Abstract
Background
Long-QT syndrome (LQTS) is an inherited disorder associated with sudden cardiac death. The cytoskeletal protein syntrophin-α1 (SNTA1) is known to interact with the cardiac sodium channel (hNav1.5), and we hypothesized that SNTA1 mutations might cause phenotypic LQTS in patients with genotypically normal hNav1.5 by secondarily disturbing sodium channel function.
Methods and Results
Mutational analysis of SNTA1 was performed on 39 LQTS patients (QTc≥480 ms) with previously negative genetic screening for the known LQTS-causing genes. We identified a novel A257G-SNTA1 missense mutation, which affects a highly conserved residue, in 3 unrelated LQTS probands but not in 400 ethnic-matched control alleles. Only 1 of these probands had a preexisting family history of LQTS and sudden death with an additional intronic variant in KCNQ1. Electrophysiological analysis was performed using HEK-293 cells stably expressing hNav1.5 and transiently transfected with either wild-type or mutant SNTA1 and, in neonatal rat cardiomyocytes, transiently transfected with either wild-type or mutant SNTA1. In both HEK-293 cells and neonatal rat cardiomyocytes, increased peak sodium currents were noted along with a 10-mV negative shift of the onset and peak of currents of the current-voltage relationships. In addition, A257G-SNTA1 shifted the steady-state activation (Vh) leftward by 9.4 mV, whereas the voltage-dependent inactivation kinetics and the late sodium currents were similar to wild-type SNTA1.
Conclusion
SNTA1 is a new susceptibility gene for LQTS. A257G-SNTA1 can cause gain-of-function of Nav1.5 similar to the LQT3.
Keywords: arrhythmia, death, sudden (if surviving, use heart arrest), ion channels, long-QT syndrome
Long-QT syndrome (LQTS) is an inherited disorder that can cause sudden cardiac death. To date, hundreds of genetic mutations and single nucleotide polymorphisms in 11 distinct LQTS-susceptibility genes have been reported. Electrophysiological studies using in vitro cell expression systems and genetically engineered animal models have suggested that “gain of function” or “loss of function” in the ion channels that are essential to generate action potentials may account for the LQTS phenotypes.1 The functional modifications of the ion channels are primarily caused by defects in the genes encoding the pore-forming subunits (KCNQ1, KCNH2, SCN5A, KCNJ2, and CACNA1C) or the β subunits (KCNE1, KCNE2, and SCN4B) of the ion channels except for the gene responsible for LQT4.2 Unlike the other genes, LQT4 is caused by the malfunction of ankyrin B that is involved in the cellular organization of the sodium/calcium exchanger and inositol-1,4,5-triphosphate receptors.3
However, in 20% to 30% of cases, genetic analysis fails to identify the responsible gene for the LQTS phenotypes in affected patients.4 Recently, Vatta et al and Cronk et al reported that mutations in caveolin-3 identified in the patients with LQTS or the infants succumbing to sudden infant death syndrome can affect human cardiac sodium channel (hNav1.5) gating kinetics and generate sustained currents, probably by direct protein–protein interactions.5,6 In addition, mutations in the A-kinase anchoring protein 9 gene (AKAP9) have been identified in LQTS.7 AKAP9 determines the subcellular localization of protein kinase A and the phosphorylation of the IKs potassium channel α subunit (KCNQ1) to which it assembles.8 These studies proposed a novel concept that the defects of non–ion channel proteins or channel-interacting proteins can affect ion-channel gating kinetics, thereby causing secondary channel dysfunction leading to LQTS. Hence, this concept uncovers a cascade or domino effect that disturbs the “final common pathway”9 that causes arrhythmias, ion channels, and focuses attention on a novel class of proteins and candidate genes to explain the residual 25% of LQTS that remain genotype negative.
Syntrophins are cytoplasmic submembranous proteins that are components of the dystrophin-associated protein complex.7 Several syntrophin isoforms (α1, β1, β2, γ1, and γ2) have been identified in the heart, skeletal muscle, and neurons.10–14 The PDZ domain of syntrophin-α1 (SNTA1), the most abundant isoform in the heart, has been reported to bind to the C-terminal domain of murine cardiac voltage-gated sodium channels (SkM2).15 Ou et al demonstrated that syntrophin-γ2 (SNTG2) affects hNav1.5 gating kinetics by protein–protein interaction via PDZ [postsynaptic density protein (PSD95), drosophila disc large tumor suppressor (DlgA), and zonula occludens-1 protein (zo-1)] domain in the distal C-terminus of the SCN5A-encoded sodium channel pore-forming subunit.16 Notably, SNTG2 shares structural similarity with SNTA1. These observations led us to hypothesize that SNTA1 might be a new candidate gene responsible for the LQTS in patients whose genetic screenings were negative for the already known subtypes operating by secondary disruption of sodium channel function.
Methods
Patients Demographics and Genetic Screening
We previously enrolled 364 unrelated probands clinically diagnosed with LQTS.5 Among them, 39 genotype-negative LQTS patients (26 females; 66.7%) presenting with a resting QTc≥480 ms and a LQTS diagnostic score ≥3 were selected for further investigation.17 All patients underwent physical examination, family history, and ECG analysis. The average age at diagnosis was 23.6±6.3 years (range, 1–84 years), and the average QTc was 537±19 ms (range, 480–670 ms).
Blood was obtained after written informed consent was obtained from all subjects. Genomic DNA was extracted from peripheral blood lymphocytes as previously described.5 Using polymerase chain reaction and direct DNA sequencing, open reading frame/splice site mutational analysis was performed on the SNTA1 gene (chromosome 20q11.2; 8 exons). Polymerase chain reaction amplification was performed using primers designed to flank the regions of this gene. All patients were, also, screened for the known LQTS-susceptibility genes (LQT1–9). All SNTA1 genetic variants regarded as putative LQTS-associated mutations were required to change a conserved residue or splice site, altering the primary amino acid structure of the encoded protein. In addition, these genetic variants were required to be absent in at least 400 ethnic-matched reference alleles to be considered as mutations. Synonymous single nucleotide polymorphisms were excluded from consideration.
