1 unclear (Kim et al, 2005; Zhu et al, 2005; Weake & Workman, 2008). Thus, we tested whether both RNF20 and RNF40 regulated H2Bub1. RNF40 knockdown decreased H2Bub1, and this effect was rescued by overexpression of mouse Rnf40 (supplementary Fig S1F online). Similarly, the knockdown of either RNF20 or RNF40 using multiple siRNAs decreased global levels of H2Bub1 (supplementary Fig S1G,H online).
CDK9 and RNF20/40 control histone mRNA processing
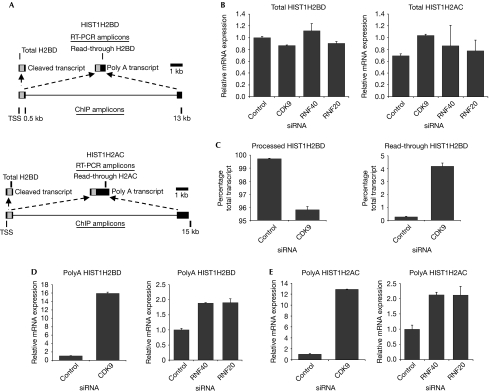
The CDK9 dependence of H2Bub1 suggested that these molecules might influence the same nuclear processes. As histone genes are targets of both H2Bub1 (RNF20) and NELF-E (Narita et al, 2007; Shema et al, 2008), we tested whether CDK9 or RNF20/40 controls replication-dependent histone gene transcription. However, none of these molecules significantly affected the mRNA levels of HIST1H2BD, HIST1H2AC or HIST2H2AA (Fig 3B; supplementary Fig S2A online).
Figure 3.
CDK9 and H2Bub1 direct replication-dependent histone messenger RNA 3′ end processing but not transcription. (A) Schematic representation of the human HIST1H2BD and HIST1H2AC genes showing both normally processed and polyadenylated transcripts resulting from the splicing of a longer transcript. (B) Total HIST1H2BD and HIST1H2AC expression is not significantly affected by CDK9, RNF20 or RNF40 knockdown. Random-primed cDNAs were analysed for total HIST1H2BD or HIST1H2AC expression after transfection of HEK293 cells with control, CDK9, RNF20 or RNF40 siRNAs. The expression of total histone transcripts was normalized to a control gene, 36B4, and is represented as relative mRNA expression; mean values+s.d., n=3. (C) CDK9 knockdown increases the formation of read-through HIST1H2BD transcripts. Total RNA from HEK293 cells transfected with control or CDK9 siRNA was analysed by random-primed qRT–PCR using specific primers for total and read-through HIST1H2BD transcripts. The expression of processed (left panel) and read-through transcripts (right panel) was normalized to 36B4 and is shown as the percentage total transcript; mean values+s.d., n=3. (D,E) CDK9 and RNF40/20 knockdown increase the formation of (D) polyadenylated HIST1H2BD and (E) HIST1H2AC transcripts. Total mRNA from cells transfected with control, CDK9, RNF40 or RNF20 siRNA was analysed by oligo-dT-primed qRT–PCR. Expression levels were normalized and expressed as in (A); mean values+s.d., n=3. CDK9, cyclin-dependent kinase 9; cDNA, complementary DNA; ChIP, chromatin immunoprecipitation; H2Bub1, histone H2B monoubiquitination; HEK, human embryonic kidney; mRNA, messenger RNA; RNF, ring finger protein; siRNAs, small interfering RNAs, qRT–PCR, quantitative reverse transcription PCR; RNF, ring finger protein; siRNAs, small interfering RNAs.
