Abstract
The repair enzyme 8-oxoguanine glycosylase/apyrimidinic/apurinic lyase (OGG) removes 8-hydroxy-2′deoxyguanosine (oh8dG) in human cells. Our goal was to examine oh8dG-removing activity in the cell nuclei of male C57BL/6 mouse brains treated with either forebrain ischemia—reperfusion (FbIR) or sham operations. We found that the OGG activity in nuclear extracts, under the condition in which other nucleases did not destroy the oligodeoxynucleotide duplex, excised oh8dG with the greatest efficiency on the oligodeoxynucleotide duplex containing oh8dG/dC and with less efficiency on the heteroduplex containing oh8dG/dT, oh8dG/dG, or oh8dG/dA. This specificity was the same as for the recombinant type 1 OGG (OGG1) of humans. We observed that the OGG1 peptide and its activity in the mouse brain were significantly increased after 90 min of ischemia and 20–30 min of reperfusion. The increase in the protein level and in the activity of brain OGG1 correlated positively with the elevation of FbIR-induced DNA lesions in an indicator gene (the c-fos gene) of the brain. The data suggest a possibility that the OGG1 protein may excise oh8dG in the mouse brain and that the activity of OGG1 may have a functional role in reducing oxidative gene damage in the brain after FbIR.
Keywords: Base excision repair, Oxidative DNA damage, Mouse nuclear type I 8-oxoguanine glycosylase/apyrimidinic/apurinic lyase, Stroke
Ischemia—reperfusion perturbs the oxygen supply and energy metabolism of the brain, causing oxidative stress by generating free radicals (Coyle and Puttfarcken, 1993). Free radicals cause damage to proteins, lipids, and nucleic acids (Demple and Harrison, 1994; Vos, 1995). Among the lesions possible in nucleic acids (base lesions, DNA strand breaks, and DNA—protein cross-links; Kasprzak et al., 1992), four DNA base lesions [2,6-diamino-4-hydroxy-5-formamidopyrimidine, 8-hydroxyadenosine, 5-hydroxycytosine, and 8-hydroxy-2′-deoxyguanosine (oh8dG)], DNA strand breaks of various kinds, DNA without a base [apyrimidinic/apurinic (AP) sites], and one RNA base lesion [8-hydroxyguanosine (oh8G)] have been shown to increase in the brain after experimental stroke (P. K. Liu et al., 1996; Chen et al., 1997; Cui et al., 1999b; Huang et al., 2000) and stress (J. Liu et al., 1996). In addition, damage to several genes has been detected in the brain of the mouse (DNA polymerase β and γ-actin genes; P. K. Liu et al., 1996) and of the rat (c-fos gene; Cui et al., 1999b). Recently, an elevation in oh8G formation has been demonstrated in the brains from patients with Alzheimer’s disease (Nunomura et al., 1999). Thus, the removal and repair of oxidative stress-induced lesions in nucleic acids play an important role in the prevention of neurological disorders (Robbins et al., 1985; Parshad et al., 1996; Kisby et al., 1997; Fujimura et al., 1999; Lovell et al., 1999) and in neuroregeneration after brain injury (LaPlaca et al., 1999; Shackelford et al., 1999).
The inability to repair damage in nucleic acids, among other risk factors (e.g., an increase in glutamate, calcium, and nitric oxide levels), may cause apoptotic cell death in the brain. Although evidence for the repair of oh8G lesions in RNA is lacking (Kamath-Loeb et al., 1997), the majority of oh8dG lesions, which are markers of oxidative DNA damage, are repaired by several glycosylases. The early presence of DNA repair activity during reperfusion after cerebral ischemia has been suggested by a gradual disappearance of Escherichia coli formamidopyrimidine DNA N-glycosylase (Fpg)-sensitive sites in several nuclear genes (P. K. Liu et al., 1996; Cui et al., 1999b, 2000). Two alternative repair pathways, in addition to the nucleotide excision pathway (Reardon et al., 1997), have been proposed to remove oxidative DNA damage: One removes base lesions (creating a single nucleotide patch), and the other removes various types of remaining oxidative DNA damage (creating a 7- to 14-nucleotide patch; Frosina et al., 1996; Wilson and Thompson, 1997). Very little is known concerning the repair of oxidative DNA damage in the brain, except that the repair of base lesions may be slower in the transcribed strand than in the nontranscribed strand during the first 30 min of reperfusion (Cui et al., 1999b).
The majority of oh8dG lesions in the mammalian cell nucleus are removed by the mammalian homologue of yeast nuclear type 1 8-oxoguanine glycosylase/AP lyase (OGG1) (Krokan et al., 1997; Monden et al., 1999). Mammalian ogg1 cDNA has been cloned from the mouse (mogg1; Rosenquist et al., 1997), the rat (rogg1; Prieto Alamo et al., 1998), and the human (hogg1; Roldan-Arjona et al., 1997; Rosenquist et al., 1997). The recombinant OGG1 proteins contain an activity that excises oh8dG from DNA and a lyase activity that nicks 3′ to the AP site via β-elimination. Mammalian OGG homologue in the mitochondria (OGG2) has been demonstrated in the liver of the rat (Croteau et al., 1997). In dividing human cells, seven alternatively spliced forms of hOGG1 are proposed to account for differential intracellular localization (Nishioka et al., 1999). The present project aimed to compare DNA repair activity in the cell nucleus of animal brains receiving a sham operation (normal control) with activity in animals treated with ischemia and reperfusion. We chose to measure the nuclear OGG1 activity because it is known to specifically repair the major oxidative marker oh8dG.