SNTA1 Gene Synthesis and Mutagenesis
Wild-type (WT) human SNTA1 was synthesized based on the previously deposited sequence (GenBank Accession No. NM_003098; GenScript Corporation, Piscataway, NJ) and subcloned into the pcDNA3.1/CT-GFP-TOPO vector (Invitrogen, Carlsbad, CA). Site-directed mutagenesis was performed with the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) by using the vector containing the WT-SNTA1 as a template. The primers used for the mutagenesis are available on request. Polymerase chain reaction and bacteria transformations were performed according to the manufacturer’s instructions. The mutated A257G-SNTA1 clones were sequenced to ensure the presence of the mutation and the absence of other substitutions introduced by DNA polymerase.
HEK-293 Cell Preparation and Transient Expression of WT and Mutant SNTA1
Stable HEK-293 cell lines expressing consistent sodium currents were established from a single cell transfected with the vector containing Flag-tagged hNav1.5 (clone cells). The clone cells were transiently transfected with the vector containing WT- or A257G-SNTA1 using the Lipofectamine 2000 transfection reagent (Qiagen, Valencia, CA). To express β1-subunit of human cardiac sodium channel (hβ1), the clone cells were transiently transfected with the pIRES vector carrying hβ1 and CD8 (kindly provided by Dr. Naomasa Makita, Hokkaido University, Sapporo, Japan) in conjunction with SNTA1. The cells were incubated at 37°C for 2 to 3 days before use.
Neonatal Rat Cardiomyocyte Isolation and Transient Expression of WT and Mutant SNTA1
All procedures were approved by the Institutional Animal Care and Use Committee at the Texas Heart Institute. Neonatal rat cardiomyocytes were isolated according to the procedures described elsewhere (see Methods in the supplemental material).
Patch-Clamp Experiments
Patch-clamp experiments were performed as previously described.18 Macroscopic sodium currents were recorded at ambient temperature (22 to 24°C). Step-pulse voltages were generated with Axopatch 200B amplifier using pClamp9.0 software (Axon Instruments, Sunnyvale, CA). Currents were filtered at 10 kHz with a built-in 4-pole Bessel filter and fed to a computer at a sampling frequency of 20 kHz. (The details are described in the supplemental material.)
Coimmunoprecipitation
Coimmunoprecipitation was performed in HEK-293 cells stably expressing Flag-tagged hNav1.5 and transfected with either green fluorescent protein (GFP)-tagged WT-SNTA1 or A257G-SNTA1 as previously reported (see Methods in the supplemental material).5
Immunohistochemistry
Immunohistochemical staining was performed by standard techniques described elsewhere. Briefly, the HEK-293 cells stably expressing hNav1.5 were transfected with GFP-tagged WT-SNTA1. The cells were fixed with 4% paraformaldehyde, incubated with 0.5% Triton X-100, then sequentially stained with anti-SCN5A sodium channel antibody (1:100 dilution, Santa Cruz sc-23174) for 1 hour at room temperature, biotinylated antigoat IgG (1:100 dilution, VECTOR BA-2000) for 30 minutes, and antistreptavidin conjugated with Texas Red (1:100 dilution, ZYMED 43 to 4317) for 30 minutes. The results were examined with TCS-SP2 confocal laser-scanning microscope (Leica Microsystems).
Data Analysis
The sodium channel gating kinetics were analyzed using Clampfit (Axon Instruments, Sunnyvale, CA) and Igor software (Wavemetrics, Lake Oswego, OR). Data were presented as mean ± SE (median), and comparisons were made using nonparametric test (Mann-Whitney test) with P<0.05 considered significant. The authors had full access to and take full responsibility for the integrity of the data. All authors have read and agree to the manuscript as written.
Results
Clinical Evaluation and Mutation Analysis
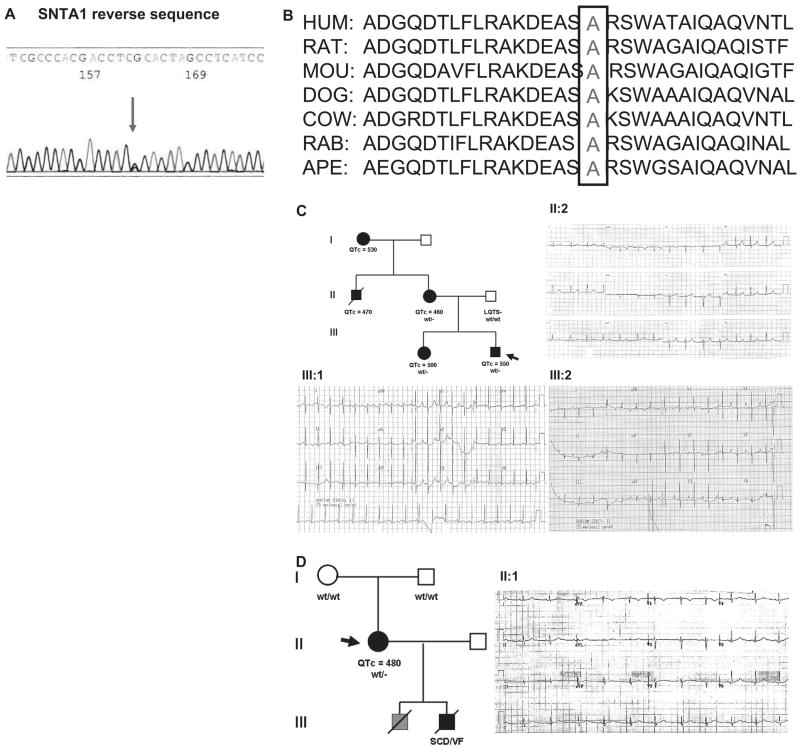
Mutational analysis of SNTA1 in 39 unrelated patients with genotype negative for the previously known LQTS subtypes was performed and one novel missense mutation (A257G) was identified in 3 unrelated patients (2 females). This mutation involves a highly conserved residue across several species (Figure 1A and 1B).
Figure 1.
The cytoskeletal protein syntrophin α1 (SNTA1) mutational analysis in long-QT syndrome (LQTS). Sequencing analysis demonstrates a novel nucleotide variant in SNTA1 leading to a nonsynonymous change in 3 patients (A) and amino acid conservation analysis of the A257G-SNTA1 variant (B), which modifies a highly conserved amino acid in SNTA1. C, LQT-249 family pedigree and ECG recording showing a pattern with features similar to LQT3 and a prolonged QT interval in proband III: 2, his sister III:1 and his mother II:2 bearing the A257G-SNTA1 mutation. The ECG of the proband demonstrates a prolonged QT interval and late onset T wave.19 D, LQT-682 family pedigree and ECG recording showing a pattern with features similar to LQT3 and a prolonged QT interval in proband II: 1 bearing the A257G-SNTA1 mutation.
The proband is a 17-year-old white male presenting with congenital LQTS. His ECG obtained at 6 hours after birth demonstrated marked QT-prolongation with late onset T-wave pattern (QTc=550 ms, Figure 1C, Table 1, case LQT-249 III: 2). The proband had a syncopal attack at 3 years of age without previous symptoms of prodromal illness, vomiting, diarrhea, fever, or upper respiratory infection. He collapsed while playing in a baby pool after excessive running in a very hot day and remained unconscious for few minutes. The patient underwent Holter monitoring, which showed the average heart rate of 128 bpm (99 to 167) and prolonged QT-intervals (260 to 280 ms). No arrhythmias were documented during the recording. The patient was treated with propranolol. In addition to the SNTA1 mutation, a second variant was identified in KCNQ1 (IVS7+5G>A), which is present in all affected members along with the SNTA1 mutation. Although this variant is of unknown significance, computer predictions analyzed in our laboratory, using the http://www.fruitfly.org/seq_tools/splice.html software,20 suggests the possible creation of a cryptic splicing site leading to an in-frame insertion of 25 amino acids in one of the KCNQ1 alleles. This suggests that the KCNQ1 variant may add to the clinical phenotype caused by SNTA1.
Table 1.
Demographics of the Long-QT (LQT) Syndrome Patients
| ECG |
|||||||
|---|---|---|---|---|---|---|---|
| Case No. | Gene Analysis | Age/Sex | Race | Symptom | Onset Age | QTc, ms | T-Wave Morphology |
| LQT-249-II:1 | NA | 25†/M | W | Syncope/SCD | UK | 470 | NA |
| LQT-249-II:2 | A257G | 45/F | W | None | 460 | Asymmetric peaked | |
| LQT-249-III:1 | A257G | 13/F | W | None | 500 | Inverted | |
| LQT-249-III:2* | A257G | 17/F | W | Syncope with pulmonary arrest | 3 | 550 | Late onset |
| LQT-682* | A257G | 37/F | W | Recurrent seizures | 480 | NA | |
| LQT-755* | A257G | 27/F | W | None | 480 | NA | |
Proband;
age at SCD.
W indicates white; SCD, sudden cardiac death; UK, unknown.
A257G mutation was also identified in the proband’s sister (subject III:1) and mother (subject II:2). Both of them have been apparently healthy. The sister’s ECG obtained at birth showed QT-prolongation (QTc: 500 ms) with normal T-wave pattern in V1 to 3 and inverted T-waves in V4 to 6. Her Holter ECG demonstrated normal sinus rhythm, no arrhythmias, and prolonged QT intervals. The mother’s ECG obtained at 27 years of age showed mild QT prolongation with asymmetrical peaked T-wave pattern (QTc=460 ms; Figure 1C). The proband’s maternal uncle died suddenly at age 25 during physical exertion. He had prior history of syncope and prolonged QT interval (QTc=470 ms). No autopsy sample was available to us. The ECG and genetic screening of the proband’s father (Figure 1C; Table 1) did not show any abnormalities.
The A257G was also identified in a 37-year-old and a 27-year-old unrelated female with unremarkable family history of either LQTS or sudden cardiac death. Both probands showed QT-prolongation at baseline ECG (QTc=480 ms). Their parents’ ECG and genetic screening analysis did not show any abnormalities, consistent with de novo genetic change.
Subject LQT-682 was identified subsequently to screening after her 11-year-old son died suddenly because of ventricular fibrillation registered at the time of the code at the hospital where he was visiting the brother admitted for a car accident. The autopsy findings were negative for structural heart diseases and led to the diagnosis of suspected LQTS, but no sample was available to us. LQT-682 proband had a longstanding history of seizure disorder with normal seizure workup and positive EEG. The proband was treated with phenytoin for her seizure since she was 12-years-old.
The ECG (Figure 1D) and Holter ECG were obtained in the absence of β-blockers but in the presence of phenytoin, and they demonstrated intermittent QT prolongation. Echocardiography assessment was normal. Analysis of 400 ethnic-matched control alleles did not identify the A257G mutation.