Replication-dependent histone mRNAs are not usually polyadenylated, but instead end in a conserved stem–loop structure. As knockdown of the CDK9-target NELF-E increased replication-dependent histone mRNA polyadenylation (Narita et al, 2007), we tested whether CDK9, RNF20 or RNF40 also influences replication-dependent histone mRNA 3′-end processing. The HIST1H2BD and HIST1H2AC genes produce not only mRNAs containing 3′-end stem–loop structures but also the polyadenylated forms (Fig 3A); their size—13 and 15 kb, respectively—also makes them amenable to ChIP analyses. Indeed, CDK9 knockdown increased the read-through of the HIST1H2BD gene approximately 15-fold (Fig 3C); the increased read-through transcript was also polyadenylated (Fig 3D). The magnitudes of these effects were similar to those observed after NELF-E or SLBP knockdown (Narita et al, 2007). The knockdown of RNF20, RNF40, CTR9, cyclinT1, SUPT5H or NELF-E also resulted in increased HIST1H2BD polyadenylation (Fig 3D; supplementary Fig S2B online). The modest effects of RNF20 and RNF40 knockdown probably reflect the pleiotropic effects of CDK9 knockdown on histone mRNA processing, which probably affects not only the CTD-dependent regulation of H2Bub1 but also the functions of SUPT5H and NELF-E. Similar results were obtained for the HIST1H2AC and HIST2H2AA genes (Fig 3E; supplementary Fig S2A online). Importantly, these effects were not due to changes in cell-cycle progression, as CDK9, RNF20 or RNF40 knockdown did not affect cell-cycle distribution (data not shown). These effects were specific for replication-dependent histone mRNAs as the levels of polyadenylated 36B4, β-actin and GAPDH were unaffected by the knockdowns (supplementary Fig S2C online; data not shown). Thus, CDK9 and H2Bub1 help to maintain the correct mode of 3′-end processing of replication-dependent histone mRNAs.
CDK9 regulates RNAPII read-through of genes
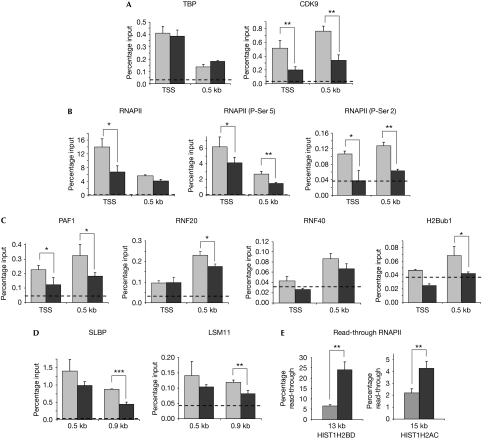
Next, we next carried out ChIP analyses to test whether CDK9 knockdown affected the recruitment of specific factors to the HIST1H2BD gene. TATA-binding protein (TBP) was recruited preferentially to the TSS and its levels were unchanged by CDK9 knockdown (Fig 4A), suggesting that the basal transcriptional apparatus was not affected by CDK9 knockdown. The knockdown of CDK9 and its presence on the HIST1H2BD gene were also verified (Fig 4A). P-Ser 2 was consistently decreased to nearly undetectable levels after CDK9 knockdown (Fig 4B). Surprisingly, the amount of total RNAPII at the TSS was decreased to about 50%, whereas the levels at the 3′-cleavage site (0.5 kb) remained unchanged (Fig 4B). In comparison to P-Ser 2, the fraction of P-Ser 5 RNAPII was only moderately affected at both positions (Fig 4B; supplementary Fig S2D online). A similar effect was observed for the recruitment of NELF-E, whereas the recruitment of cap binding protein 80 (CBP80) was unaffected by CDK9 knockdown (supplementary Fig S2E online).
Figure 4.