MATERIALS AND METHODS
Animal stroke model
Oxidative stress was induced in male C57BL/6 mice (20–25 g; Taconic Farms, Germantown, NY, U.S.A.) using a forebrain ischemia—reperfusion (FbIR) model consisting of bilateral occlusion of the common carotid arteries for 90 min followed by various lengths of reperfusion (0–4 h). A cerebral ischemic condition has been established by 80% reduction in the cerebral blood flow using bilateral occlusion of both common carotid arteries for 30 min in C57BL/6 mice (Fujii et al., 1997). Mice were anesthetized with ketamine (100 mg/kg i.p.) plus xylazine (13 mg/kg i.p.) prior to surgery. The control group underwent the same surgical procedure but without artery occlusion. The body temperature was maintained at 37 ± 0.5°C during surgery and the postoperative period until the animals recovered fully from anesthesia. Housing and anesthesia were in accordance with the NIH Guide for the Care and Use of Laboratory Animals, USDA regulations, and the American Veterinary Medical Association Panel on Euthanasia guidelines.
Preparation of nuclear extracts
After decapitation, the brain was quickly dissected on ice, and the cerebellum and the brainstem were removed. The cerebral hemispheres were quickly frozen in liquid nitrogen. The flash-frozen tissue was homogenized with a tissue grinder in a hypotonic buffer containing 10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.2 mM phenylmethylsulfonyl fluoride (PMSF), and 0.5 mM dithiothreitol (DTT) (An et al., 1993; Liu et al., 1994). After being stored on ice for 10 min, the nuclear fraction was separated from the cytoplasmic fraction by centrifugation at 3,300 g at 4°C for 15 min. The resultant nuclear pellets were disrupted with buffer A [20 mM HEPES (pH 7.9), 25% glycerol, 1.5 mM MgCl2, 20 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF, and 0.5 mM DTT] using a mechanical grinder, and then a 0.5× volume of buffer B (1.2 M KCl in buffer A) was added. After incubation on ice for 30 min, samples were separated at 25,000 g at 4°C for 30 min. The supernatant was stored in aliquots at -70°C. The protein content in each sample was determined in triplicate using the Bio-Rad protein assay.
Measurement of OGG1 activity in nuclear extracts
A synthetic deoxyoligonucleotide [oligo DNA; 5′-CATCATGGTCXTGGTTTGGGCA-3′, where X is oh8dG (oligoZ), an AP site (oligoD), or a dG (oligoG)] of the c-fos gene was custom-made by Sigma Genosys (Woodlands, TX, U.S.A.). The oligo DNA (oligoZ, oligoA, or oligoG) was labeled at the 5′ end using [γ-32P]ATP and T4 polynucleotide kinase and was purified using a Sephadex G-25 column. The oligo DNA was mixed with a complementary strand (5′-tgcccaaaccaYgaccatgatg-3′, where Y was usually a dC), then denatured by heating at 70°C for 10 min, and hybridized by gradually cooling to 25°C. To determine the substrate specificity of the OGG1 activity in nuclear extracts, the 32P-labeled oligoZ was hybridized to a complementary strand that contained dC, dG, dT, or dA in the Y position. To obtain reaction product markers, 32P-labeled oligoD or oligoZ duplex was incubated with 1 U of E. coli endonuclease IV or E. coli Fpg protein, respectively (Trevigen, Gaithersburg, MD, U.S.A.). To test for nonspecific nuclease activities, the 32P-labeled oligoG duplex was incubated with nuclear extracts under the same conditions in a separate tube. In brief, 100 fmol of oligo duplex (∼1 × 106 cpm/fmol) was incubated for 15 min at 30°C with 30 μg of nuclear extracts in 20 mM HEPES (pH 7.9), 0.75 mM MgCl2, 200 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF, and ≤25% glycerol. The reactions were terminated by adding an equal volume of stop solution [80% formamide, 50 mM Tris-HCl (pH 8.3), 1 mM EDTA, 0.1% bromophenol blue, and 0.1% xylene cyanole] and resolved in 10% denaturing polyacrylamide gel electrophoresis. The image was analyzed using a PhosphorImager SF (Molecular Dynamics, Sunnyvale, CA, U.S.A.).
Detection of DNA injury and repair in the c-fos gene
Damage to nuclear genes was determined using a fragment shift assay (Bohr et al., 1985; Mellon et al., 1987; Driggers et al., 1993; Bhagwat and Gerlt, 1996; P. K. Liu et al., 1996; Taffe et al., 1996; Cui et al., 1999b). Total DNA was isolated from the entire brain, excluding the cerebellum, of 24 FbIR animals (4 for each of the six reperfusion times). A detailed description for genomic DNA isolation technique has been previously reported (P. K. Liu et al., 1996; Cui et al., 1999b). The purified DNA was stored in Tris—EDTA buffer [10 mM Tris HCl (pH 8.0), 1 mM EDTA] and was never exposed to phenol or chloroform before analysis. Fresh DNA (within 3 months of isolation) does not contain excess or measurable base lesions (P. K. Liu et al., 1996; Cui et al., 1999b). DNA from four animals at each reperfusion time point was pooled, and a total of 24 μg of DNA from each time point was tested for the presence of DNA base lesions sensitive to digestion by E. coli Fpg protein, which removes oh8dG lesions, as well as 5-hydroxycytosine and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (open ring of dG modification lesions).
We used an mRNA or cRNA probe transcribed from the cDNA clone of mouse c-fos gene in the presence of [α-32P]UTP (3,000 Ci/mmol; New England Nuclear, Wilmington, DE, U.S.A.) using T7 or T3 RNA polymerase (Promega Corp., Madison, WI, U.S.A.), respectively. These procedures have been described previously (P. K. Liu et al., 1996; Cui et al., 1999b). The autoradiogram was developed at -70°C for 16 h. Because DNA samples were pooled from four animals, we did not calculate the frequency of DNA base lesions.
Western blot
The nuclear extract from each sample (20 μg) was resolved in 10% sodium dodecyl sulfate—polyacrylamide gel electrophoresis and blot transferred to nitrocellulose membranes. The membranes were incubated with the polyclonal antibodies against hOGG1 (Novus Biologicals, Littleton, CO, U.S.A.) at a final concentration of 1.8 μg/ml for 1 h at room temperature. This was followed by incubation with an anti-rabbit horseradish peroxidase-conjugated antibody (1:10,000 dilution). For detection of the mOGG1 protein, we used the ECL western blotting detection kit (Amersham Pharmacia Biotech, Piscataway, NJ, U.S.A.). An autoradiograph was developed, and the signal intensity of the mOGG1 protein was quantified using AlphaImager 3.2 (Alpha Innotech, San Leandro, CA, U.S.A.). Preabsorption of the anti-hOGG1 antibody with its inducing synthetic peptide eliminated the protein bands detected in western blots. After mOGG1 protein detection, the nitrocellulose membranes were submerged in the stripping buffer [100 mM 2-mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl (pH 6.7)] for 30 min at 70°C. The stripped membranes were then reprobed with an anti-actin polyclonal antibody (0.4 μg/ml; Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.) and then with an anti-goat horseradish peroxidase-conjugated secondary antibody (1:20,000 dilution; Santa Cruz Biotechnology).
PCR
Total RNA was isolated and reverse-transcribed to cDNA (Cui et al., 1999a). A 159-bp cDNA was amplified using the forward (5′-ttggccaacaaagaactgga-3′) and backward (5′-ctagccctctggcctcttag-3′) primers for the mogg1a cDNA (Rosenquist et al., 1997). The primers were from the Sigma Genosys. The mogg1 cDNA was amplified for 32 cycles in a 25-μl PCR mix [20 pmol of each primer, 200 μM of each dNTP, 50 mM KCl, 25 mM Tris-HCl (pH 8.3), 1 mM MgCl2, 0.01% gelatin, 2 mM DTT, 0.6 U of Ampli Taq DNA polymerase] with a hot-start protocol in a thermocycler (Perkin Elmer Cetus) (Cui et al., 1999a). Each PCR cycle consisted of a denaturation step at 94°C for 45 s, an annealing step at 60°C for 30 s, and an extension step at 72°C for 60 s. The reaction was terminated by a 10-min extension at 72°C. Fifteen microliters of the resulting products was resolved in agarose gel (2%) using electrophoresis and stained by ethidium bromide (1 μg/ml of H2O). The identity of the PCR product was confirmed by direct sequencing using the dideoxy NTP sequencing method (Liu, 1993).
RESULTS
oh8dG-removing activity in nuclear extracts of mouse brain
The specificity for removal of oh8dG in the oligoZ duplexes with oh8dG opposite to a dC, dT, dA, or dG was examined in the nuclear extracts from mouse brains. One major cleavage product was detected along with three faint cleavage products (Fig. 1A). The major fragment had the same mobility as the cleavage product of E. coli endonuclease IV (11-mer with a 3′-OH end). Figure 1B shows that the nuclear extracts excised oh8dG with the greatest efficiency on the DNA duplex with oh8dG/dC (100%, or 20 fmol/mg of protein), with less efficiency on the DNA duplex with oh8dG/dT (28% of the efficiency on oh8dG/dC), and with little efficiency on oh8dG/dG (10% of the efficiency on oh8dG/dC) and oh8dG/dA (5% of the efficiency on oh8dG/dC). Increasing concentrations of MgCl2 in the reaction mix reduced this enzyme activity (data not shown). This specificity to remove oh8dG from oligoZ duplex suggests that OGG1 accounts for the primary activity in the nuclear extract (Monden et al., 1999).
FIG. 1.
Endogenous OGG1 activity from normal mouse brain. The specificity of endogenous OGG1 activity on its substrates was examined by incubating nuclear extracts from four non-FbIR brains (30 μg of protein per reaction, one animal per lane) with 100 fmol of 32P-labeled oligoZ duplexes with various bases in the complementary oligo DNA (i.e., dG, dA, dT, and dC opposite to oh8dG). Lane labeled with “O” was the uncut 22-mer containing oligoZ duplex without adding nuclear extracts. The AP site containing 32P-labeled oligoD duplex was incubated with 1 U of E. coli endonuclease IV and resolved in the lane labeled with “E4+D.” The results of A were quantitatively analyzed and presented in B.
Elevation of DNA base lesions in nuclear genes
We have previously used a fragment shift assay to show an elevation of DNA lesions in indicator gene coding for DNA polymerase β and actin from the mouse brain after cerebral ischemia—reperfusion (P. K. Liu et al., 1996). In the current study, we used the assay to show DNA lesions in another indicator gene (the c-fos gene). The signal intensities indicated the amount of intact c-fos gene. In the current assay, we noted that the signal intensity in samples without Fpg were weaker at early reperfusion time points than at late reperfusion time points (Fig. 2). This could be the result of strand breaks in the nuclear DNA of the mouse brain that occur during early reperfusion following 90 min of ischemia (Huang et al., 2000). Because we were interested in knowing if nuclear genes contained base modification damage, we compared samples with and without Fpg protein at each reperfusion time point. A decrease in intensity in the sample with Fpg protein treatment suggests the presence of Fpg protein-sensitive sites (FPGSS). Figure 2 shows that the intensity of the c-fos gene treated with Fpg protein was decreased during the first 10–30 min of reperfusion after 90 min of ischemia, indicating the presence of increasing FPGSS in the c-fos gene. At 30 min of reperfusion, we observed no signal in either strand of the mouse c-fos gene with Fpg protein treatment (Fig. 2). Therefore, the c-fos gene at 30 min of reperfusion contained at least one hit per strand, that is, two base lesions per gene. We observed the reappearance of the c-fos signal in DNA samples at 60 and 120 min after 90 min of ischemia. Our results suggest that a resistance to Fpg protein, or a reduction of DNA base lesions, occurs beginning between 30 and 60 min of reperfusion. Therefore, the data suggest an active DNA repair process in the mouse brain with FbIR.
FIG. 2.

DNA lesions detected as FPGSS in the c-fos gene. DNA base lesions were detected in genomic DNA from 24 mouse brains that underwent 90 min of ischemia and various durations (min, except 2 h) of reperfusion as indicated on top of each lane. Each lane contains DNA from a pool of four animals having the same reperfusion time. The Fpg protein-treated DNA samples (+) were resolved in parallel with the same DNA samples without Fpg protein treatment (-). The blot was hybridized to a cRNA probe (for the nontranscribed strand) from the c-fos gene, and the autoradiagraph was developed as previously described (P. K. Liu et al., 1996; Cui et al., 1999b). The blot was then boiled to remove 32P-cRNA and was hybridized to an mRNA probe of the same gene. The signal intensities in the normal brain are similar to those at the end of ischemia, that is, “0” time point (P. K. Liu et al., 1996), and are not shown.
Increase in nuclear OGG1 activities after FbIR
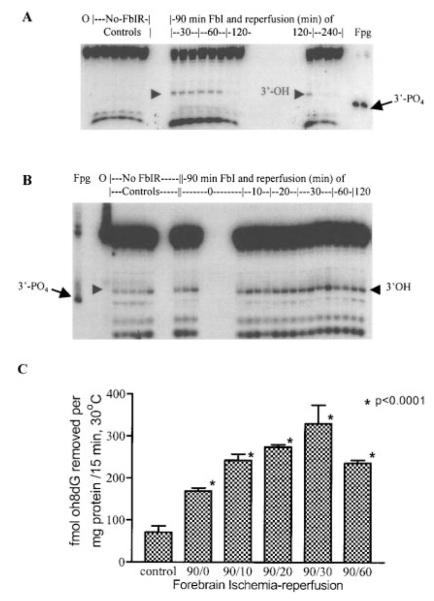
The gradual increase of FPGSS in the c-fos gene before 30 min of reperfusion, followed by a decrease of DNA damage observed at 60 min of reperfusion, suggests that DNA lesions can be repaired after cerebral ischemia. To determine if the observed reappearance of the c-fos gene at 60 and 120 min of reperfusion after FbIR correlated with an increase in the base excision repair activity in the nucleus, we examined the level of mOGG1 activity in nuclear extracts isolated from normal and ischemic mouse brains. The endogenous nuclear OGG1 activity was measured as the ability of the nuclear extracts to nick the oligoZ duplex containing a single oh8dG lesion opposite the dC base. We chose to use the oligoZ duplex containing an oh8dG/dC base pair for this experiment because FPGSS (i.e., oh8dG) have been detected after oxidative stress in rat and mouse brains (J. Liu et al., 1996; P. K. Liu et al., 1996; Cui et al., 1999b). One major cleavage product of 11-mer and several less intense products were detected (Fig. 3). The major OGG1 product did not appear to have the same mobility as the cleavage product using E. coli Fpg protein (3′-PO4 ends; Fig. 3). Moreover, that the three less intense fragments present in samples after mouse nuclear extracts were smaller than the 11-mer product suggests that enzymes other than mOGG1 may have further digested the product. Furthermore, the nuclear extracts did not significantly cleave the DNA duplex containing dG/dC under the condition in which OGG1 excised oh8dG from the oligoZ duplex (data not shown). We found that the endogenous nuclear OGG1 activity was significantly higher in all FbIR groups than in the non-FbIR control groups. The peak activity occurred with 90 min of ischemia followed by 30 min of reperfusion; a two- to threefold increase over the control level was observed (Fig. 3C). The elevated nuclear OGG1 activity was observed in samples from individual animals (Fig. 3B) as well as in pooled samples from two animals (Fig. 3A), except at the longer reperfusion times of 120 and 240 min. When oligoZ was replaced with oligoD, which contains one AP site in the place of oh8dG, we found that the AP site-removing activity was already high in the normal brain, and that there was no significant increase in the ischemic brain (data not shown).
FIG. 3.
Activation of endogenous OGG1 activities in the nuclear extracts of mouse brain after FbIR. The endogenous nuclear OGG1 activity was measured by incubating 100 fmol of 32P-labeled oligoZ (oh8dG/dC) duplex with 30 μg of nuclear protein from the control (sham-operated without FbIR) and FbIR animals. The oligoZ duplex incubated with Fpg protein (lane marked “Fpg,” 1 U/assay) or without enzymes (lane labeled as “O”) was also included in each experiment. Results from control animals (n = 6) and FbIR animals (n = 3 each for 90/30, 90/60, 90/120, and 90/240) are shown in A (reaction in the sample from one animal per lane). Results from a second experiment in which pooled nuclear extracts from two animals were used in each reaction and then resolved in each lane are presented in B. A total of 46 animals were used in B: 12 animals in the control group, 6 animals each with FbIR of 90/0, 90/10, or 90/20, 8 animals with FbIR of 90/30, and 4 animals each with FbIR of 90/60 or 90/120. Arrowheads indicate the positions of the major 11-mer digested products of the mouse nuclear extracts; arrows indicate the 11-mer with 3′-PO4 end (product of Fpg protein). The results in B (excluding the data at 120 min due to the variation in A) were quantitatively analyzed and presented in C. Asterisks indicate statistically significant (p < 0.001) differences from the control samples (one-way ANOVA followed by Tukey analysis).
Increase of idiotype of OGG1 from ischemic mouse brain after FbIR
To address whether DNA repair activity correlates with an increase in the amount of protein in the nuclear extract, we examined the levels of the mOGG1 protein in the nuclear extracts using an antibody raised against human OGG1 protein. The synthetic peptide used to induce the OGG1 antibody was derived from the hOGG1 amino acids 65–79 and shared high homology with the corresponding sequences in mOGG1 protein. Using the anti-hOGG1 antibody, we detected one protein with an apparent molecular mass of 40 kDa in nuclear extracts from mouse brains (Fig. 4A). This peptide had a molecular mass similar to the recombinant OGG1 protein, which had a molecular mass of 39–40 kDa (Prieto Alamo et al., 1998; Monden et al., 1999). The 40-kDa protein signal could be eliminated by preabsorption of the anti-hOGG1 antibody with its inducing peptide (Fig. 4A). Moreover, we found that the level of the 40-kDa protein was significantly higher in animals that received 90 min of ischemia and 20 or 30 min of reperfusion than in the non-FbIR control ( p < 0.05; Fig. 4A and B). On the other hand, the levels of the γ-actin gene in the same blot were the same in control and ischemic brain samples (Fig. 4B and C).
FIG. 4.
Increase of mOGG1 protein in nuclear extracts of the brain after FbIR. Nuclear extracts (20 μg of protein per animal per lane) from control animals and animals with FbIR of 90/0, 90/20, or 90/30 were resolved in sodium dodecyl sulfate—polyacrylamide gel electrophoresis (A and B) and analyzed using anti-hOGG1 polyclonal antibodies in western blot. In A, an additional blot was probed with anti-hOGG1 antibodies that had been preadsorbed with its inducing synthetic peptide. In another experiment, the nitrocellulose membrane was stripped after mOGG1 protein detection and then reprobed with anti-actin antibodies (B, bottom). The level of mOGG1 protein in B was analyzed using the AlphaImager and was found to be significantly (t test, p < 0.05) increased in the 90/30 FbIR group compared with the control (C).
To determine if the elevation of the mOGG1 peptide was correlated with activation of the mogg1 mRNA transcript, we measured the expression of mogg1 mRNA in 12 animals before and after FbIR. We observed no increase of the mogg1 mRNA level in 12 FbIR animals using RT-PCR and primers that amplified a 159-bp fragment on the 3′ terminus of the mogg1 cDNA (Fig. 5). The data suggest that no activation of mogg1 mRNA transcription occurs in the mouse brain after FbIR.
FIG. 5.

Expression of mOGG1 mRNA in nuclear extracts of the brain after FbIR. A 159-bp mOGG1 cDNA from the cortices of 12 animals (2 each for control lanes 1–2 and 90/0, 90/30, 90/60, 90/120, or 90/240 FbIR lanes 3–12, respectively) was individually amplified by RT-PCR and resolved in agarose gel (2%).
DISCUSSION
In the current study, we found that the temporal up-regulation in oh8dG repair activity correlated positively with the appearance of oxidative damage on the nuclear genes. We also observed that the increase in the OGG1 protein (40-kDa peptide), detected using anti-hOGG1 antibodies, correlated positively with an elevation of base excision repair activity in the mouse brains after ischemia—reperfusion treatment. Base excision repair activity in the mouse brain has a substrate preference order of oh8dG/dC > oh8dG/dT > oh8dG/dG ≥ oh8dG/dA, suggesting that the repair activity for oh8dG detected in the mouse brain is most likely from mOGG1. The data suggest that this enhanced nuclear mOGG1 activity may be secondary to the increase of oh8dG damage in the nuclear genes of the brain. In addition, the up-regulated mOGG1 activity is likely to account for the observed disappearance of base lesions on nuclear genes (DNA polymerase β, actin, and c-fos genes), especially after 30 min of reperfusion (P. K. Liu et al., 1996; Cui et al., 1999b).
Three different DNA repair enzymes, mutT, mutY, and mutM, have been found to be involved in the removal of the majority of oh8dG in E. coli. Mammalian homologues of mutT, mutY, and mutM have been cloned; they are named 8-oxo-dGTPase, MYH, and OGG1/MMH, respectively (Sakumi et al., 1993; McGoldrick et al., 1995; Roldan-Arjona et al., 1997). 8-Oxo-dGTPase, which hydrolyzes free oh8dGTP nucleotides in the dNTP pool, prevents incorporation of oh8dGTP nucleotides into the DNA patch during excision repair. MYH protein is a DNA mismatch glycosylase that cleaves adenines from dA/oh8dG, dA/dG, and dA/dC mismatches. MutM, usually known as Fpg protein in E. coli, is another DNA glycosylase. Fpg protein cleaves oh8dG from DNA with oh8dG/dC, oh8dG/dT, or oh8dG/dG lesions in addition to other lesions such as FappyG and oh5dC (Rabow and Kow, 1997). Although the OGG1/MMH protein is the mammalian equivalent of E. coli Fpg protein, it is different from Fpg protein in terms of structure and substrate specificity. Unlike Fpg, the OGG1 protein selectively cleaves the DNA duplex containing the oh8dG/dC pair and acts only weakly on oh8dG/dT, oh8dG/dG, and oh8dG/dA (Roldan-Arjona et al., 1997; Rosenquist et al., 1997; Prieto Alamo et al., 1998). Although we did not purify nuclear mOGG1 from the mouse brain, the substrate preference shown by the mOGG1 in the nuclear extract of mouse brain closely agreed with the substrate preference of hOGG1. This finding is supported by previous studies that demonstrated that the human OGG1/MMH type 1a protein is the major enzyme responsible for the endogenous OGG activity detected in the whole-cell extract of human cells (Aburatani et al., 1997; Monden et al., 1999).
Furthermore, the increase in OGG1 activity and the amount of OGG1 peptide in nuclear extracts from mouse brain after FbIR suggest that the mOGG1 protein may play a significant role in base excision repair after FbIR. At least seven hOGG1 mRNAs and peptides (i.e., type 1a, 1b, 2a, 2b, 2c, 2d, and 2e) have been identified as a result of alternative splicing (Kohno et al., 1998; Nishioka et al., 1999). All seven OGG peptides contain a putative mitochondria-targeting signal on the N-terminus. Type 1a of OGG1 protein also has a unique nuclear location signal on its C-terminus. The type 1a OGG1 peptide has an apparent molecular mass of 36–40 kDa (Aburatani et al., 1997; Prieto Alamo et al., 1998; Monden et al., 1999; Nishioka et al., 1999). We detected one 40-kDa peptide in mouse brain nuclear extracts using antibodies induced by a synthetic peptide derived from hOGG1. The elimination of the 40-kDa peptide signal after preabsorption of hOGG1 antibodies with the inducing synthetic peptide suggests that this nuclear protein from the mouse brain is a homologue of hOGG1.
The increases in mOGG1 activity and its protein in nuclear extracts after the FbIR treatment coincide with the elevation in DNA base lesions found in the nuclear genes of the brain. We attempted to detect if there was an increase in the expression of mogg mRNA by amplifying a constant region of mogg mRNA using RT-PCR, but we observed no significant increase in the ischemic brain compared with the control brain. The accumulation of the mOGG1 protein in brain nuclei after FbIR treatment is unlikely to be due to the transcription activation. Moreover, we detected very little OGG activity in the cytoplasmic fraction. Thus, it is likely that the increase in mOGG1 protein in the nucleus is a result of an alternative splicing and/or posttranslational modification after FbIR (Nishioka et al., 1999). We are currently carrying out experiments to address this issue.
Increases in enzymes that are associated with DNA repair activity after oxidative stress have been reported (Fornance et al., 1992). Recent studies suggest that the base excision repair pathway may utilize enzymes, such as poly(ADP-ribose) polymerase, in addition to the glycosylases and proteins that are involved in nucleotide excision repair, such as the proliferating cell nuclear antigen (Wilson and Thompson, 1997). This alternative base excision repair pathway probably repairs oh8dG and could be defective in humans with certain genetic defects, as described by Reardon et al. (1997). Several previous studies have shown that the levels of DNA repair proteins such as APE/Ref-1, GADD45, poly-(ADP-ribose) polymerase, and Ku80 are either up- or down-regulated in ischemic brain (Jin et al., 1996; Gillardon et al., 1997; Love et al., 1998; Fujimura et al., 1999; Kawase et al., 1999; Shackelford et al., 1999), in brain injury (LaPlaca et al., 1999), and in cell culture (Fornance et al., 1992). The direction of regulation appears to depend on the cell types. For instance, APE/Ref-1 protein in the nucleus is increased in the granular cells of the ischemia-resistant dentate gyrus but decreased in the pyramidal cells of the ischemia-vulnerable CA1 area (Gillardon et al., 1997; Kawase et al., 1999). We have also shown that the cells that die as a result of oxidative DNA damage are mostly nonastrocytes (neurons), even when the astrocytes and neurons contain the same intensity of oxidative DNA damage immediately following 90 min of forebrain ischemia (Cui et al., 2000; Huang et al., 2000). These findings suggest that either astrocytes are more resistant to oxidative DNA damage or astrocytes use a different and more efficient pathway for DNA repair. A better understanding of DNA damage and repair in the brain may yield new and important information concerning the mechanisms of neuronal death and provide opportunities for the development of novel and effective means to reduce or prevent CNS injury.
Acknowledgment
This work was supported in part by an Established Investigator Award (9640202N) from the American Heart Association and by a grant from NINDS (NS34810).
Abbreviations used
- AP
apyrimidinic/apurinic
- DTT
dithiothreitol
- FbIR
forebrain ischemia—reperfusion
- Fpg
formamidopyrimidine DNA N-glycosylase
- FPGSS
Fpg protein-sensitive sites
- OGG1
hOGG1, and mOGG1, 8-oxoguanine glycosylase/AP lyase and human and mouse forms, respectively
- oh8dG
8-hydroxy-2′-deoxyguanosine
- oh8G
8-hydroxyguanosine
- PMSF
phenylmethylsulfonyl fluoride.
REFERENCES
- Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M. Cloning and characterization of mammalian 8-hydroxyguaninespecific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- An G, Lin T, Liu JS, Xue JJ, He YY, Hsu CY. Expression of c-fos and c-jun family genes after focal cerebral ischemia. Ann. Neurol. 1993;33:457–464. doi: 10.1002/ana.410330508. [DOI] [PubMed] [Google Scholar]
- Bessho T, Tano K, Kasai H, Ohtsuka E, Nishimura S. Evidence for two DNA repair enzymes for 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) in human cells. J. Biol. Chem. 1993;268:19416–19421. [PubMed] [Google Scholar]
- Bhagwat M, Gerlt JA. 3′-and 5′-strand cleavage reactions catalyzed by the Fpg protein from Escherichia coli occur via successive β- and δ-elimination mechanisms, respectively. Biochemistry. 1996;35:659–665. doi: 10.1021/bi9522662. [DOI] [PubMed] [Google Scholar]
- Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- Chen J, Jin K, Chen M, Pei W, Kawaguchi K, Greenberg DA, Simon RP. Early detection of DNA strand breaks in the brain after transient focal ischemia: implications for the role of DNA damage in apoptosis and neuronal cell death. J. Neurochem. 1997;69:232–245. doi: 10.1046/j.1471-4159.1997.69010232.x. [DOI] [PubMed] [Google Scholar]
- Coyle JT, Puttfarcken P. Oxidative stress, glutamate, and neurodegenerative disorders. Science. 1993;262:689–694. doi: 10.1126/science.7901908. [DOI] [PubMed] [Google Scholar]
- Croteau DL, Rhys CMJ, Hudson EK, Dianov GL, Hansford RG, Bohr VA. An oxidative damage-specific endonuclease from rat liver mitochondria. J. Biol. Chem. 1997;272:27338–27344. doi: 10.1074/jbc.272.43.27338. [DOI] [PubMed] [Google Scholar]
- Cui JK, Hsu CY, Liu PK. Suppression of postischemic hippocampal NGF expression by a c-fos antisense oligodeoxynucleotide. J. Neurosci. 1999a;19:1335–1344. doi: 10.1523/JNEUROSCI.19-04-01335.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Holmes EH, Liu PK. Oxidative damage to the c-fos gene and reduction of its transcription after focal cerebral ischemia. J. Neurochem. 1999b;73:1164–1174. doi: 10.1046/j.1471-4159.1999.0731164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JK, Holmes EH, Cao S, Greene TG, Liu PK. In situ detection of Fpg protein sensitive sites in the rat brain after focal cerebral ischemia—reperfusion. FASEB J. 2000 (in press) [Google Scholar]
- Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- Driggers WJ, LeDoux SP, Wilson GL. Repair of oxidative damage within the mitochondrial DNA of RINr 38 cells. J. Biol. Chem. 1993;268:22042–22045. [PubMed] [Google Scholar]
- Fornance AJ, Jackman J, Hollander MC, Hoffman-Liebermann B, Liebermann DA. Genotoxic-stress-response genes and growth-arrest genes. Ann. NY Acad. Sci. 1992;663:139–159. doi: 10.1111/j.1749-6632.1992.tb38657.x. [DOI] [PubMed] [Google Scholar]
- Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J. Biol. Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- Fujii M, Hara H, Meng W, Vonsattel JP, Huang Z, Moskowitz MA. Strain-related differences in susceptibility to transient forebrain ischemia in SV-129 and C57black/6 mice. Stroke. 1997;28:1805–1810. doi: 10.1161/01.str.28.9.1805. [DOI] [PubMed] [Google Scholar]
- Fujimura M, Morita-Fujimura Y, Kawase M, Chan PH. Early decrease of apurinic/apyrimidinic endonuclease expression after transient focal cerebral ischemia in mice. J. Cereb. Blood Flow Metab. 1999;19:495–501. doi: 10.1097/00004647-199905000-00003. [DOI] [PubMed] [Google Scholar]
- Gillardon F, Bottiger B, Hossmann K-A. Expression of nuclear redox factor ref-1 in the rat hippocampus following global ischemia induced by cardiac arrest. Mol. Brain Res. 1997;52:194–200. doi: 10.1016/s0169-328x(97)00237-4. [DOI] [PubMed] [Google Scholar]
- Huang D, Shenoy A, Huang W, Cao S, Cui J, Liu PK. Nitric oxide mediates oxidative DNA damage in cell nuclei and neuronal death in cerebral cortex after experimental forebrain ischemia. FASEB J. 2000 (in press) [Google Scholar]
- Jin K, Chen J, Kawaguchi K, Zhu RL, Stetler RA, Simon RP, Graham SH. Focal ischemia induces expression of the DNA damage-inducible gene GADD45 in the rat brain. Neuroreport. 1996;7:1797–1802. doi: 10.1097/00001756-199607290-00022. [DOI] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Hizi A, Tabone J, Solomon MS, Loeb LA. Inefficient repair of RNA DNA hybrids. Eur. J. Biochem. 1997;250:492–501. doi: 10.1111/j.1432-1033.1997.0492a.x. [DOI] [PubMed] [Google Scholar]
- Kasprzak KS, Diwan BA, Rice JM, Misra M, Riggs CW, Olinski R, Dizdaroglu M. Nickel(II)-mediated oxidative DNA base damage in renal and hepatic chromatin of pregnant rats and their fetuses. Possible relevance to carcinogenesis. Chem. Res. Toxicol. 1992;5:809–815. doi: 10.1021/tx00030a013. [DOI] [PubMed] [Google Scholar]
- Kawase M, Fujimura M, Morita-Fujimura Y, Chan PH. Reduction of apurinic/apyrimidinic endonuclease expression after transient global cerebral ischemia in rats: implication of the failure of DNA repair in neuronal apoptosis. Stroke. 1999;30:441–448. doi: 10.1161/01.str.30.2.441. [DOI] [PubMed] [Google Scholar]
- Kisby GE, Milne J, Sweatt C. Evidence of reduced DNA repair in amyotrophic lateral sclerosis brain tissue. Neuroreport. 1997;8:1337–1340. doi: 10.1097/00001756-199704140-00004. [DOI] [PubMed] [Google Scholar]
- Kohno T, Shinmura K, Tosaka M, Tani M, Kim SR, Sugimura H, Nohmi T, Kasai H, Yokota J. Genetic polymorphisms and alternative splicing of the hOGG1 gene, that is involved in the repair of 8-hydroxyguanine in damaged DNA. Oncogene. 1998;16:3219–3225. doi: 10.1038/sj.onc.1201872. [DOI] [PubMed] [Google Scholar]
- Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997;325:1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlaca MC, Raghupathi R, Verma A, Pieper AA, Saatman KE, Snyder SH, McIntosh TK. Temporal patterns of poly(ADP-ribose) polymerase activation in the cortex following experimental brain injury in the rat. J. Neurochem. 1999;73:205–213. doi: 10.1046/j.1471-4159.1999.0730205.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang X, Shigenage MK, Yeo HC, Mori A, Ames BN. Immobilization stress causes oxidative damage to lipid, protein and DNA in the brain of rats. FASEB J. 1996;10:1532–1538. [PubMed] [Google Scholar]
- Liu PK. Enhanced expression of alpha-type DNA polymerase genes reduces AZT cytotoxicity in hamster tr5 cells. Somat. Cell Mol. Genet. 1993;19:211–220. doi: 10.1007/BF01233069. [DOI] [PubMed] [Google Scholar]
- Liu PK, Salminen A, He YY, Jiang MH, Xue JJ, Liu JS, Hsu CY. Suppression of ischemia-induced Fos expression and AP-1 activity by an antisense oligodeoxynucleotide to c-fos mRNA. Ann. Neurol. 1994;36:566–576. doi: 10.1002/ana.410360405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PK, Hsu CY, Dizdaroglu M, Floyd RA, Kow YW, Karakaya A, Rabow LE, Cui JK. Damage, repair, and mutagenesis in nuclear genes after mouse forebrain ischemia—reperfusion. J. Neurosci. 1996;16:6795–6806. doi: 10.1523/JNEUROSCI.16-21-06795.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love S, Barber R, Wilcock GK. Apoptosis and expression of DNA repair proteins in ischemic brain injury in man. Neuroreport. 1998;9:955–959. doi: 10.1097/00001756-199804200-00001. [DOI] [PubMed] [Google Scholar]
- Lovell MA, Gabbita SP, Markesbery WR. Increased DNA oxidation and decreased levels of repair products in Alzheimer’s disease ventricular CSF. J. Neurochem. 1999;72:771–776. doi: 10.1046/j.1471-4159.1999.0720771.x. [DOI] [PubMed] [Google Scholar]
- McGoldrick JP, Yeh YC, Solomon M, Essigmann JM, Lu AL. Characterization of a mammalian homolog of the Escherichia coli MutY mismatch repair protein. Mol. Cell. Biol. 1995;15:989–996. doi: 10.1128/mcb.15.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Monden Y, Arai T, Asano M, Ohtsuka E, Aburatani H, Nishimura S. Human MMH (OGG1) type 1a protein is a major enzyme for repair of 8-hydroxyguanine lesions in human cells. Biochem. Biophys. Res. Commun. 1999;258:605–610. doi: 10.1006/bbrc.1999.0649. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Ohtsubo T, Oda H, Fujiwara T, Kang D, Sugimachi K, Nakabeppu Y. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol. Biol. Cell. 1999;10:1637–1652. doi: 10.1091/mbc.10.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunomura A, Perry G, Pappolla MA, Wade R, Hirai K, Chiba S, Smith MA. RNA oxidation is a prominent feature of vulnerable neurons in Alzheimer’s disease. J. Neurosci. 1999;19:1959–1964. doi: 10.1523/JNEUROSCI.19-06-01959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parshad R, Sanford KK, Price FM. Fluorescence light-induced chromatid breaks distinguish Alzheimer disease cells from normal cells in tissue culture. Proc. Natl. Acad. Sci. USA. 1996;93:5146–5150. doi: 10.1073/pnas.93.10.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo M. J. Prieto, Jurado J, Francastel E, Laval F. Rat 7,8-dihydro-8-oxoguanine DNA glycosylase: substrate specificity, kinetics and cleavage mechanism at an apurinic site. Nucleic Acids Res. 1998;26:5199–5202. doi: 10.1093/nar/26.22.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabow LE, Kow YW. Mechanism of action of base release by Escherichia coli Fpg protein: role of lysine 155 in catalysis. Biochemistry. 1997;36:5084–5096. doi: 10.1021/bi963005a. [DOI] [PubMed] [Google Scholar]
- Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurode-generation in xeroderma pigmentosum patients. Proc. Natl. Acad. Sci. USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J, Brumback RA, Polinsky RJ, Wirtschafter JD, Tarone RE, Scudiero DA, Otsuka F. Hypersensitivity to DNA-damaging agents in abiotrophies: a new explanation for degeneration of neurons, photoreceptors, and muscle in Alzheimer, Parkinson, and Huntington disease, retinitis pigmentosa, and Duchenne muscular dystrophy. Basic Life Sci. 1985;35:315–344. doi: 10.1007/978-1-4899-2218-2_20. [DOI] [PubMed] [Google Scholar]
- Roldan-Arjona T, Wei YF, Carter KC, Klungland A, Anselmino C, Wang RP, Augustus M, Lindahl T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl. Acad. Sci. USA. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist TA, Zharkov DO, Grollman AP. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl. Acad. Sci. USA. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakumi K, Furuichi M, Tsuzuki T, Kakuma T, Kawabata S, Maki H, Sekiguchi M. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem. 1993;268:23524–23530. [PubMed] [Google Scholar]
- Shackelford DA, Tobaru T, Zhang S, Zivin JA. Changes in expression of the DNA repair protein complex DNA-dependent protein kinase after ischemia and reperfusion. J. Neurosci. 1999;19:4727–4738. doi: 10.1523/JNEUROSCI.19-12-04727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taffe BG, Larminat F, Laval J, Croteau DJ, Anson RM, Bohr VA. Gene-specific nuclear and mitochondrial repair of formamidopyrimidine DNA glycosylase-sensitive sites in Chinese hamster ovary cells. Mutat. Res. 1996;364:183–192. doi: 10.1016/s0921-8777(96)00031-6. [DOI] [PubMed] [Google Scholar]
- Vos JMH. DNA Repair Mechanism: Impact on Human Diseases and Cancer. R. G. Landes; Austin: 1995. [Google Scholar]
- Wilson DM, Thompson LH. Life without DNA repair. Proc. Natl. Acad. Sci. USA. 1997;94:12754–12757. doi: 10.1073/pnas.94.24.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]