Immunostaining
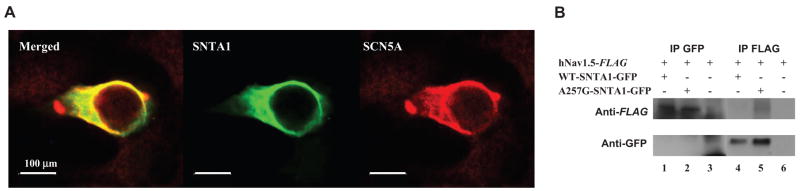
HEK-293 cells stably expressing Flag-tagged hNav1.5 and GFP-tagged WT-SNTA1 were fixed and stained with anti-Flag antibody. Figure 2A demonstrates that the green fluorescence (SNTA1, middle panel) and red fluorescence (hNav1.5, right panel) when merged give the orange-yellow areas, suggesting colocalization of hNav1.5 and SNTA1, consistent with possible association in a protein complex.
Figure 2.
hNav1.5 and cytoskeletal protein syntrophin-α1 (SNTA1) may form a protein complex in HEK-293 cells. A, Immunohistochemical staining: The left panel shows the merged images of green fluorescence (mid) and red fluorescence (right) images. The green fluorescence depicts the GFP-tagged SNTA1 and the red fluorescence depicts the Flag-tagged hNav1.5. B, Coimmunoprecipitation assays. IP indicates immunoprecipitants; (+), transfected with the genes indicated in the left column; (−) nontransfected with the genes.
Coimmunoprecipitation Assay of hNav1.5 and SNTA1
Previous studies showed that the PDZ domain of SNTG2, highly homologous to SNTA1, binds the C-terminal domain of Nav1.5 and affect its gating kinetics.16 Therefore, we performed coimmunoprecipitation assay using HEK-293 cells stably expressing Flag-tagged hNav1.5 and transiently transfected with either GFP-tagged WT- or A257G-SNTA. The anti-GFP antibody detected SNTA1 (87 kDa) in the coimmunoprecipitants with the anti-Flag antibody (Figure 2B, lanes 4 and 5), whereas the anti-Flag antibody detected hNav1.5 (260 kDa in the coimmunoprecipitants with the anti-GFP (Figure 2B, lanes 1 and 2) for both WT-SNTA1 and A257G-SNTA1, suggesting that human SNTA1 can physically interact with hNav1.5 and that perturbations in SNTA1 might alter hNav1.5 function.
Electrophysiological Experiments
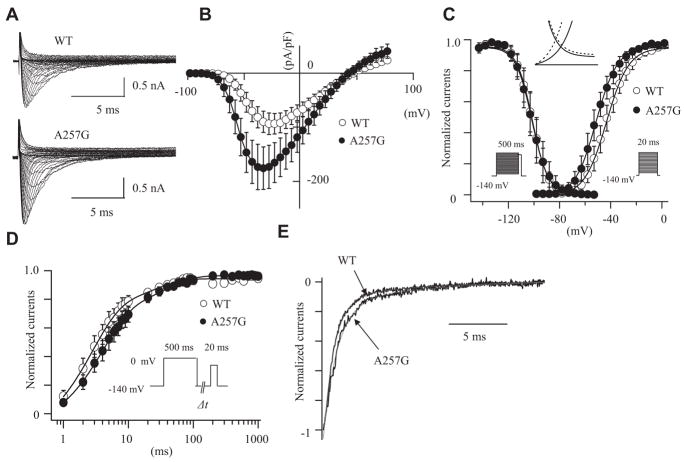
Figure 3A shows the superimposed whole-cell current traces obtained from HEK-293 cells stably expressing consistent hNav1.5 currents and transiently transfected with WT- or A257G-SNTA1. Figure 3B shows the current–voltage (I-V) relationships and demonstrates peak current densities nearly twice as large in the cells expressing A257G-SNAT1 compared with WT. In addition, A257G-SNAT1 shifted the onset and peak of the currents toward more negative potentials by 10 mV compared with the WT. Figure 3C shows the effects of A257G-SNAT1 on the voltage-dependent kinetics of steady-state activation and inactivation. The half maximal voltage of the steady-state activation (Vh) was shifted leftward by 8.3 mV in the cells expressing A257G-SNAT1. The slope factor (k) was not affected by A257G-SNAT1. Steady-state inactivation was examined by a prepulse protocol. The normalized currents were plotted as a function of the membrane potentials and fitted by the Boltzman equation. Voltage-dependent inactivation kinetics were not significantly affected by A257G-SNAT1. The leftward shift of steady-state activation without shift of the inactivation kinetics increased the window currents (Figure 3C, inset). These biophysical parameters were also studied in the presence of β1-subunit of the human cardiac sodium channel (hβ1); hβ1 did not significantly modulate the effects of A257G-SNTA1 on the hNav1.5 gating kinetics (Table 2).
Figure 3.
The effects of wild-type cytoskeletal protein syntrophin-α1 (WT-SNTA1) and A257G-SNTA1 on hNav1.5 in HEK-293 cells. A, Superimposed whole-cell current traces induced by a step-pulse protocol from a holding potential of −140 mV. B, I-V relationships. C, Voltage-dependence of peak conductance and steady-state fast inactivation. Conductance G(V) was calculated by the equation: G(V) = I/(Vm − Erev), where I is the peak currents, Erev is the measured reversal potential, Vm is the membrane potential. The normalized peak conductance was plotted as a function of membrane potentials. Steady-state inactivation was estimated by prepulse protocols (500 ms) from a holding potential of −140 mV. The normalized peak currents were plotted as a function of membrane potentials. Steady-state activation and inactivation were fitted with the Boltzmann equation: y = [1 + exp ((Vh − Vm)/k)]−1, where y represents variables; Vh, midpoint; k, slope factor; Vm, membrane potential. The inset indicates the magnified illustration of the window currents. The solid lines represent WT-SNTA1, and the dotted lines represent A257G-SNTA1. D, Recovery from the fast-inactivation estimated by a double pulse protocol. Cells were depolarized at 0 mV for 500 ms from a holding potential of −140 mV, then stepped to −140 mV for various durations before the second pulse (20 ms at −20 mV). The fractional recovery was calculated as the ratio of peak currents at the second pulse. The recovery time course was fitted with a double exponential function: I(t)/Imax = C − Af × exp (−t/τf) −As × exp (−t/τs), where t is the recovery time, Af and As are the fractions of fast and slow components, τf and τs are the time constants of fast and slow components of recovery. E, Macroscopic current decay was fit with a double exponential function: I(t)/Imax = C − Af exp(−t/τf) − As exp (−t/τs), where Imax is the peak current, t is the time, Af and As are the fractions of fast and slow components, τf and τs are the time constants of fast and slow components, respectively. The data were represented as mean±SE.
Table 2.
Effects of Wild Type (WT) and A257G-SNTA1 on the Nav1.5 Gating Kinetics
| HEK Without β1 Subunit |
HEK With β1 Subunit |
Neonatal Cardiomyocyte |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | A257G | P | WT | A257G | P | WT | A257G | P | |
| Current density (pA/pF) | −97.8±9.1 (−95.4) | −179.6±18.4 (−182.0)** | 0.000 | −151.0±25.4 (−140.7) | −336.9±70.0 (−356.7)* | 0.018 | −316.3±51.1 (−305.7) | −517.5±70.7 (3567.5)* | 0.041 |
| n | 15 | 13 | 12 | 9 | 6 | 6 | |||
| Activation | |||||||||
| Vh, mV | −43.5±1.5 (−44.0) | 351.7±1.6 (−51.5)** | 0.002 | −43.1±1.3 (−42.7) | −50.8±1.5 (−48.9)** | 0.001 | −47.9±1.7 (−48.4) | −57.3±3.1 (−55.0)* | 0.015 |
| k | 7.87±0.31 (7.47) | 7.91±0.62 (7.49) | 0.892 | 8.80±0.47 (8.98) | 8.22±0.24 (8.10) | 0.464 | 7.56±0.37 (7.54) | 7.06±0.14 (7.09) | 0.24 |
| n | 15 | 13 | 12 | 9 | 6 | 6 | |||
| Fast-inactivation | |||||||||
| Vh, mV | −97.4±1.8 (−96.7) | −101.3±2.0 (−102.1) | 0.190 | −104.6±7.7 (−102.8) | −105.9±5.8 (−106.0) | 0.430 | −91.9±2.3 (−91.2) | −91.4±3.1 (−92.1) | 0.943 |
| k | 5.18±0.19 (5.39) | 5.44±0.19 (5.49) | 0.740 | 5.47±0.17 (5.59) | 5.59±0.23 (5.76) | 0.650 | 6.43±0.26 (6.71) | 7.10±0.13 (6.92) | 0.065 |
| n | 12 | 11 | 13 | 14 | 8 | 5 | |||
| Recovery from fast-inactivation | |||||||||
| τf, ms | 2.92±0.30 (2.59) | 4.33±0.56 (3.63)* | 0.033 | ||||||
| τs, ms | 33.4±34.57 (29.86) | 41.34±2.14 (40.93) | 0.094 | ||||||
| Af, % | 93.4±3.6 (91.6) | 87.2±2.8 (87.46) | 0.169 | ||||||
| As, % | 20.4±2.7 (19.8) | 23.4±1.8 (22.09) | 0.169 | ||||||
| n | 13 | 14 | |||||||
| Current decay | |||||||||
| τf, ms | 0.92±0.04 (0.94) | 1.06±0.04 (1.01)* | 0.033 | 0.80±0.07 (0.74) | 1.12±0.08 (1.12)* | 0.026 | |||
| τs, ms | 8.57±0.99 (7.76) | 8.97±1.00 (9.99) | 0.586 | 5.90±1.78 (3.53) | 4.23±0.43 (3.96) | 0.589 | |||
| n | 15 | 13 | 6 | 6 | |||||
Vh, indicates midpoint voltage of maximal activation/inactivation; k, slope factor; τf and τs, time constants of fast and slow components of recovery; Af and As, fractions of fast and slow components; n, No. of patches. Numbers in parentheses represent the median of data.
P<0.05;
P<0.01 versus WT.
The recovery time from fast inactivation was studied using a 2-pulse protocol. Figure 3D shows the fraction of channels that recovered from the fast inactivation. The data were fitted by a double-exponential equation; the fast components of the time constant (τf) were significantly slower in the cells expressing A257G-SNTA1 than WT. The slow components of the time constant (τs) and the fraction of fast and slow components were comparable between the WT-SNTA1 and the A257G-SNTA1.
Delay of sodium current decay and generation of sustained sodium currents have been proposed as the pathophysiological mechanisms responsible for the LQT3 phenotype. We estimated the time course of whole-cell current decay and the late sodium currents. Because the I-V curve was shifted, the comparison of the current decay at a same membrane potential might not simply express the time-dependent kinetics. The decay of the peak sodium currents was analyzed with a double exponential fit. Figure 3E shows that A257G-SNTA1 slightly delayed the macroscopic current decay compared with the WT-SNTA1. Although the slow component of time constant was comparable between WT-SNTA1 and A257G-SNTA1, A257G-SNTA1 significantly slowed the fast component of time constant (Table 2). The late sodium currents were evaluated by a long depolarization pulse (2000 ms at −20 mV from a holding potential of −140 mV) using 30 μmol/L tetrodotoxin (TTX). No TTX-sensitive sustained currents were detected in the cells expressing WT-SNTA1 and A257G-SNTA1 (data not shown). Table 2 summarizes the effects of WT-SNTA1 and A257G-SNTA1 on the Nav1.5 gating kinetics.
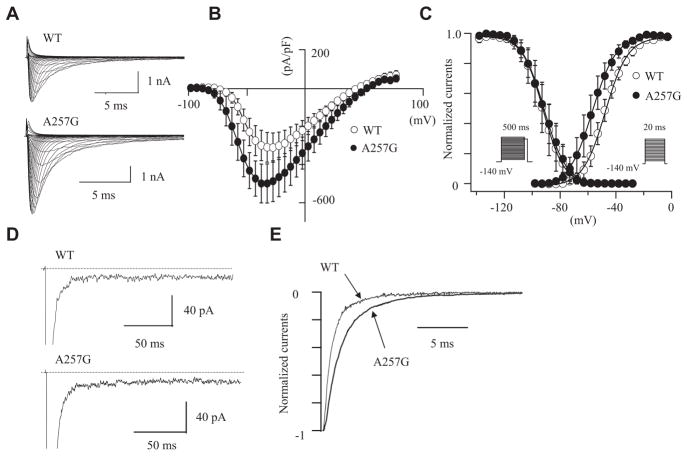
Because the cellular environment of HEK-293 cells might be far different from myocardial cells, we studied the effects of WT-SNTA1 and A257G-SNTA1 on Nav1.5 in neonatal rat cardiomyocytes. The cardiomyocytes were isolated from neonatal rats (3 to 5 days old) and transfected with GFP-tagged WT-SNTA1 and the A257G-SNTA1 by using the Nucleofector. The cardiomyocytes showing green fluorescence were used for the patch-clamp experiments. In native cardiomyocytes, A257G-SNTA1 increased the peak sodium currents and altered the kinetics consistently with that observed in HEK-293 cells. Figure 4A shows the representative whole-cell current traces obtained from the cardiomyocytes transiently transfected with either WT-SNTA1 or A257G-SNTA1. Figure 4B shows the I-V relationships. The peak current densities were significantly larger (1.6 fold) in cells expressing A257G-SNTA1 compared with the WT. The onset and peak of the currents of the I-V relationships were shifted toward negative by approximately 10 mV in cells expressing the A257G-SNTA1 compared with WT. Figure 4C shows the effects of A257G-SNTA1 on the voltage-dependency of the steady-state activation and the inactivation. The Vh of activation curve was shifted leftward by 9.4 mV in the cells transfected with A257G-SNTA1; on the contrary, voltage-dependent inactivation kinetics were not significantly affected by A257G-SNTA1. The persistent sodium currents were examined using a long depolarization pulse protocol (Figure 4D). The 30 μmol/L TTX-sensitive persistent sodium currents were comparable between the WT and the A257G mutant (percentage over the peak currents: WT, 0.25±0.09%, n=4; A257G, 0.22±0.04%, n=4, NS). Figure 4E demonstrates that A257G-SNTA1 delayed the macroscopic current decay (peak currents) compared to the WT-SNTA1. Although the slow component of time constant was comparable between WT and mutant SNTA1, A257G-SNTA1 significantly increased the fast component of the time constant (Table 2).
Figure 4.
The effects of wild-type cytoskeletal protein syntrophin-α1 (WT-SNTA1) and A257G-SNTA1 on Nav1.5 in neonatal rat cardiomyocytes. A, Superimposed whole-cell current traces induced by a step-pulse protocol from a holding potential of −140 mV. B, I-V relationships. C, Voltage-dependence of peak conductance and steady-state fast inactivation. D, Magnified current traces of the tetrodotoxin-sensitive late sodium currents induced by a long depolarization-pulse protocol (2000 ms at −20 mV from a holding potential of −140 mV). The dotted lines indicate zero current levels. E, Macroscopic current decay was fit with a double exponential function. The data were represented as mean±SE.
Discussion
Until recently, the pathophysiological mechanisms of inherited arrhythmias were thought to stem merely from primary alterations in ion channels leading to the term “ion channelopathies.” However, this paradigm has been challenged by the identification of altered channel-interacting proteins, which can directly modify ion channel functions. In particular, our recent finding demonstrating that caveolin-3 (which cofractionates with dystrophin and the dystrophin glycoprotein complex; DGC) binds Nav1.5 in human heart and if mutated leads to LQTS5 has focused our attention on the other DGC proteins such as SNTA. SNTA1 is an integral part of the DGC, and regulates the Nav1.5 by forming a multiprotein complex through its direct binding to dystrophin.21 Our study, for the first time, demonstrates that alteration of SNTA1 can affect the Nav1.5 gating kinetics, leading to the LQTS phenotype.
How Does A257G-SNTA1 Affect the Gating Kinetics of Nav1.5?
A257G-SNTA1 caused gain-of-function of the Nav1.5 through the leftward shift of the activation curve by 8 to 9 mV without shift of the inactivation curve, which might increase the channel availability. The extent of this shift is equal to or larger than previously reported values for SCN5A mutations (N1325S, L619F; Figures 3C and 4C).22,23 In addition, the subtle but significant delay of the fast component of whole-cell current decay (Figures 3E and 4E) and the significantly larger peak current density for A257G-SNTA1 might contribute to an increase of total inward currents.
Ou et al reported that SNTG2, which is highly homologous to SNTA1, can attenuate sodium current densities and shift the voltage-dependent activation kinetics compared with the control.16 Therefore, it is possible that WT-SNTA1 also attenuates sodium currents, whereas A257G-SNTA1 abolished the ability of WT-SNTA1 to decrease the currents. To prevent the endogenous gene expression level affecting the comparison of the current densities in a transiently transfection system, we established stable HEK-293 cell lines that generate consistent sodium current densities from one single clone, and we transfected them with WT-SNTA1 and A257G-SNTA1 by consistent protocols. Therefore, it is reasonable to speculate that A257G-SNTA1 might increase the open-probability of the channel compared with the WT. Although single-channel recordings need to be studied to elucidate the detail underlying mechanisms, our data indicate that these biophysical modifications by the A257G-SNTA1 can cause gain of function of the cardiac sodium channels in comparison with the WT-SNTA1.
How Does SNTA1 Interact With Nav1.5?
Previous studies demonstrated that the PDZ domain of syntrophins (human SNTG2 and rodent SNTA1) can bind to the C-terminal domain of cardiac sodium channels (hNav1.5 and rodent Skm2).15,16 Our study demonstrated that human SNTA1 can directly interact with hNav1.5 (Figure 2B), which is consistent with these reports and suggests direct protein-protein interaction as the mechanism by which A257G-SNTA1 modifies the hNav1.5 function.
The previous study using the dystrophin-deficient mdx mice suggested that the syntrophin-dystrophin complex plays critical roles in the regulation of the sodium channel gating kinetics, as well as the ECG phenotype (conduction disturbances and QT-prolongation).24 Therefore, even secondary alterations of the DGC may result in primary hNav1.5 defects via SNTA1 in cardiomyocytes. In addition, the effect of A257G-SNTA1 on hNav1.5 in HEK-293 cells demonstrates that SNTA1 plays a key role in hNav1.5 regulation. Further studies using genetically engineered animal models and addressing the detailed mechanism of Nav1.5 function regulation by SNTA1 and the involvement of other syntrophin associated proteins, such as caveolin-3, dystrobrevin, and nitric oxide synthase 1,24 are warranted.
Study Limitations
This study has several significant limitations : (1) the study population is small and linkage analysis could not be performed; (2) we could not perform functional studies on the KCNQ1 intronic variant, and therefore, we cannot exclude that this intronic variant or other unknown genes can be accountable for the patients’ phenotypes although we examined and excluded all known LQTS genes; and (3) the data were obtained by in vitro experiments using cell lines expressing only the α- and β1-subunit of the cardiac sodium channels or neonatal rat cardiomyocytes overexpressing human SNTA1, which is far different from the physiological conditions in the actual human heart. Although further studies are warranted to explore the links between our data and the phenotypes of the affected patients, our study does indicate that SNTA1 now includes ANKB, CAV3, and AKAP9 (yotiao) as “nonprimary channelopathic” mechanisms for LQTS.2,3,5
Conclusions
We identified a novel mutation of SNTA1, A257G, in 3 unrelated LQTS patients whose genetic screenings were negative for LQT1–9. Electrophysiological study suggests that biophysical modifications of the Nav1.5 by A257G-SNTA1 can lead to “gain-of-function,” through 3 mechanisms: (1) increase of the channel availability by leftward shift of activation kinetics; (2) delay of the current decay; and (3) increase in the current density. The molecular and electrophysiological evidences implicate A257G-SNTA1 in the pathogenesis of LQTS as a novel, albeit rare, LQTS-susceptibility gene. Although further studies are warranted to understand the detailed mechanisms leading to LQTS phenotypes in patients with the A257G-SNTA1, our study demonstrated that SNTA1, as an integral part of the DGC, plays an important role in regulating hNav1.5 function. Mutation in SNTA1 may not only account for the development of cardiac arrhythmias in affected patients but also potentially serve as novel target(s) of therapeutic interventions.
Supplementary Material
The online-only Data Supplement is available with this article at http://circ.ahajournals.org/cgi/content/full/CIRCEP.108.769224/DCI.
Acknowledgments
The authors are grateful to the patients and their family members who participated in this study. In addition, we are grateful to Jennifer L. Robinson, LQTS Study Coordinator for registry information assistance.
Sources of Funding
This work was supported by the National Heart, Lung, and Blood Institute (HL078807; M.V.), and the National Institute of Child and Health Development HD42569 (M.J.A.). T.A. and J.C. were recipients of the Roderick D. MacDonald General Research Fund Awards. J.C. was in part supported by a Cardiovascular Initiative Grant from the St. Luke’s Episcopal Hospital/Texas Heart Institute. J.A.T. was supported by the Texas Children’s Foundation Chair in Pediatric Cardiac Research.
Footnotes
Presented in part at the 2007 American Heart Association (AHA) Annual Scientific Sessions, Orlando, Fla.
Disclosures
None.
References
- 1.Shah M, Akar FG, Tomaselli GF. Molecular basis of arrhythmias. Circulation. 2005;112:2517–2529. doi: 10.1161/CIRCULATIONAHA.104.494476. [DOI] [PubMed] [Google Scholar]
- 2.Saenen JB, Vrints CJ. Molecular aspects of the congenital and acquired long QT syndrome: clinical implications. J Mol Cell Cardiol. 2008;44:633–646. doi: 10.1016/j.yjmcc.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Mohler PJ, Schott JJ, Gramolini AO, Dilly KW, Guatimosim S, duBell WH, Song LS, Haurogne K, Kyndt F, Ali ME, Rogers TB, Lederer WJ, Escande D, Le Marec H, Bennett V. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature. 2003;421:634–639. doi: 10.1038/nature01335. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz PJ. Management of long QT syndrome. Nat Clin Pract Cardiovasc Med. 2005;2:346–351. doi: 10.1038/ncpcardio0239. [DOI] [PubMed] [Google Scholar]
- 5.Vatta M, Ackerman MJ, Ye B, Makielski JC, Ughanze EE, Taylor EW, Tester DJ, Balijepalli RC, Foell JD, Li Z, Kamp TJ, Towbin JA. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation. 2006;114:2104–2112. doi: 10.1161/CIRCULATIONAHA.106.635268. [DOI] [PubMed] [Google Scholar]
- 6.Cronk LB, Ye B, Kaku T, Tester DJ, Vatta M, Makielski JC, Ackerman MJ. Novel mechanism for sudden infant death syndrome: persistent late sodium current secondary to mutations in caveolin-3. Heart Rhythm. 2007;4:161–166. doi: 10.1016/j.hrthm.2006.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guglieri M, Magri F, Comi GP. Molecular etiopathogenesis of limb girdle muscular and congenital muscular dystrophies: boundaries and contiguities. Clin Chim Acta. 2005;361:54–79. doi: 10.1016/j.cccn.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Marquardt ML, Tester DJ, Sampson KJ, Ackerman MJ, Kass RS. Mutation of an A-kinase-anchoring protein causes long-QT syndrome. Proc Natl Acad Sci U S A. 2007;104:20990–20997. doi: 10.1073/pnas.0710527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Towbin JA. Cardiac arrhythmias: the genetic connection. J Cardiac Electrophysiol. 2000;11:601–602. doi: 10.1111/j.1540-8167.2000.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 10.Adams ME, Butler MH, Dwyer TM, Peters MF, Murnane AA, Froehner SC. Two forms of mouse syntrophin, a 58 kd dystrophin-associated protein, differ in primary structure and tissue distribution. Neuron. 1993;11:531–540. doi: 10.1016/0896-6273(93)90157-m. [DOI] [PubMed] [Google Scholar]
- 11.Adams ME, Dwyer TM, Dowler LL, White RA, Froehner SC. Mouse alpha 1- and beta 2-syntrophin gene structure, chromosome localization, and homology with a discs large domain. J Biol Chem. 1995;270:25859–25865. doi: 10.1074/jbc.270.43.25859. [DOI] [PubMed] [Google Scholar]
- 12.Ahn AH, Yoshida M, Anderson MS, Feener CA, Selig S, Hagiwara Y, Ozawa E, Kunkel LM. Cloning of human basic A1, a distinct 59-kDa dystrophin-associated protein encoded on chromosome 8q23–24. Proc Natl Acad Sci U S A. 1994;91:4446–4450. doi: 10.1073/pnas.91.10.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang B, Ibraghimov-Beskrovnaya O, Moomaw CR, Slaughter CA, Campbell KP. Heterogeneity of the 59-kDa dystrophin-associated protein revealed by cDNA cloning and expression. J Biol Chem. 1994;269:6040–6044. [PubMed] [Google Scholar]
- 14.Piluso G, Mirabella M, Ricci E, Belsito A, Abbondanza C, Servidei S, Puca AA, Tonali P, Puca GA, Nigro V. Gamma1- and gamma2-syntrophins, two novel dystrophin-binding proteins localized in neuronal cells. J Biol Chem. 2000;275:15851–15860. doi: 10.1074/jbc.M000439200. [DOI] [PubMed] [Google Scholar]
- 15.Gee SH, Madhavan R, Levinson SR, Caldwell JH, Sealock R, Froehner SC. Interaction of muscle and brain sodium channels with multiple members of the syntrophin family of dystrophin-associated proteins. J Neurosci. 1998;18:128–137. doi: 10.1523/JNEUROSCI.18-01-00128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ou Y, Strege P, Miller SM, Makielski J, Ackerman M, Gibbons SJ, Farrugia G. Syntrophin gamma 2 regulates SCN5A gating by a PDZ domain-mediated interaction. J Biol Chem. 2003;278:1915–1923. doi: 10.1074/jbc.M209938200. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz PJ, Moss AJ, Vincent GM, Crampton RS. Diagnostic criteria for the long QT syndrome: an update. Circulation. 1993;88:782–784. doi: 10.1161/01.cir.88.2.782. [DOI] [PubMed] [Google Scholar]
- 18.Shuraih M, Ai T, Vatta M, Sohma Y, Merkle EM, Taylor EW, Li Z, Razavi M, Towbin JA, Cheng J. A common SCN5A variant alters its pharmacological responsiveness to class I antiarrhythmic agents. J Cardiovasc Electrophysiol. 2007;18:434–440. doi: 10.1111/j.1540-8167.2007.00777.x. [DOI] [PubMed] [Google Scholar]
- 19.Moss AJ, Zareba W, Benhorin J, Locati EH, Hall WJ, Robinson JL, Schwartz PJ, Towbin JA, Vincent GM, Lehmann MH. ECG T-wave patterns in genetically distinct forms of the hereditary long QT syndrome. Circulation. 1995;92:2929–2934. doi: 10.1161/01.cir.92.10.2929. [DOI] [PubMed] [Google Scholar]
- 20.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in genie. J Comp Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 21.Gavillet B, Rougier JS, Domenighetti AA, Behar R, Boixel C, Ruchat P, Lehr HA, Pedrazzini T, Abriel H. Cardiac sodium channel Nav1.5 is regulated by a multiprotein complex composed of syntrophins and dystrophin. Circ Res. 2006;99:407–414. doi: 10.1161/01.RES.0000237466.13252.5e. [DOI] [PubMed] [Google Scholar]
- 22.Wang DW, Yazawa K, George AL, Jr, Bennett PB. Characterization of human cardiac Na+ channel mutations in the congenital long QT syndrome. Proc Natl Acad Sci U S A. 1996;93:13200–13205. doi: 10.1073/pnas.93.23.13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wehrens XH, Rossenbacker T, Jongbloed RJ, Gewillig M, Heidbuchel H, Doevendans PA, Vos MA, Wellens HJ, Kass RS. A novel mutation L619F in the cardiac Na+ channel SCN5A associated with long-QT syndrome (LQT3): a role for the I-II linker in inactivation gating. Hum Mutat. 2003;21:552. doi: 10.1002/humu.9136. [DOI] [PubMed] [Google Scholar]
- 24.Aarnoudse AJ, Newton-Cheh C, de Bakker PI, Straus SM, Kors JA, Hofman A, Uitterlinden AG, Witteman JC, Stricker BH. Common NOS1AP variants are associated with a prolonged QTc interval in the Rotterdam Study. Circulation. 2007;116:10–16. doi: 10.1161/CIRCULATIONAHA.106.676783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The online-only Data Supplement is available with this article at http://circ.ahajournals.org/cgi/content/full/CIRCEP.108.769224/DCI.