CDK9 activity recruits proteins involved in H2B ubiquitination and histone messenger RNA 3′-end formation, and decreases RNAPII read-through. ChIP analysis of the HIST1H2BD (or HIST1H2AC in (E)) gene in HEK293 cells after transfection with control (grey bars) or CDK9 (black bars) siRNAs using the indicated antibodies. (A) TBP is specifically enriched at the TSS and is not affected by CDK9 knockdown, whereas CDK9 is significantly enriched both at the TSS and at 0.5 kb and these levels decrease on knockdown. (B) CDK9 knockdown similarly decreases RNAPII and P-Ser 5-RNAPII levels at the TSS without affecting elongation to the 3′-end cleavage site. By contrast, P-Ser 2 is markedly decreased to near-background levels both at the TSS and at 0.5 kb. (C) The H2B-ubiquitinating complex (PAF1, RNF20 and RNF40) is enriched at 0.5 kb of the HIST1H2BD gene together with H2Bub1, and their levels decrease following CDK9 knockdown. (D) ChIP analysis of SLBP and the U7 snRNP component LSM11 shows decreased recruitment to the 3′-cleavage site. (E) CDK9 knockdown increases RNAPII read-through past the normal 3′-end cleavage site of the HIST1H2BD and HIST1H2AC genes. ChIP analysis of total RNAPII levels at the polyadenylation site (13 kb for HIST1H2BD or 15 kb for HIST1H2AC) were compared with the amount of total initiating RNAPII (at the TSS) and plotted as the percentage read-through of RNAPII. ChIP experiments are shown as percentage input as in Fig 1; mean values+s.d., n=3. Statistically significant differences are indicated: *P⩽0.05; **P⩽0.01; ***P⩽0.001. The experimental background is shown in each graph as a dotted line. CDK9, cyclin-dependent kinase 9; ChIP, chromatin immunoprecipitation; H2Bub1, histone H2B monoubiquitination; HEK, human embryonic kidney; LSM11, Sm-like protein 11; PAF, RNA polymerase II associated factor; RNAPII, RNA polymerase II; RNF, ring finger protein; SLBP, stem–loop (histone) binding protein; siRNAs, small interfering RNAs; snRNP, small nuclear ribonucleoprotein; TBP, TATA-binding protein; TSS, transcriptional start site, 0.5 kb is equal to 3′ end cleavage site, 0.9 kb is equal to 400 bp 3′ to the cleavage site, 13 kb or 15 kb is equal to polyadenylation site of the read-through transcripts for HIST1H2BD and HIST1H2AC, respectively.
Given that CDK9 was essential for global levels of H2Bub1, and as both CDK9 and H2Bub1 regulated the replication-dependent histone mRNA processing, we tested whether H2Bub1 and the PAF1/RNF20/40 complex were also present on the HIST1H2BD gene. Indeed, RNF20, RNF40 and H2Bub1 showed a preferential enrichment at the 3′-cleavage site, with very low levels at the TSS (Fig 4C). Importantly, the presence of PAF1 and all of the H2Bub1-associated factors decreased after CDK9 knockdown. This was most apparent for PAF1 and H2Bub1, as PAF1 enrichment at 0.5 kb was decreased to 50% above background levels and H2Bub1 was nearly undetectable following CDK9 knockdown.
The increased polyadenylation of replication-dependent histone mRNAs might be attributable to incorrect recognition of the 3′ stem–loop structure, which might result in read-through of RNAPII beyond the normal 3′-processing site. Thus, we carried out ChIP analyses of SLBP and the U7 snRNP component LSM11 on the HIST1H2BD gene to test whether their recruitment is affected by CDK9 knockdown. Indeed, both SLBP and LSM11 were enriched at the 3′-cleavage position, as well as 400 bp downstream of the cleavage position, and decreased following CDK9 knockdown (Fig 4D). Most notably, SLBP recruitment at 0.9 kb decreased nearly 50% following CDK9 knockdown.
