Abstract
In recent years, electron microscopy (EM) has allowed the generation of three-dimensional structure maps of several spliceosomal complexes. However, owing to their limited resolution, little is known at present about the location of the pre-mRNA, the spliceosomal small nuclear ribonucleoprotein or the spliceosome's active site within these structures. In this work, we used EM to localise the intron and the 5′ and 3′ exons of a model pre-mRNA, as well as the U2-associated protein SF3b155, in pre-catalytic spliceosomes (i.e. B complexes) by labelling them with an antibody that bears colloidal gold. Our data reveal that the intron and both exons, together with SF3b155, are located in specific regions of the head domain of the B complex. These results represent an important first step towards identifying functional sites in the spliceosome. The gold-labelling method adopted here can be applied to other spliceosomal complexes and may thus contribute significantly to our overall understanding of the pre-mRNA splicing process.
Keywords: electron microscopy, localisation, pre-mRNA, SF3b155, spliceosome
Introduction
The task of the spliceosome is to excise the non-translated regions (introns) of a pre-messenger RNA and ligate its exons to form mRNA. To achieve this, the spliceosome must recognise the ends of the intron and subsequently juxtapose its chemically reactive sites—the 5′ splice site (SS), branch site (BS) and 3′ SS—in a manner leading to catalysis of the splicing reaction (Moore and Sharp, 1993). This task requires great precision, as an incorrectly spliced mRNA is not only of no use for the cell, but can even be deleterious. Introns vary enormously in length, and many can be spliced in one of several possible alternative patterns, generating unique mRNAs and ultimately different translation products. Thus, the spliceosome plays a major part in expanding the diversity of the cell's protein content (Graveley, 2001).
The composition and conformation of the spliceosome vary substantially during its work cycle, making studies of its ultrastructure especially challenging. Spliceosomes form anew in a stepwise manner on each intron (Will and Lührmann, 2006). Spliceosome assembly starts with the binding of the small nuclear ribonucleoproteins (snRNPs) U1 and U2 to the 5′ SS and the BS, respectively, yielding the prespliceosome (A complex). The U4/U6.U5 tri-snRNP—in which the U4 and U6 snRNAs are base-paired with one another—then joins the prespliceosome, generating the B complex. The latter has no catalytic centre and must be activated to a catalytically competent state. Activation involves major rearrangements including destabilisation or loss of the U1 and U4 snRNPs and the formation of an intricate network of interactions among U2, U6 and the pre-mRNA, which together make up part of the catalytic core of the spliceosome (Nilsen, 2007). The first catalytic step of splicing, during which the 5′ SS is cleaved and a lariat intron intermediate is formed, leads to the spliceosomal C complex. This complex then catalyses the second step, yielding the excised intron and ligated 5′ and 3′ exons. Recent proteomic studies of purified spliceosomes at defined stages of assembly and function have shown that numerous proteins join and leave the spliceosome throughout its life cycle (Hartmuth et al, 2002; Jurica et al, 2002; Makarov et al, 2002; Makarova et al, 2004; Deckert et al, 2006; Bessonov et al, 2008).
Although the molecular structures of many of the spliceosome's individual proteins or small protein subcomplexes have been determined at high resolution by X-ray crystallography or NMR spectroscopy, the method of choice for elucidating the structure of spliceosomal complexes is electron microscopy (EM; Stark and Lührmann, 2006). In recent years, three-dimensional (3D) structure maps of spliceosomal A (Behzadnia et al, 2007) and C (Jurica et al, 2004) complexes assembled in vitro, as well as of B complexes treated with heparin that lack the U1 snRNP (Boehringer et al, 2004), have been obtained through EM analyses, yielding first insights into the overall size and shape of these complexes. However, the resolution of the structural data presently available for these spliceosomal complexes (∼3 nm) is too low to allow a functional interpretation of the observed structure. At present it is not possible to conclude with confidence where (i) the major building blocks of the spliceosome (i.e. the snRNPs), (ii) the pre-mRNA or (iii) the active site are located within these 3D maps.
Human and drosophila B complexes were recently isolated under native conditions and contained all of the spliceosomal snRNPs (U1, U2 and U4/U6.U5; Deckert et al, 2006; Herold et al, 2009). Two-dimensional EM analyses of these complexes revealed that they have an overall rhombic shape ∼40 nm long that includes a ‘head' and a ‘body' region, each of which contains characteristic features (Deckert et al, 2006; see below). The B complex represents the fully assembled spliceosome and is the substrate for the major remodelling events that lead to catalytic activation of the splicing machinery. Within the B complex, the pre-mRNA is bound in what is believed to be a functionally committed manner—that is, the intron to be excised has been selected among all possible alternatives, and its 5′ and 3′ ends are in place. This would be expected from a functional standpoint, as one would anticipate that in the pre-catalytic B complex the reaction partners for the activation of the spliceosome—that is, the 5′ SS, the BS, the U2 snRNP and the functionally relevant regions of U6 and U5 snRNA (Nilsen, 1998)—should be in reasonably close proximity to one another, to facilitate the rearrangements required for catalytic activation of the spliceosome. Indeed, biochemical studies, especially site-directed RNA-probing with hydroxyl radicals, have shown that the reactive nucleotides of the intron are close to one another already in the A complex (Kent and MacMillan, 2002) and that the U1 and U2 snRNPs are likewise in proximity to one another (Dönmez et al, 2007). However, it remains to be determined where they and other spliceosomal components are located in the overall structure of the spliceosomal B complex.
Here, we have addressed these questions by locating the 5′ and 3′ exons and the intron of the pre-mRNA, as well as the U2-associated protein SF3b155, at the surface of affinity-purified, human B complexes. This was accomplished by labelling them specifically with antibodies, either directly or, in the case of RNA, indirectly by insertion of an MS2 stem-loop and subsequent binding of an MS2–MBP fusion protein, and examining them under the electron microscope. To facilitate visualisation of the bound antibody, the latter was additionally labelled with colloidal gold coated with protein A. Our data reveal that the intron and both exons are located in the head domain of the B complex, together with the U2-associated protein SF3b155. On the basis of our results, a 2D model showing the putative path of the pre-mRNA within the head of the B complex could be generated. These results represent a first step towards mapping functional sites within the spliceosome.
Results
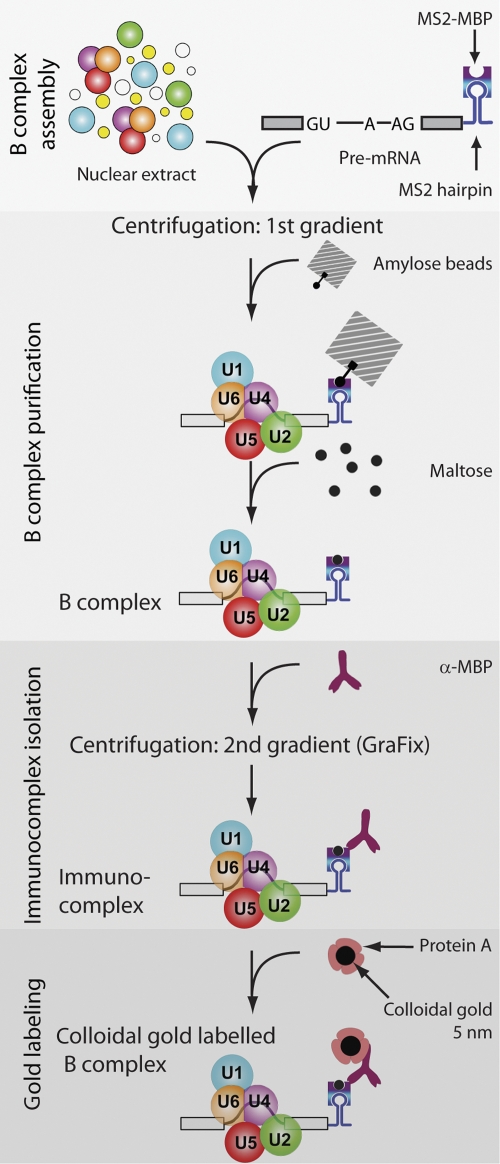
To map various regions of the pre-mRNA within the B complex, adenovirus-derived MINX pre-mRNA (Zillmann et al, 1988) was first modified by insertion of two or three MS2 hairpins at either its 5′ or 3′ end, or within its intron. For labelling the 5′end, the 5′ exon was shortened to 36 nucleotides (Supplementary Figure S1A). All three pre-mRNAs undergo splicing in vitro with similar kinetics (data not shown). After incubating with MS2–MBP (a fusion of the MS2 and maltose-binding proteins (MBPs); Zhou et al, 2002b), human spliceosomal B complexes were allowed to form on the MINX pre-mRNA by incubating it under splicing conditions in HeLa nuclear extract (Figure 1; Deckert et al, 2006). The splicing reaction was subjected to glycerol-gradient centrifugation (‘first gradient' in Figure 1), and B complexes peaking in the 40S region of the gradient were bound to amylose beads and subsequently eluted under native conditions with maltose (Figure 1); under these conditions, the MS2–MBP remains associated with the MS2 hairpins of the MINX substrate. Affinity-purified B complexes, with MS2 hairpins at either the 5′ or 3′ end of the pre-mRNA or within its intron, all contained nearly stoichiometric amounts of U1, U2, U4, U5 and U6 snRNA and unspliced pre-mRNA (see Supplementary Figure S1B), as well as a protein pattern typical for human B complexes (data not shown).
Figure 1.
Scheme of B complex purification and labelling. Spliceosomal B complexes were assembled on MS2–MBP-tagged pre-mRNA and purified by glycerol-gradient centrifugation (‘first gradient') followed by affinity chromatography with amylose beads (for details see text). Immunocomplexes were then formed, isolated by glycerol-gradient centrifugation (‘second gradient') and labelled with gold.
Specific antibody labelling of pre-mRNA sites in the B complex
To localise the intron or ends of the pre-mRNA in the B complex under the electron microscope, we used a polyclonal rabbit IgG antibody against the MBP. To confirm that the antibody is specific, we first performed western blotting using nuclear extract, either alone or mixed with MS2–MBP; a single band that corresponded in size to the MS2–MBP protein was only observed when MS2–MBP was added (Supplementary Figure S2A). More importantly, glycerol-gradient-fractionated B complexes containing MS2–MBP (bound at the 3′ end), were efficiently precipitated by α-MBP, whereas B complexes without MS2–MBP were precipitated only at background levels similar to those found in the control with no antibody (Supplementary Figure S2B). Similar results were obtained when the MS2 hairpins were located at the 5′ end or in the intron (data not shown). This confirms that the α-MBP antibody binds specifically under native conditions and that the target epitope is accessible in assembled spliceosomal complexes.
To produce immunocomplexes for EM localisation studies, affinity-purified B complexes were incubated with the α-MBP antibody, and immunocomplexes were then separated from unbound antibodies by glycerol-gradient centrifugation (Figure 1; ‘second gradient' under GraFix conditions). The GraFix procedure (Kastner et al, 2008), in which glutaraldehyde is included in the glycerol gradient, was used to stabilise the immunocomplexes during their separation from free IgG. Immunocomplex formation was monitored by ELISA analysis of the gradient fractions and optimised to maximise the number of antibody:B-complex dimers relative to unwanted larger aggregates, while keeping antibody excess as low as possible. In the presence of B complexes, ELISA revealed a large shift for a significant portion of the antibody from the top of the gradient to fractions containing B complex (Supplementary Figure S2C), demonstrating efficient immunocomplex formation and stable association of the bound α-MBP under centrifugation conditions.
EM of affinity-purified B complexes
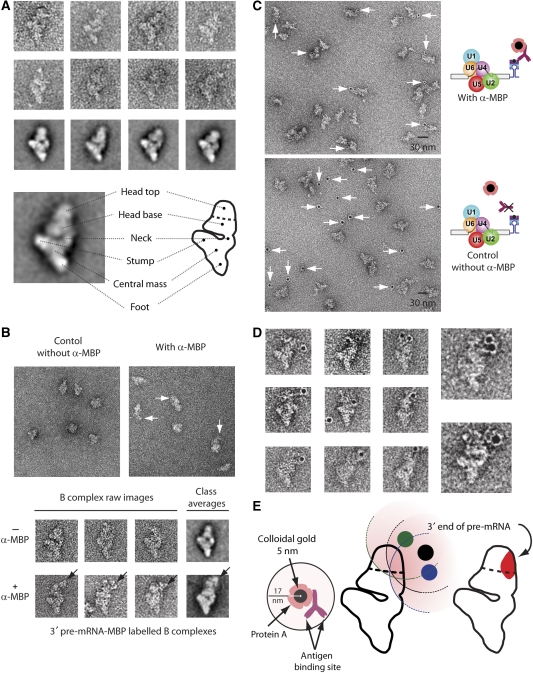
B complexes formed on 3′ MS2-tagged, MINX pre-mRNA (3′ExMS2) were affinity-purified as described above. After the second centrifugation step (Figure 1), fractions containing B complexes were prepared for EM and negatively stained. We first analysed complexes prepared in the absence of α-MBP antibodies. Figure 2A shows a gallery of typical views of single-particle images, with corresponding class averages, obtained by multivariate statistical analysis (MSA) and classification techniques (see Materials and methods), of a dataset containing 6000 individual raw images. The topographical features of the particle are shown below the gallery. Consistent with earlier EM analyses of B complexes purified by the gentle MS2–MBP method (Deckert et al, 2006), our purified 3′ExMS2 B complexes appear more or less rhombic in most views. In the preferred orientation two domains can be recognised: a roughly triangular body and a more globular head positioned at one side of the triangular body. The triangular body is made up of three branches connected to a central mass. The lower branch is named the ‘foot', the massive upper one the ‘stump' and the thinner one the ‘neck'. The head, which is located on the same side of the body as the neck and the stump can be divided into an elongated element close to the triangular body (head base) and a globular domain positioned further away from the body (head top; Figure 2A). The head domain shows some degree of structural variability, whereas the triangular body appears very well defined. Highly similar 2D structures of affinity-purified B complexes were observed if the MS2 tag was located within the intron or at the 5′ end of the 5′ exon (see also below).
Figure 2.
Electron microscopy of negatively stained human B complexes with a tag at the 3′ end of the pre-mRNA. (A) Gallery of images (first and second rows) and class averages (third row) of affinity-purified human spliceosomal B complexes formed on MINX pre-mRNA with MS2 aptamers at its 3′ end. At the bottom, an interpretative sketch (right) of the most frequently found view of the B complex (left) is shown. Structural designations are according to Boehringer et al (2004) and Deckert et al (2006). (B) Overview of particles, with galleries of corresponding views below. MS2–MBP-bound B complexes were incubated without (left-hand panel) or with (right-hand panel) α-MBP antibodies and subjected to a second glycerol-gradient centrifugation before EM analysis. The additional mass ascribed to the antibody is indicated by arrows. (C) Overview of B complexes with and without α-MBP incubated with colloidal gold coated with Protein A before EM analysis. (D) Gallery of gold-labelled B complexes shown in the most frequently observed orientation; two enlarged images are also shown. The foot–stump axis (running from the bottom to the left in the images) of the body is seen clearly and is used to position particles in the same orientation. (E) The 3′ end of the 3′ exon maps to an area in the head domain of the B complex. The diagram illustrates the maximum distance (˜17 nm) from the centre of the gold label to the antibody-binding site (left diagram). Three different positions of the gold label (green, black and blue) taken from separate EM images of labelled B complexes are shown (middle), and a 17-nm radius around each gold label is shaded pink (see description in the Materials and methods section and in Supplementary Figure S3). The overlapping area of the 17-nm circles determined by analysis of several EM images is shown in red (right), and indicates the region wherein the 3′ end of the 3′ exon is located.
Location of the 3′ end of the pre-mRNA in the B complex
We next compared directly EM images of purified B complexes incubated with and without α-MBP antibodies after each of these had been subjected to a second glycerol gradient under GraFix conditions (Figure 2B). B complexes incubated with antibody sometimes appear to possess an extra protrusion of mass compared with the control without antibody, so this mass probably represents bound antibody. This additional density was frequently observed in the upper head region of the B complex and was orientated more towards the ‘neck' side of the particle (Figure 2B). After averaging 2500 single-particle images, we still observed an additional mass in some classes in a similar position at the top of the globular head (Figure 2B).
Because of variation in the density distribution of the head domain of the B complex, even in unlabelled particles, and because of the failure to discern the typical Y-shape of the bound antibody, we performed an additional labelling step to allow the unequivocal assignment of the apparent extra density to bound antibody. Thus, to facilitate detection of the bound antibody, we attached a gold label to it. For this purpose, we used 5-nm colloidal gold particles coated with Protein A (Figure 2E, left). These large heavy-metal clusters (Bendayan, 2000) can be identified unambiguously at any position at the negatively stained particles (see Supplementary Figure S4 for micrographs of antibody-Protein A-gold complexes). To prevent clustering, and to ensure specific binding, we added carefully optimised amounts of Protein A-coated colloidal gold to the gradient-purified, antibody-labelled B complexes and prepared negatively stained EM specimens. The specificity of the gold labelling was always checked by a parallel experiment in which antibody was omitted. To maximise the labelling efficiency, we optimised not only the amount of antibody added to the B complex, but also the fixation conditions in the subsequent gradient (GraFix) to preserve the Protein A-binding capacity of the antibodies bound to the B complex (see Materials and methods).
When antibody was present, many of the complexes were associated with gold, and most gold colloids were found directly adjacent to spliceosomal complexes (Figure 2C, upper field). In contrast, if antibodies were omitted, almost no gold colloids were observed directly next to B complexes, demonstrating the specificity of the labelling procedure (Figure 2C, lower field). Images of ∼200 labelled particles were sorted into several groups according to the appearance of the B complex and some typical views in the preferred orientation are shown in Figure 2D. The typical morphological domains of the B complex, such as the foot, stump and head, were used to orient the particles in a similar manner. Here, the often predominant and sometimes slightly kinked foot–stump axis allows a straightforward identification of the orientation of each B complex.
The position of the gold label was found closest to the head domain (Figure 2D), confirming the presence of bound antibody in this region of the B complex as predicted above by direct visualisation of the antibody (Figure 2B). By analysing all the 57 images of B complexes obtained in the preferred orientation using the 17-nm distance constraint (see Materials and methods; Figure 1E, left; Supplementary Figures S3 and S4) an area at the right towards the top of the head was identified as the region in which the 3′ end of the pre-mRNA is most likely to be located (shaded in the diagram in Figure 2E, rightmost diagram).
Location of the 5′ end and intron of the pre-mRNA in the B complex
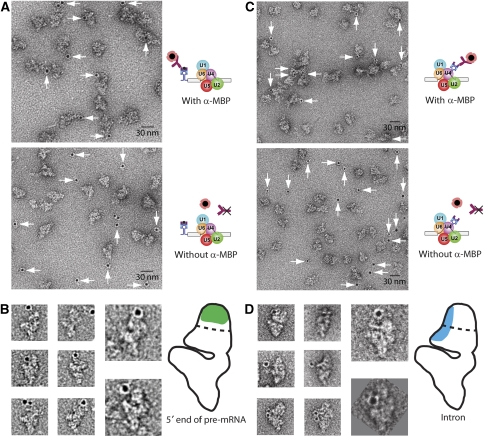
To locate the 5′ end of the pre-mRNA and its intron, we allowed B complexes to form on MINX pre-mRNA with two MS2 stem-loops either at its 5′ end (‘5′ExMS2'; Supplementary Figure S1A) or within the intron, between the 5′ SS and the branch point adenosine (i.e. after nucleotide 44 of the 120-nt intron, Supplementary Figure S1A; ‘InMS2'). Spliceosomal B complexes were affinity-purified as described above (Figure 1) and exhibited compositions (Supplementary Figure S1B), sedimentation behaviours (Supplementary Figure S1C) and morphologies (data not shown) similar to B complexes formed on MINX pre-mRNA with MS2 hairpins at its 3′ end. Thus, in each case B complexes in structurally very similar states were obtained. Purified B complexes with MS2–MBP tags at the 5′ end or in the intron were labelled with α-MBP antibodies. Here—and in the labelling of SF3b155 (see below)—EM analyses of the immunocomplexes failed to identify any possible additional density in the original images or class averages that could be attributed with certainty to the bound antibody (data not shown). Thus, labelling with colloidal gold, as described above, was required to visualise the bound antibody. EM analyses of negatively stained particles revealed in both cases a high degree of labelling with gold when the antibody was added, whereas in the absence of antibody hardly any gold colloids were found directly adjacent to the B complexes (Figure 3A and C, compare upper and lower panels). This demonstrates again the high specificity of the gold label for the bound IgG molecule.
Figure 3.
Location of the intron and the 5′ end of the pre-mRNA within the head domain of the B complex. B complexes were assembled on a pre-mRNA with MS2–MBP protein bound to MS2 hairpins positioned at either the 5′ end of the 5′ exon (A, B) or within the intron (C, D). After incubation with or without α-MBP and a second centrifugation step, gold-tagged Protein A was added. EM fields (A, C) with (upper field) or without (lower field) α-MBP are shown, as well as selected images (B, D). Illustrative drawings (B, D, on the right) are provided, in which the α-MBP binding-site area determined using the 17-nm criterion (see text) is shaded.
In both complexes (5′ExMS2 and InMS2), the gold colloids were found close to the head of the B complex. However, initial inspection of single particles of both complexes indicated that there are differences among the 5′ExMS2, InMS2 and 3′ExMS2 B complexes in the general position of the gold label in the head domain. Application of the 17-nm distance constraint (see Materials and methods; Supplementary Figure S3) on all the particle images showing the B complex in the preferred orientation (57 images for 5′ExMS2 and 27 images for InMS2), the 5′ end of the pre-mRNA was mapped to an area at the top of the globular head region, while the intron was located more in the middle towards the stump side of the head, extending into the base of the head (Figure 3B and D). This is just opposite the site where the 3′ end of the pre-mRNA was found. Thus, the 5′ exon, the 3′ exon and the intron are positioned at somewhat different regions within the head domain of the B complex. However, the 5′ and 3′ exons seem to be relatively close in the 2D projection, as the two localisation areas overlap somewhat. Further, the intron may also be close to the 5′ exon, but is clearly remote from the 3′ end of the 3′ exon.
Location of U2 snRNP and the pre-mRNA BS through labelling of U2 SF3b155
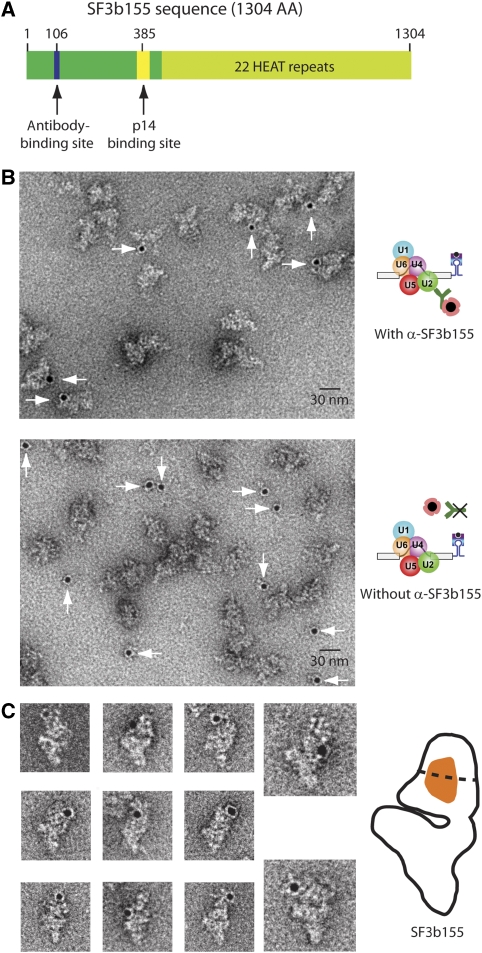
The protein SF3b155 is a member of the spliceosome's functionally important protein heteromeric complex SF3b, which associates with the U2 snRNP (Will and Lührmann, 2001). SF3b proteins help stabilise the association of U2 with the BS, and they are in direct contact with the pre-mRNA at or near the branch adenosine (Query et al, 1996; Gozani et al, 1998; Will et al, 2001). To locate SF3b155, and thus also the U2 snRNP, we labelled B complexes with a peptide antibody specific for the SF3b155 protein (α-SF3b155). This antibody is highly specific for an epitope on SF3b155 (Figure 4A; see western blot, immunoprecipitation and ELISA in Supplementary Figure S2) and has already been used for immuno-EM localisation of the N terminus of SF3b155 in the U11/U12 di-snRNP of the minor spliceosome (Golas et al, 2005). 3′ExMS2 B complexes were affinity-purified and incubated with α-SF3b155 antibodies; the optimum amount for the formation of monomeric B complex bound by an antibody was first determined by ELISA (Supplementary Figure S2C). Immunocomplexes were purified by density-gradient centrifugation, and colloidal gold labelling and EM were subsequently performed. As with the α-MBP antibodies, efficient and specific gold labelling was observed (Figure 4B). In the EM images, all the gold labels were found positioned around the head structure, indicating that SF3b155 is also located in this region (Figure 4C). In comparison with the results obtained above for complexes labelled at the pre-mRNA, the gold colloid was always found much closer to the B complex, often within the particle's outline. Because of this, identifying the particles showing the preferred orientation is here more challenging. To exclude any uncertainty concerning the orientation, we subjected the entire dataset to a reference-free alignment procedure (Supplementary Figure S5), which confirmed the orientation of the labelled particles. By applying the 17-nm distance constraint to all 41 immunocomplexes that had the B complex in the preferred orientation, an area spanning from the lower part of the globular domain to the base of the head resulted (Supplementary Figures S3 and 4C). This area is surrounded by the sites mapped for the intron and the 5′ and 3′ exon ends and should roughly demark also the BS position (Figure 5A). Taken together, these data indicate that not only the pre-mRNA, but also the U2 snRNP is located in the head of the spliceosomal B complex.
Figure 4.
EM localisation of the SF3b155 protein in the head of the B complex. (A) Schematic diagram of SF3b155. The epitope for the SF3b155-specific antibody, the binding site for the SF3b protein p14 and the HEAT repeats are shown. (B) Purified B complexes assembled on 3′ MS2 hairpin-tagged pre-mRNA (3′ExMS2) were incubated with (upper panel) or without (lower panel) α-SF3b155, subjected to glycerol-gradient centrifugation and labelled by incubating with colloidal gold coated with Protein A. (C) Complexes in the most frequently observed view are shown. The sketch at the right shows the α-SF3b155 binding-site area (orange) determined using the 17-nm criterion (see text).
Figure 5.
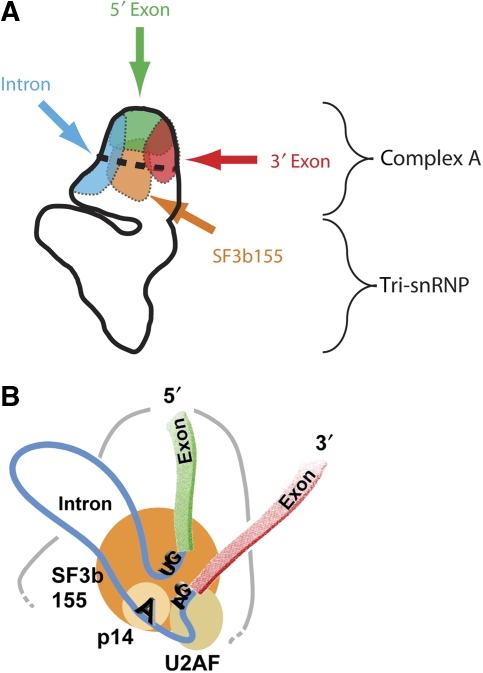
The pre-mRNA and SF3b155 are located in the head domain of the B complex. (A) Comparison of the mapped position of the three pre-mRNA positions and SF3b155, as indicated. The likely location of components of the A complex and the tri-snRNP (see Discussion) are indicated. (B) Interpretative model of the position of the pre-mRNA in the B complex head domain. SF3b155 and its binding partners, p14 and U2AF65, are shown schematically. GU sequence represents the 5′ splice site, AG the 3′ splice site and A the branch site.
Discussion
Here, we have mapped the location of various regions of the pre-mRNA, as well as a functionally important protein of the U2 snRNP, in the EM structure of the human spliceosomal B complex. The 5′ and 3′ exons and the intron of the pre-mRNA, and also the SF3b155 protein, were found in distinct areas of the head domain of the B complex by immunolabelling and enhancement of the antibody label by additional labelling with colloidal gold particles coated with Protein A. As the spliceosomal B complex is poised to undergo catalytic activation, our data provide important insights into the spatial organisation of the spliceosome at a crucial point in its assembly. The method used here can be used in the future to study the organisation of other spliceosomal complexes, and may therefore be expected ultimately to lead to a better understanding of the structural dynamics of the spliceosome during its assembly and of its catalytic activity.
Mapping of antibody-binding sites on the B complex using colloidal gold
The B complex is particularly amenable to mapping studies as it has an asymmetric shape, allowing the precise location of a label bound to a specific position on the particle. In the main 2D projection of the B complex, distinct morphological features can be clearly discerned—for example, the foot–stump axis—and the latter was used to position particles in the same orientation. Here, we have performed classical immuno-EM techniques to locate specific components of the spliceosome by antibody labelling. As bound antibodies were often not discernable under the electron microscope, we additionally labelled them with colloidal gold to allow/enhance their visualisation. Colloidal gold has commonly been used as an electron-opaque label in the visualisation of cellular structures (Bendayan, 2000). Here, we have adapted this method for single-particle labelling and have increased the precision of mapping by applying the <17 nm distance constraint. This method assumes a maximum distance of 17 nm between the visualised gold and the actual antibody-binding site (MS2–MBP bound to the MS2 aptamer or the SF3b155 protein). The gold-labelling method appears to be highly efficient and specific, and also flexible (i.e. both proteins and RNA structures can be located), and it can, thus, potentially be used to map components in other RNP complexes that are amenable to structural determination by EM. In principle, proteins larger than IgGs can be used to enhance the visibility of a label bound to a large complex, as was recently demonstrated for the spliceosomal C complex (Alcid and Jurica, 2008). The use of a heavy-metal marker, as described here, has the added advantage that all labelled sites, including those within the outline of the EM image, can readily be visualised.
Spatial organisation of spliceosomal snRNPs in the B complex
EM of immunolabelled B complexes allowed us to locate topographical labels introduced into the pre-mRNA (indirectly, through bound MS2–MBP protein). All of the regions of the pre-mRNA that we analysed were found in the head domain of the B complex (Figure 5A). Therefore, protein factors and snRNPs directly bound to the pre-mRNA may also be expected to be located in this domain or at least close by. This was indeed the case for the U2 snRNP, which we located in the head domain through its associated protein SF3b155 (Figure 5A). During spliceosome assembly, initially the U1 and U2 snRNPs bind to the pre-mRNA, forming the A complex. Our results thus suggest that the head domain is mainly formed already during A complex assembly. As the B complex is subsequently generated on association of the U4/U6.U5 tri-snRNP with the A complex, this implies in turn that the triangular body of the B complex is made up largely of the tri-snRNP. This idea is supported by current structural information available for several spliceosomal complexes or subcomplexes. First, the size and overall shape of the human A complex as recently determined by EM (Behzadnia et al, 2007) is similar to that of the head domain of the B complex. Further, the BΔU1 spliceosome, which differs from the B complex in that the U1 snRNP and several non-snRNP proteins are absent (Makarova et al, 2004), possesses a smaller head domain than the B complex, as determined by EM (Boehringer et al, 2004). Finally, like the body of the B complex, both human and yeast U4/U6.U5 tri-snRNPs have a characteristic triangular or Y-shaped structure (Fabrizio et al, 1994; Sander et al, 2006; Häcker et al, 2008) and it has already been suggested (Stark and Lührmann, 2006) that the B complex body may harbour the tri-snRNP. Using our method for locating the 5′ m3G-cap of the snRNAs (U1, U2, U4 and U5 snRNAs; Sander et al, 2006) we also observed labels at the B complex body, and these might indicate the position of U5 snRNA (data not shown). In summary, our data strongly support the contention that components of complex A (the pre-mRNA, the U1 and U2 snRNPs, and other associated proteins) are present in the head of complex B, and that components of the tri-snRNP are present in its body.
Location of functional sites of the pre-mRNA in the B complex
The localisation of the SF3b155 protein in a central area of the B complex head domain suggests strongly that the SSs and the branch point are likewise located in the head region. Earlier data indicate that the U2 snRNP takes part in structurally organising the SSs (Dönmez et al, 2007). SF3b155 can be cross-linked directly to the pre-mRNA 6 nucleotides downstream and 5 nucleotides upstream of the branch-point adenosine in complex A (Gozani et al, 1998), whereas SF3b14a (p14) cross-links directly to the branch point (MacMillan et al, 1994; Will et al, 2001) and is at the same time in direct contact with SF3b155 (Will et al, 2001). Thus, the branch point must be located near the region mapped for SF3b155. SF3b155 also interacts directly with U2AF65 (Gozani et al, 1998), a factor binding the pre-mRNA's polypyrimidine tract that connects the BPS with the 3′ SS (Valcarcel et al, 1996); thus U2AF65 and the polypyrimidine tract are also very probably nearby. The 5′ SS must also be in the vicinity, as hydroxyl-radical footprinting experiments revealed a close proximity (<3 nm) not only between the branch point and the 3′ SS, but also between both of these sites and the 5′ SS in the E and A complexes (Kent and MacMillan, 2002; Dönmez et al, 2007).
The ends of the exons (5′ end of the 5′ exon and 3′ end of the 3′ exon) were also located in the head domain, but clearly more in the upper part (Figure 5A). The distance of the 5′ SS from the 5′ end of the exon in the shortened MINX substrate used for 5′-end labelling is 36 nucleotides. If this stretch were in an A-form helical conformation (which is one possible conformation) it would be ∼10 nm long (about half the length of the head domain). The 52 nucleotides between the 3′ SS and the 3′ end of the exon would be ∼14 nm long in this conformation. If the exons were in a fully extended RNA conformation, they would have double this length. These numbers show that a stacked (A-form) and an extended conformation of the exons are both compatible with a location of the SSs at the centre of the head of the B complex. The 5′ and 3′ exons of the pre-mRNA are probably close to each other within the B complex. A close spatial relationship of the two exon ends were also found in the C complex (Alcid and Jurica, 2008).
The MINX intron (without MS2 tags) is about 120 nucleotides in length, and would have a length of ∼32 nm if it adopted a stacked A-form conformation or about double this length when maximally extended. To localise the intron, we placed the MS2 tag halfway (about 50 nucleotides, corresponding to 14 nm in the A form and about twice this if extended) between the 5′ SS and the branch point. Our finding that the intron site is also located in the head of the B complex fits in very well with the assumption that the SSs and the branch point are all located in the head region of the B complex.
A model of the possible arrangement of pre-mRNA in the B complex
An interpretative view of a possible arrangement of the pre-mRNA in the B complex is shown in Figure 5B. This model takes into account information gained in this work and additional biochemical data from earlier studies as described above. In our interpretation, the SF3b155 protein, together with p14, U2AF65, and the branch adenosine and the SSs, are located near the centre of the head domain, as suggested by our SF3b155 labelling data (Figure 4). The 5′ exon would then run from the top of the particle to the centre of the head, whereas the 3′ exon would extend from the centre towards the right of the head (Figure 5B). The intron would loop out, away from the rest of the particle, thus allowing the necessary flexibility in respect of its length.
Our data also suggest that the emerging catalytic centre that is formed on activation of the B complex is located near the head. After transition to an active spliceosome, components of the tri-snRNP—that is, U5 and U6—contribute to the catalytically active core RNP of the spliceosome (Sontheimer and Steitz, 1993; Yean et al, 2000). A functionally important RNA duplex forms between the 5′ end of U2 (most probably located in the head domain) and the 3′ end of U6 (Madhani and Guthrie, 1992), which is likely to be present in the body of the B complex (see above). Thus, it seems very plausible that these components are at the interface between the head and the body of the B complex, either in the neck or spanning the cleft between the head and body (Figure 2A). If this is indeed the case, then the emerging catalytic centre may also be expected to be found here.
The colloidal gold-labelling method that we describe here should be applicable to other target sites as well, if suitable antibodies are available. The labelled site was found unambiguously, independently of the target structure and irrespectively of whether the label was at the edge or in the middle of the particle image. Our experimental approach should thus be highly suited for other spliceosomal complexes, such as the C complex. Mapping of the pre-mRNA sites examined here in other spliceosomal complexes should shed light on the extensive structural rearrangements accompanying the transition from the pre-catalytic B complex to the catalytically active C complex. Thus, an initial functional view of spliceosome structure may now be attainable.
Materials and methods
Synthesis of pre-mRNAs tagged with MS2 aptamers
All pre-mRNA constructs used in this work were derived from the pMINX plasmid (Zillmann et al, 1988) by using standard PCR techniques (see Supplementary data for PCR primers used). The construct encoding MINX pre-mRNA with three MS2 RNA hairpins at its 3′ end (3′ExMS2, shown schematically in Supplementary Figure S1B) was generated as described earlier (Deckert et al, 2006). The 5′ end-tagged template (5′ExMS2, Supplementary Figure S1B) was produced by inserting two MS2 RNA hairpins from the pAdML-M3 plasmid (Zhou et al, 2002a) upstream of a shortened 5′ exon (36 nucleotides) of the MINX construct. The construct encoding intron-tagged MINX pre-mRNA (InMS2; Supplementary Figure S1B) was derived by inserting two MS2 RNA hairpins 44 nucleotides downstream of the 5′ SS and 52 nucleotides upstream of the branch-point adenosine. Uniformly 32P-labelled m7G(5′)ppp(5′)G-capped pre-mRNAs (Hartmuth et al, 2002) were synthesised in vitro by T7 run-off transcription (Milligan and Uhlenbeck, 1989) in the presence of [α32P]-UTP.
MS2 affinity selection of spliceosomal B complexes
HeLa nuclear extract was prepared as described elsewhere (Behzadnia et al, 2006). Spliceosomal B complexes were assembled and purified essentially as described earlier (Deckert et al, 2006). Briefly, MINX pre-mRNA containing MS2 RNA aptamers was complexed with a fusion protein that comprised the MS2 coat protein and MBP (MS2–MBP; Zhou et al, 2002b). After incubation for 8 min at 30°C under splicing conditions in the presence of 50% (v/v) HeLa nuclear extract, the splicing reaction was loaded onto a linear 10–30% (v/v) glycerol gradient to separate B complexes from other spliceosomal complexes and unbound MS2-MBP protein. 40–45S gradient fractions containing the B complex were pooled and affinity selection was performed with amylose beads, followed by elution with maltose. The RNA composition of the affinity-purified B complexes was analysed by denaturing PAGE; bands were visualised by staining with silver and by autoradiography. The protein composition of the purified complexes (data not shown) was determined by mass spectrometry after separating proteins by SDS–PAGE as described earlier (Deckert et al, 2006).
Antibody labelling and immunocomplex preparation for EM
For immunocomplex preparation, 5 pmol of affinity-purified B complexes (in 200–400 μl of elution buffer) were incubated for 90 min at 4°C with a 10–20-fold molar excess of affinity-purified rabbit polyclonal antibodies against MBP (ab9084, Abcam) or SF3b155 (raised against a peptide of SF3b155; Will et al, 2001). The antibody:B-complex ratio was initially optimised to maximise the number of [antibody · complex B] dimers relative to unwanted oligomeric complexes, while at the same time avoiding spurious, non-specific binding of the antibody to the B complex by ELISA (see below). The optimum antibody:B-complex ratio was found to be approximately 15:1. For EM analysis, immunocomplexes were separated from free IgG by gradient centrifugation (‘second gradient', Figure 1). A measure of 400 μl of the antibody-B complex mixture were loaded onto a 4.4-ml linear 10–30% (v/v) glycerol gradient and centrifuged for 107 min at 60 000 r.p.m. in a TH-660 rotor (Thermo). The glycerol gradient contained a linear gradient of 0–0.1% glutaraldehyde (25% aqueous solution, EM grade, EMS, Hatfield, USA), to stabilise the complex by chemical cross-linking (GraFix method; Kastner et al, 2008). Under these conditions, the Protein A-binding capacity of the IgG antibody bound to the B complex is maintained. The gradient was harvested from the top for analytical purposes (24 175-μl fractions) or from the bottom for EM preparation (35 ∼120-μl fractions). The cross-linking reaction was stopped immediately after fractionation by adding 10 μl of 1 M glycine-NaOH, pH 7.9, to each fraction and incubating for 30 min at 4°C.
Immunoblotting, immunoprecipitation and ELISA
For immunoblot analysis, proteins from HeLa nuclear extract were fractionated by SDS–PAGE, transferred to nitrocellulose, immunostained with α-MBP or α-SF3b155 antibodies, and bound antibody was visualised with an enhanced chemiluminescence detection kit (Pierce). For immunoprecipitation analysis, 12 μg of affinity-purified antibody was bound to 30 μl Protein A Sepharose beads (GE Healthcare) in the presence of 0.5 mg/ml BSA and 0.5 mg/ml Escherichia coli tRNA (Sigma). After washing, 15 μl beads were incubated with 400 μl of peak gradient fractions (first gradient) containing 3′ExMS2 B complexes in gradient buffer (20 mM HEPES/KOH, pH 7.9, 150 mM NaCl, 1.5 mM MgCl2), supplemented with 0.5 mg/ml BSA, 0.5 mg/ml E. coli tRNA and 0.05% NP-40 (final concentrations) plus an additional 400 μl gradient buffer. Beads were washed three times with 600 μl of gradient buffer.
ELISA was performed with B complexes subjected to the second gradient-centrifugation step (Figure 1). After stopping the cross-linking reaction with glycine (see above), 20 μl of each gradient fraction was pipetted into the wells of a pre-coated goat anti-rabbit ELISA plate (Pierce) and incubated overnight at 4°C in the presence of 3% (w/v) BSA and 0.05% (w/v) Tween-20. After washing, the ELISA plate was incubated with goat anti-rabbit antibody conjugated to horseradish peroxidase (Dianova) for 1 h at 37°C, followed by TMB solution (Pierce) to quantify the antibody (free or bound to B complexes). The colour reaction was stopped by adding an equal volume of 2 M H2SO4 and absorption at 450 nm was subsequently measured.
Colloidal gold labelling and EM
Gradient fractions (second gradient) containing antibody-labelled B complexes (or unlabelled complexes as a control), as determined by the distribution of 32P-marked pre-mRNA, were pooled and incubated with colloidal gold coated with Protein A (G Posthuma, Cell Microscopy Center, University Medical Center, Utrecht). The amount of colloidal gold required for optimum labelling depended on the concentrations of B complex; therefore, this amount was optimised in each experiment. Approximately 0.1 μl of colloidal gold solution was mixed with 100 μl of B complexes (∼200 fmol B complexes, ∼100 fmol gold colloids). This mixture was incubated at 4°C for 90 min and then used directly for EM-grid preparation, which was performed as described earlier (Golas et al, 2003), but with a prolonged particle-adsorption period of 2 h. Images were obtained at room temperature at a magnification of 55 000 on a calibrated CM120 electron microscope (FEI) in eucentric height at a defocus of approximately 2 μm. The microscope was equipped with a TemCam F224A digital camera (TVIPS). Particles were analysed and grouped into similarity classes by visual inspection.
For direct analysis of bound antibody, EM grids were prepared before incubating affinity-purified B complexes with colloidal gold; images were obtained under the same conditions as described above and were subsequently analysed by published image-analysis techniques (van Heel et al, 1996). After performance of a ‘reference-free' alignment procedure (Dube et al, 1993), images were subjected to MSA (van Heel and Frank, 1981) and classification (Van Heel, 1989).
Localisation of labelling targets
To determine more precisely the area in which the targeted pre-mRNA site (3′ end, 5′ end, intron) is located, we anticipated a maximum distance of 17 nm from the centre of the gold colloid to the epitope targeted by the antibody, that is, the MS2–MBP protein bound to the respective site on the pre-mRNA (Figure 2E). This distance was obtained by assuming a gold-colloid particle radius of 2.5 nm, a Protein A diameter of 2.5 nm, and a greatest distance of 12 nm between the Protein A and the antigen-binding site of the IgG molecule. Indeed, measurements of purified antibody-gold-conjugates confirmed ca. 17 nm as the maximum length (Supplementary Figure S4). As the antibody arms are flexible, and superimposition of gold and antibody is possible, shorter distances are also observed in the EM images (Supplementary Figure S4C). Consequently, for every labelled particle, the MBP sites are taken to be located within a circle of radius 17 nm from the gold label. For each labelling experiment, all the complexes showing the B complex in the preferred orientation (Supplementary Figure S3) were then combined and the area was determined where all 17-nm circles overlapped (see Figure 2E). A single area where all such circles overlapped was found in each experiment; this defines—or, more precisely, delimits—the site of antibody binding at the B complex.
The true position of the pre-mRNA—that is, the site at which the MS2 hairpin that binds to the fusion protein is inserted—may differ somewhat from this location, as the MS2 hairpins (2–3 nm; Keryer-Bibens et al, 2008) and the bound MS2–MBP fusion protein (50 kDa; Zhou et al, 2002b) bridge the antibody-binding site to the MS2 hairpin insertion site at the pre-mRNA. To take this potential uncertainty into account, a more generously dimensioned area (enlarged by ∼3 nm) was also considered. This area is the one shown in Figures 2E, 3B and C).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Information
Acknowledgments
We are grateful to Thomas Conrad, Hossein Kohansal and Irene Öchsner for excellent technical assistance. We also thank Dietmar Riedel and Prakash Dube for help with the EM, Michael Grote and Evgeny Tonevitsky for productive discussions about experiments and Cindy L Will for very helpful comments on the manuscript. This work was supported by a grant from the Studienstiftung des deutschen Volkes to E.W. and grants from the Deutsche Forschungsgemeinschaft, the EU network EURASNET, the Fonds der Chemischen Industrie and the Ernst-Jung-Stiftung to RL.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alcid EA, Jurica MS (2008) A protein-based EM label for RNA identifies the location of exons in spliceosomes. Nat Struct Mol Biol 15: 213–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will CL, Urlaub H, Stark H, Lührmann R (2007) Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J 26: 1737–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadnia N, Hartmuth K, Will CL, Lührmann R (2006) Functional spliceosomal A complexes can be assembled in vitro in the absence of a penta-snRNP. RNA 12: 1738–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendayan M (2000) A review of the potential and versatility of colloidal gold cytochemical labelling for molecular morphology. Biotech Histochem 75: 203–242 [PubMed] [Google Scholar]
- Boehringer D, Makarov EM, Sander B, Makarova OV, Kastner B, Lührmann R, Stark H (2004) Three-dimensional structure of a pre-catalytic human spliceosomal complex B. Nat Struct Mol Biol 11: 463–468 [DOI] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R (2008) Isolation of an active step I spliceosome and composition of its RNP core. Nature 452: 846–850 [DOI] [PubMed] [Google Scholar]
- Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Lührmann R (2006) Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol 26: 5528–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dönmez G, Hartmuth K, Kastner B, Will CL, Lührmann R (2007) The 5′ end of U2 snRNA is in close proximity to U1 and functional sites of the pre-mRNA in early spliceosomal complexes. Mol Cell 25: 399–411 [DOI] [PubMed] [Google Scholar]
- Dube P, Tavares P, Lurz R, van Heel M (1993) The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J 12: 1303–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Esser S, Kastner B, Lührmann R (1994) Isolation of S. cerevisiae snRNPs: comparison of U1 and U4/U6.U5 to their human counterparts. Science 264: 261–265 [DOI] [PubMed] [Google Scholar]
- Golas MM, Sander B, Will CL, Lührmann R, Stark H (2003) Molecular architecture of the multiprotein splicing factor SF3b. Science 300: 980–984 [DOI] [PubMed] [Google Scholar]
- Golas MM, Sander B, Will CL, Lührmann R, Stark H (2005) Major conformational change in the complex SF3b upon integration into the spliceosomal U11/U12 di-snRNP as revealed by electron cryomicroscopy. Mol Cell 17: 869–883 [DOI] [PubMed] [Google Scholar]
- Gozani O, Potashkin J, Reed R (1998) A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol 18: 4752–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet 17: 100–107 [DOI] [PubMed] [Google Scholar]
- Häcker I, Sander B, Golas MM, Wolf E, Karagöz E, Kastner B, Stark H, Fabrizio P, Lührmann R (2008) Localisation of PRP8, Brr2, Snu114 and U4/U6 proteins in the yeast tri-snRNP by electron microscopy. Nat Struct Mol Biol 15: 1206–1212 [DOI] [PubMed] [Google Scholar]
- Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Lührmann R (2002) Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc Natl Acad Sci USA 99: 16719–16724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Lührmann R (2009) Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol Cell Biol 29: 281–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ (2002) Purification and characterisation of native spliceosomes suitable for three-dimensional structural analysis. RNA (8): 426–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Sousa D, Moore MJ, Grigorieff N (2004) Three-dimensional structure of C complex spliceosomes by electron microscopy. Nat Struct Mol Biol 11: 265–269 [DOI] [PubMed] [Google Scholar]
- Kastner B, Fischer N, Golas MM, Sander B, Dube P, Boehringer D, Hartmuth K, Deckert J, Hauer F, Wolf E, Uchtenhagen H, Urlaub H, Herzog F, Peters JM, Poerschke D, Lührmann R, Stark H (2008) GraFix: sample preparation for single-particle electron cryomicroscopy. Nat Methods 5: 53–55 [DOI] [PubMed] [Google Scholar]
- Kent OA, MacMillan AM (2002) Early organization of pre-mRNA during spliceosome assembly. Nat Struct Biol 9: 576–581 [DOI] [PubMed] [Google Scholar]
- Keryer-Bibens C, Barreau C, Osborne HB (2008) Tethering of proteins to RNAs by bacteriophage proteins. Biol Cell 100: 125–138 [DOI] [PubMed] [Google Scholar]
- MacMillan AM, Query CC, Allerson CR, Chen S, Verdine GL, Sharp PA (1994) Dynamic association of proteins with the pre-mRNA branch region. Genes Dev 8: 3008–3020 [DOI] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C (1992) A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71: 803–817 [DOI] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Lührmann R (2002) Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208 [DOI] [PubMed] [Google Scholar]
- Makarova OV, Makarov EM, Urlaub H, Will CL, Gentzel M, Wilm M, Lührmann R (2004) A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J 23: 2381–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan JF, Uhlenbeck OC (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol 180: 51–62 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Sharp PA (1993) Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature 365: 364–368 [DOI] [PubMed] [Google Scholar]
- Nilsen TW (1998) RNA–RNA interactions in nuclear pre-mRNA splicing. In RNA Structure and Function, Simons RW (ed). pp 279–308. Cold Spring Harbor, New York: Cold Spring Harbor Press [Google Scholar]
- Nilsen TW (2007) RNA 1997–2007: a remarkable decade of discovery. Mol Cell 28: 715–720 [DOI] [PubMed] [Google Scholar]
- Query CC, Strobel SA, Sharp PA (1996) Three recognition events at the branch-site adenine. EMBO J 15: 1392–1402 [PMC free article] [PubMed] [Google Scholar]
- Sander B, Golas MM, Makarov EM, Brahms H, Kastner B, Lührmann R, Stark H (2006) Organisation of core spliceosomal components U5 snRNA loop I and U4/U6 di-snRNP within U4/U6.U5 tri-snRNP as revealed by electron cryomicroscopy. Mol Cell 24: 267–278 [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ, Steitz JA (1993) The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262: 1989–1996 [DOI] [PubMed] [Google Scholar]
- Stark H, Lührmann R (2006) Cryo-electron microscopy of spliceosomal components. Annu Rev Biophys Biomol Struct 35: 435–457 [DOI] [PubMed] [Google Scholar]
- Valcarcel J, Gaur RK, Singh R, Green MR (1996) Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA [corrected]. Science 273: 1706–1709 [DOI] [PubMed] [Google Scholar]
- Van Heel M (1989) Classification of very large electron microscopical image data sets. Optik 82: 114–126 [Google Scholar]
- van Heel M, Frank J (1981) Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy 6: 187–194 [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M (1996) A new generation of the IMAGIC image processing system. J Struct Biol 116: 17–24 [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 13: 290–301 [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R (2006) Spliceosome structure and function. In The RNA World, Gesteland RF, Cech TR, Atkins AJF (eds) 3rd edn, pp 369–400. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Will CL, Schneider C, MacMillan AM, Katopodis NF, Neubauer G, Wilm M, Lührmann R, Query CC (2001) A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J 20: 4536–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yean SL, Wuenschell G, Termini J, Lin RJ (2000) Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408: 881–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R (2002a) Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Sim J, Griffith J, Reed R (2002b) Purification and electron microscopic visualisation of functional human spliceosomes. Proc Natl Acad Sci USA 99: 12203–12207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillmann M, Zapp ML, Berget SM (1988) Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol Cell Biol 8: 814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Information