The decrease in the recruitment of histone mRNA 3′-end processing factors also correlated with an increase in read-through of RNAPII beyond the normal 3′-end cleavage position on the HIST1H2BD gene (Fig 4E); similar results were obtained for HIST1H2AC (Fig 4E). This is consistent with the increase in polyadenylated transcripts detected by reverse transcription PCR (RT–PCR; Fig 3D,E) and suggests that CDK9 is important for the correct positioning of 3′-end processing proteins. This is the first report to show that the increase in the read-through and polyadenylation of replication-dependent histone gene transcripts is associated with an increased detection of RNAPII at the downstream polyadenylation site. The non-linear relationship between the increases in the read-through of RNAPII, as detected by ChIP analysis (Fig 4E), and polyadenylated mRNA levels, as detected by RT–PCR (Fig 3D,E), are common in transcriptional studies and might be due to factors such as mRNA stability, RNAPII processivity and differences in RNA processing.
Although CDK9 is known to regulate transcription-associated nuclear processes such as splicing and polyadenylation, the significance of CDK9-directed chromatin modifications in these or other nuclear processes have remained unexplored. We have shown that H2Bub1 is a CDK9-dependent histone modification that directs replication-dependent histone mRNA 3′ end processing by the SLBP–U7 snRNP complex. We propose that CDK9 regulates this, at least in part, by directing H2Bub1, which might influence the accessibility of the newly synthesized mRNA or the recruitment of 3′-end processing machinery by modifying the chromatin structure. Consistent with a role for chromatin structure in histone mRNA 3′-end processing, the histone variants H3.3 and H2Av and the histone chaperone Anti-silencing function 1 (Asf1) are necessary for correct histone mRNA 3′-end formation in Drosophila (Wagner et al, 2007). However, the presence of these histone variants on the replication-dependent histone genes and their roles in mammals remain unknown. In our model, specific CDK9-dependent phosphorylations and changes in H2Bub1 would act as markers for the correct recognition of the histone mRNA 3′-end cleavage site. The presence of these markers might be regulated by altering CTD phosphorylation or H2Bub1. In the absence of P-Ser 2 or H2Bub1, RNAPII would continue to transcribe until another termination site (that is, a downstream polyadenylation signal) is reached. Thus, we have revealed a new and unexpected role for CDK9, and a unique cross talk between CDK9 and the histone code in the regulation of histone mRNA 3′-end processing.
Speculation
In support of the hypothesis that CDK9 and/or H2Bub1 regulation of histone mRNA processing might have a role in human disease, it is interesting to note that changes in the expression of polyA+ replication-dependent histone mRNAs have been revealed by microarray analyses in various subtypes of breast cancer (Zhao et al, 2004; Abba et al, 2005) and oropharyngeal squamous-cell carcinoma (Martinez et al, 2007). Although the function of these transcripts is unclear, it is possible that they provide an advantage to rapidly dividing cells by supplying an additional source of histones outside of the S-phase owing to their cell-cycle-independent stability and translation (Kirsh et al, 1989). Future experiments will specifically address this hypothesis.
Methods
Plasmids and siRNAs. CDK9 and RNF40 were cloned into pcDNA3.1-Hygro (Invitrogen, Heidelberg, Germany). The α-amanitin-resistant RNAPII constructs were described previously (Chapman et al, 2007). The siRNAs used are listed in supplementary Table S4 online.
Cell culture and transfection. H1299, HEK293 and U-2 OS cells were grown in high-glucose DMEM containing 10% fetal bovine serum (FBS) and 1 × penicillin–streptomycin. Cells were treated with KM05283 (Thermo Fisher Scientific International, Waltham, MA, USA), DRB (Sigma, Munich, Germany), flavopiridol (Sigma), CDK9 inhibitor II (Merck KGaA, Darmstadt, Germany) or nutlin 3 (Sigma) as indicated. Transfections were carried out using Lipofectamine 2000 or Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Studies using the α-amanitin-resistant RNAPII constructs were carried out as described previously (Chapman et al, 2007).
Western blot analysis. Protein samples were analysed by Western blot analysis using the antibodies listed in supplementary Table S1 online.
ChIP and gene expression analysis. ChIP analyses were carried out essentially as described previously (Nelson et al, 2006; Johnsen et al, 2009) using the antibodies and dilutions listed in supplementary Table S1 online. RNA was harvested with TRIZOL (Invitrogen) according to the manufacturer's instructions and reverse transcribed using either oligo-dT or random nonamer primers as indicated in the respective figure legends. DNA samples (ChIP or cDNA) were analysed by using real-time PCR using the iQ SYBR Green Supermix (BioRad, Munich, Germany) with the primers listed in supplementary Tables S2 and S3 online. Experimental background was determined by carrying out a control ChIP with either a non-specific IgG and/or no primary antibody. The averages of the background signals for all primer pairs were similar and are graphically represented by the dotted line (Fig 1C,D; Fig 4A–D). The percentage of total transcripts for the processed and read-through HIST1H2BD transcripts shown in Fig 3C was determined by RT–PCR using random primed cDNA from control or CDK9-siRNA-transfected cells and dividing the signal for the ‘read-through' transcript by the total amount of transcript. The remaining fraction of total transcript was interpreted as the processed fraction. All quantitative RT–PCRs were normalized to 36B4 or MTRNR2 before additional normalization and statistical analyses.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Materials
Acknowledgments
We thank H. Handa, I. Mattaj, H. Clarke and R. Lührmann for antibodies, and T. Prenzel and S. Murphy for advice and scientific discussions.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abba MC, Hu Y, Sun H, Drake JA, Gaddis S, Baggerly K, Sahin A, Aldaz CM (2005) Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics 6: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D (2007) Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318: 1780–1782 [DOI] [PubMed] [Google Scholar]
- Davie JR, Murphy LC (1990) Level of ubiquitinated histone H2B in chromatin is coupled to ongoing transcription. Biochemistry 29: 4752–4757 [DOI] [PubMed] [Google Scholar]
- Egloff S, Murphy S (2008) Cracking the RNA polymerase II CTD code. Trends Genet 24: 280–288 [DOI] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM (2006) Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev 20: 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen SA et al. (2009) Regulation of estrogen-dependent transcription by the LIM cofactors CLIM and RLIM in breast cancer. Cancer Res 69: 128–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hake SB, Roeder RG (2005) The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell 20: 759–770 [DOI] [PubMed] [Google Scholar]
- Kirsh AL, Groudine M, Challoner PB (1989) Polyadenylation and U7 snRNP-mediated cleavage: alternative modes of RNA 3′ processing in two avian histone H1 genes. Genes Dev 3: 2172–2179 [DOI] [PubMed] [Google Scholar]
- Martinez I, Wang J, Hobson KF, Ferris RL, Khan SA (2007) Identification of differentially expressed genes in HPV-positive and HPV-negative oropharyngeal squamous cell carcinomas. Eur J Cancer 43: 415–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ (2008) Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M (2008) Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol 10: 483–488 [DOI] [PubMed] [Google Scholar]
- Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H (2007) NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol Cell 26: 349–365 [DOI] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K (2006) Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc 1: 179–185 [DOI] [PubMed] [Google Scholar]
- Shema E et al. (2008) The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev 22: 2664–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Workman JL (2008) Crosstalk among histone modifications. Cell 135: 604–607 [DOI] [PubMed] [Google Scholar]
- Sullivan E, Santiago C, Parker ED, Dominski Z, Yang X, Lanzotti DJ, Ingledue TC, Marzluff WF, Duronio RJ (2001) Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev 15: 173–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Burch BD, Godfrey AC, Salzler HR, Duronio RJ, Marzluff WF (2007) A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol Cell 28: 692–699 [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL (2008) Histone ubiquitination: triggering gene activity. Mol Cell 29: 653–663 [DOI] [PubMed] [Google Scholar]
- Wood A et al. (2007) Ctk complex-mediated regulation of histone methylation by COMPASS. Mol Cell Biol 27: 709–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD (2005) Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol 25: 637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H et al. (2004) Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell 15: 2523–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D (2005) Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell 20: 601–611 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials