Abstract
The gonad in C. elegans is an important model system for understanding complex morphogenetic processes including cellular movement, cell fusion, cell invasion and cell polarity during development. One class of signaling proteins known to be critical for the cellular events underlying morphogenesis is the Rho family GTPases, particularly RhoA, Rac and Cdc42. In C. elegans orthologues of these genes have been shown to be important for gonad development. In our current study we have extended those findings by examining the patterns of 5’ cis-regulatory element (5’CRE) activity associated with nineteen putative guanine nucleotide exchange factors (GEFs) encoded by the C. elegans genome predicted to activate Rho family GTPases. Here we identify thirteen RhoGEF genes that are expressed during gonadogenesis and characterize the cells in which their 5’CREs are active. These data provide the basis for designing experiments to examine Rho GTPase activation during morphogenetic processes central to normal gonad development.
Keywords: C. elegans, gonadogenesis, RhoGEF, morphogenesis, organogenesis, anchor cell invasion, gonad development, Rho GTPase, rho, rac, cdc42, mig-2, ced-10, cdc-42, rho-1
1. Results and Discussion
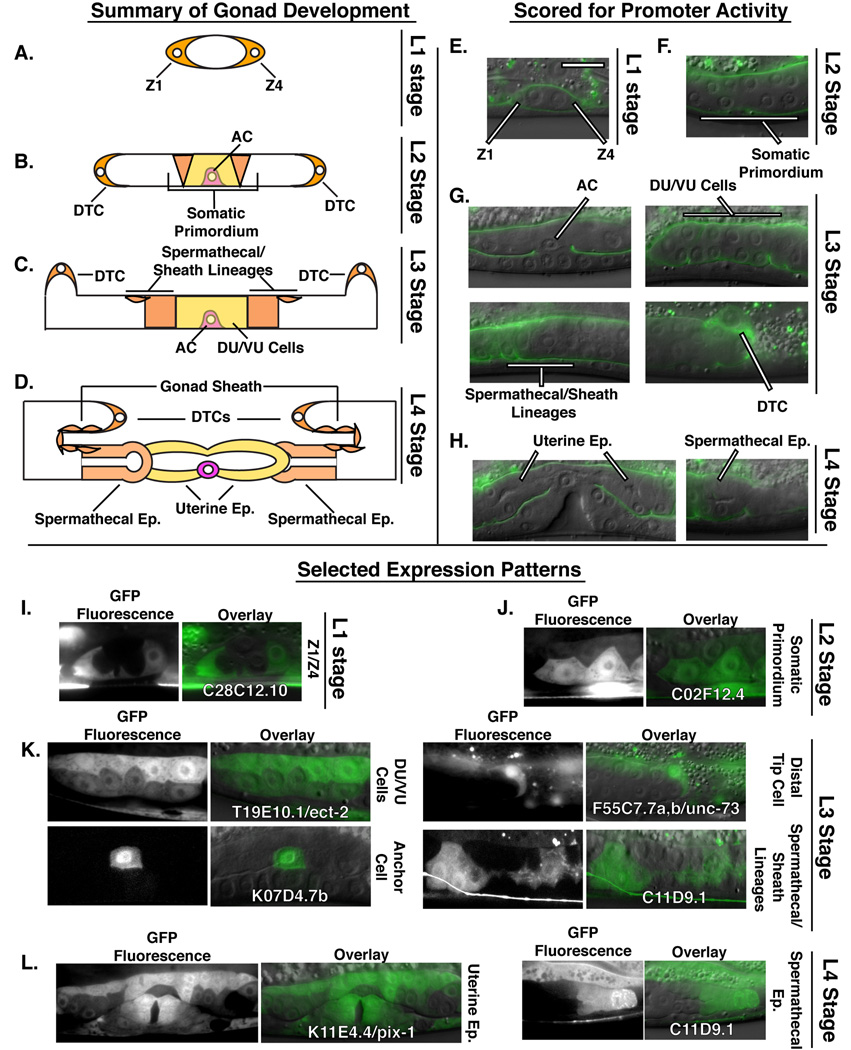
The gonad in C. elegans is an excellent model for elucidating the genetic and cell-biological mechanisms underlying organogenesis (Hubbard and Greenstein, 2000). The somatic tissues of the adult hermaphrodite gonad are generated through stereotyped division, rearrangement and patterning of the progeny of two somatic gonadal cells known as Z1 and Z4 (Kimble and Hirsh, 1979). These cells encircle the primordial germ cells near the end of embryogenesis to form the gonad primordium (Figure 1A and E) (Sulston et al., 1983). After hatching, development in C. elegans proceeds through four distinct larval growth stages (L1–L4) separated by cuticular molts. During the L1 stage, Z1 and Z4 begin a process of oriented cell division that generates the cells whose progeny give rise to the tissues of the gonad (Figure 1A and E) (Kimble and Hirsh, 1979). Cell division continues during the L2 stage, but this developmental window is also characterized by early morphogenetic changes within the gonad. Of particular importance are the movements of cells that give rise to the uterine and spermathecal epithelia as well as the gonad sheath cells. During the late L2 stage, these ten cells come to completely occupy the central region of the gonad, forming the somatic gonad primordium in a process coincident with exclusion of the germ cells into the gonad arms (Figure 1B and F) (Kimble and Hirsh, 1979). Also at this time, elongation of the gonad arms along the anterior-posterior axis occurs and is guided by two specialized cells known as distal tip cells (DTCs). The DTCs are located at the extreme anterior and posterior ends of the gonad (one on each gonad arm) and migrate along the body wall during larval development (Figure 1B–D and G). During this process gonad sheath cells flatten and migrate along the extending gonad, ultimately encirlcling the adult gonad arms (Figure 1C,D, G and K) (Kimble and Hirsh, 1979). Migration of the DTCs continues through the L3 and L4 larval stages, during which they execute a stereotypic series of movement and turns to generate the reflexed, fully extended arms of the adult hermaphrodite gonad (Figure 1C, D, G and H) (Hedgecock et al., 1987).
Figure 1.
(A–D) Schematic representation of larval gonad development in C. elegans. (A) After hatching, during the L1 stage, the somatic gonad precursor cells Z1 and Z4 surround the primordial germ cells and are located on the anterior and posterior sides of the early gonad. (B) During the L2 stage these cells give rise to somatic lineages of the gonad including the distal tip cells (DTCs) and the cells of the somatic gonad primordium. (C) During the L3 stage the cells of the somatic gonad primordium divide to form the dorsal and ventral uterine cells (DU/VU cells) as well as the gonad sheath and spermathecal cells. (D) During the L4 stage, the uterine and spermathecal epithelia form mature lumens. The DTCs and the gonad sheath cells complete their migrations. (E–H) DIC micrographs overlaid with LAM-1::GFP fluorescence labeling the basement membrane which surrounds and outlines the gonad. RhoGEF 5’CRE activity was scored in: (E) Z1 and Z4 at the L1 stage; (F) the somatic gonad primordium at the L2 stage; (G) the AC, VU/DU cells, the spermathecal and sheath lineage and the DTCs during the L3 stage and (F) the uterine and spermathecal epithelia at the L4 stage. (I–L) Selected examples of RhoGEF 5’CRE activity during gonadogenesis: (I) 5’CREs from C28C12.10 were active in Z1 and Z4; (J) C02F12.4 was detected in the somatic gonad primordium; (K) T19E10.1/ect-2, K07D4.7b, F55C7.7a,b/unc-73 and C11D9.1 5’CRE activity detected in the DU/VU cells, AC, DTC and spermathecal and sheath lineages respectively; (L) K11E4.4/pix-1 5’CREs were active broadly within the uterine epithelium, whereas elements from C11D9.1 were active specifically within the spermatheca. Scale bar in (E) represents 20 microns. Abbreviations: epithelium, Ep; distal tip cell, DTC; anchor cell, AC; dorsal uterine cell, DU; ventral uterine cell, VU in this and all subsequent figures.
Successful passage of fertilized eggs out of the adult hermphrodite requires connection of the uterus to the vulva, which forms as a separate epithelial tube in the hypodermis directly ventral to the uterine precursors. A specialized uterine cell, the anchor cell (AC), is central to the development of both tissues (Newman et al., 1995; Sherwood and Sternberg, 2003; Sundaram, 2004). During the L2–L3 molt, the AC induces vulval tissue by secreting the EGF-like ligand LIN-3 (Sundaram, 2004). Later, during the mid-L3 stage, the AC plays a second critical role in establishing the initial connection between the uterus and vulva by invading through the basement membranes separating the gonad and hypodermis (Sherwood et al., 2005; Sherwood and Sternberg, 2003; Ziel et al., 2009). Following basement membrane removal and attachment to the central vulval cells, the AC fuses with surrounding uterine cells, which, during the late L3 and L4 stages, begin to form the mature epithelium characteristic of adult worms (Figure 1D and H) (Newman and Sternberg, 1996; Sharma-Kishore et al., 1999). This event occurs at the same time as maturation of the spermathecal epithelia and their connection to the uterus (Figure 1D and H) (Hubbard and Greenstein, 2000).
The Rho family of small GTPases (Rho GTPases) are known to be critical for key steps in hermaphroditic gonadogenesis (Blelloch et al., 1999; Welchman et al., 2007; Ziel et al., 2009). The C. elegans genome contains clear orthologues of vertebrate racs (mig-2, ced-10 and rac-2), rhoA (rho-1) and cdc42 (cdc-42) (Lundquist, 2006). Both mig-2 and ced-10 function during DTC migration and AC invasion, while cdc-42 has been shown to regulate early gonad organization (Blelloch et al., 1999; Welchman et al., 2007; Ziel et al., 2009). However, the early embryonic lethality associated with complete loss-of-function mutations in ced-10 and cdc-42 or with rho-1 RNAi treatment, have complicated functional analysis of these genes during gonadogenesis.
An alternative approach to further understand Rho GTPase signaling during development is to examine the regulatory proteins that control their activity. Rho family members are molecular switches that interact with their effector proteins in the GTP-bound state. Activation of Rho GTPases is controlled by guanine nucleotide exchange factors (GEFs), while inactivation is regulated by GTPase activating proteins (GAPs) (Narumiya, 1996). Of particular interest are the GEF proteins, as these molecules are thought to couple Rho GTPase activity to specific signaling events, such as those controlling developmental timing or directional migration (Schiller, 2006). Thus, mutations in individual GEF genes may be used to uncover functions for Rho GTPases in a specific subset of developmental or cellular events. Because most genomes contain a much larger complement of RhoGEFs than Rho-family GTPases, designing experiments to determine the individual or combinatorial contribution of specific RhoGEFs to Rho GTPase regulation is difficult without prior documentation of expression patterns (Schmidt and Hall, 2002). To this end, we have characterized 5’ cis-regulatory element (5’CRE) activity from nineteen putative RhoGEF genes during gonadogenesis in C. elegans. Our results provide evidence for a wide range of tissue and temporal specificity amongst these 5’CREs. We anticipate that the profiles described here will aid in designing experiments to determine the genetic requirements for Rho GTPase regulation by GEF proteins during organogenesis in vivo.
1.1 Identification of likely RhoGEF proteins in C. elegans
To identify likely RhoGEF-encoding genes in C. elegans we searched the SMART database for C. elegans proteins predicted to contain a dbl-homology (DH) domain, a characteristic feature of GEF proteins for Rho-type GTPases (Letunic et al., 2009). From this search we identified sixteen C. elegans genes likely to encode typical RhoGEF proteins (Figure 2). As expected, most of the predicted proteins encoded by these genes also contained a pleckstrin-homology (PH) domain in close proximity to the DH domain, which aids in the nucleotide exchange reaction (Schmidt and Hall, 2002). In addition, a second family of proteins related to vertebrate DOCK180 and C. elegans CED-5 have been shown to function as GEFs for some Rho family members (Cote and Vuori, 2007). For this reason, we examined 5’CRE activity from the three genes in this family (C02F4.1/ced-5, F22G12.5 and F46H5.4) that are encoded by the C. elegans genome (Cote and Vuori, 2007). While some of these genes (C02F4.1/ced-5, C33D9.1/exc-5, T19E10.1/ect-2, K11E4.4/pix-1, F32F2.1/uig-1, F55C7.7/unc-73, C09D1.1/unc-89 and C35B8.2/vav-1) have been identified by genetic screens or other loss-of-function approaches, eleven have not been previously characterized (Benian et al., 1996; Hikita et al., 2005; Lucanic and Cheng, 2008; Morita et al., 2005; Steven et al., 1998; Steven et al., 2005; Yoo and Greenwald, 2005). Supplemental Figure 2 contains a phylogenetic tree examining the evolutionary relationships between the DH-domain containing RhoGEFs from C. elegans, C. briggsae and C. remanei as well as a schematic representation of the domain architecture for all of the examined RhoGEF proteins. Our phylogenetic analysis revealed strong support for putative orthologous genes between the three nematode species, but failed to recover well-supported relationships between RhoGEF family members, likely due to poor sequence conservation and/or large divergence times between family members (Supplemental Figure 2).
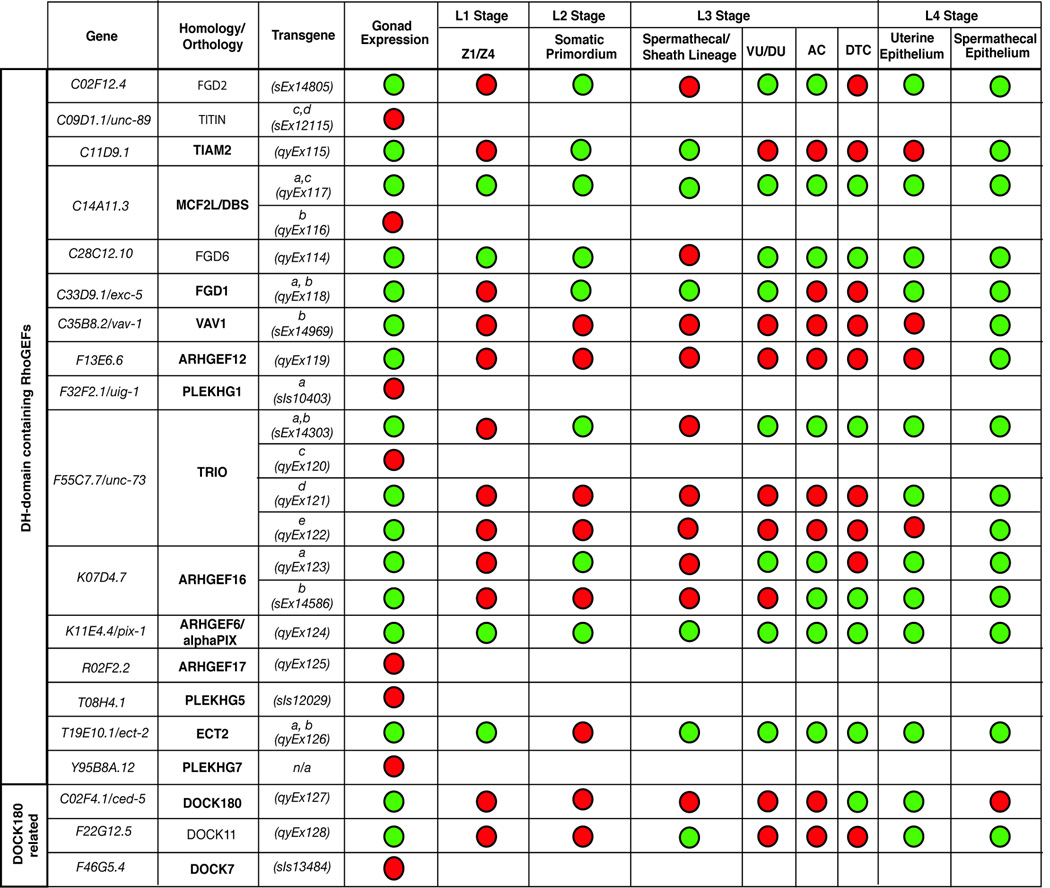
Figure 2.
Tabular summary of RhoGEF expression patterns during gonad development. Activity from sixteen 5’CREs derived from thirteen genes was detected during larval gonadogenesis. Red circles indicate an absence of activity whereas green circles indicate 5’CREs active within a particular cell type. Homology/orthology to human proteins was determined using BLAST, the best hit for the longest isoform of the C. elegans protein in the human genome is indicated in column two. Bolded entries indicate circumstances in which the human and C. elegans proteins are reciprocal best BLAST matches. Where shown, lowercase letters in column three indicate specific transcripts predicted to be regulated by each 5’CRE.
1.2 Identification of RhoGEF 5’CRE activity during gonadogenesis
To determine which RhoGEF proteins might be important during gonadogenesis, we sought to examine their spatial and temporal expression during larval development. For this purpose, we took advantage of the fact that 5’CRE activity can be efficiently assayed in C. elegans by linking putative regulatory elements to the coding sequences for the green fluorescent protein (GFP) in transgenic worms (Hunt-Newbury et al., 2007). In this way we examined 5’CREs from each of the nineteen RhoGEF genes identified due to the presence of a DH domain or similarity to CED-5 and DOCK180. In total we examined 24 transgenic lines representing 5’CREs from all 19 genes, encompassing 28 of the 34 total RhoGEF 5’CREs based upon current transcript predictions in Wormbase (WS199). A schematic view of each RhoGEF gene and detailed information regarding each associated reporter construct is available in Supplemental Figure 1 and Supplemental Table 1.
In our preliminary screen (see methods section) we identified 5’CREs from thirteen of these nineteen RhoGEFs that were active during gonadogenesis (Figure 2). Importantly, all 5’CREs examined drove GFP expression in at least some cells during larval development with the exception of those from Y95B8A.12 (data not shown). The lack of expression across multiple transgenic lines indicated that this gene may not be expressed in larvae or that our construct lacked the complete cis-regulatory architecture necessary to drive expression. For those RhoGEF 5’CREs that were active during gonadogenesis, we conducted a more detailed analysis of their expression patterns by focusing on critical developmental landmarks during larval gonad formation (examples of 5’CRE activity in each cell type examined are shown in Figure 1 I–L and summarized in Figure 2; all raw scoring data is provided in Supplemental Table 2).
1.3 RhoGEF 5’CRE activity during early gonadogenesis
To examine RhoGEF 5’CRE activity during early events in the organization of the somatic gonad, we scored GFP expression in Z1 and Z4 and in the somatic primordium during the L1 and L2 larval stages, respectively (Figure 1A, B, E, F and I). RhoGEFs expressed during these stages could contribute to early cell movements required for general somatic gonad organization and exclusion of the germ-cells into the gonad arms (Hubbard and Greenstein, 2000; Welchman et al., 2007). 5’CREs from four genes (C14A11.3, C28C12.10, K11E4.4/pix-1 and T19E10.1/ect-2) were active in Z1 and Z4 (Figure 1I; Figure 2). Interestingly, only the construct corresponding to the short transcripts of C14A11.3 (C14A11.3a,c) was active during gonadogenesis, indicating the potential for isoform-specific activity during gonad development (Figure 2). During the L2 stage we detected continued GFP expression directed by 5’CREs from C14A11.3a,c, C28C12.10 and K11E4.4/pix-1 within the somatic gonad primordium. In addition, regulatory regions corresponding to C02F12.4, C11D9.1, C33D9.1/exc-5 and K07D4.7a became active at this time (Figure 1J; Figure 2). Notably, the 5’CRE from T19E10.1/ect-2 was silent within these cells, but was reactivated within their progeny during the L3 stage, highlighting the potential for dynamic regulation of RhoGEF genes during gonadogenesis (Figure 2).
1.4 RhoGEF 5’CRE activity during DTC migration
We also identified RhoGEF 5’CREs that were active within the DTCs, where Rho GTPase function has been clearly demonstrated by loss-of-function studies (Blelloch et al., 1999). Six RhoGEF 5’CREs (C14A11.3a,c, C28C12.10, F55C7.7a,b/unc-73, K11E4.4/pix-1, K07D4.7b and C02F4.1/ced-5) drove GFP expression within the DTCs during the L3 stage, when mig-2 and ced-10 regulate DTC turning and directional migration (Figure 1K; Figure 2)(Reddien and Horvitz, 2000; Zipkin et al., 1997). Importantly, these data were consistent with previous reports of DTC phenotypes caused by mutations in C02F4.1/ced-5 and double-stranded RNA targeting F55C7.7/unc-73, which show defects DTC migration and guidance, and mutations in K11E4.4/pix-1, which manifest DTC morphology defects as well as guidance defects. (Cram et al., 2006; Lucanic and Cheng, 2008; Wu and Horvitz, 1998).
1.5 RhoGEF 5’CRE activity during AC invasion
We have shown previously that mig-2 and ced-10 function to promote basement membrane removal by the AC (Ziel et al., 2009). cdc-42 also plays an uncharacterized role in this process (J.W.Z., unpublished observations). How these proteins might be activated in response to the pathways that promote the timing and targeting of invasion is not known. Analysis of RhoGEF 5’CRE activity at the time of AC invasion in the mid-to-late L3 stage identified eight RhoGEF 5’CREs from seven genes capable of driving AC expression (Figure 1K; Figure 2; Figure 3A–H). In most cases (C02F12.4, C14Al1.3a,c, C28C12.10, F55C7.7a,b/unc73, K07D4.7a, K11E4.4/pix-1 and T19E10.1/ect-2) GFP expression was also detected amongst the dorsal and ventral uterine cells and thus might reflect enhancer elements broadly active within the central somatic cells of the gonad (Figure 2; Figure 3A–G). In contrast, a 5’CRE construct from K07D4.7b, highly similar to Ephexin and a human gene ARHGEF16, showed a high degree of specificity within the gonad. This 5’CRE, corresponding to the smaller transcript of K07D4.7, was only expressed within the AC and the DTCs (Figure 2; Figure 3H).
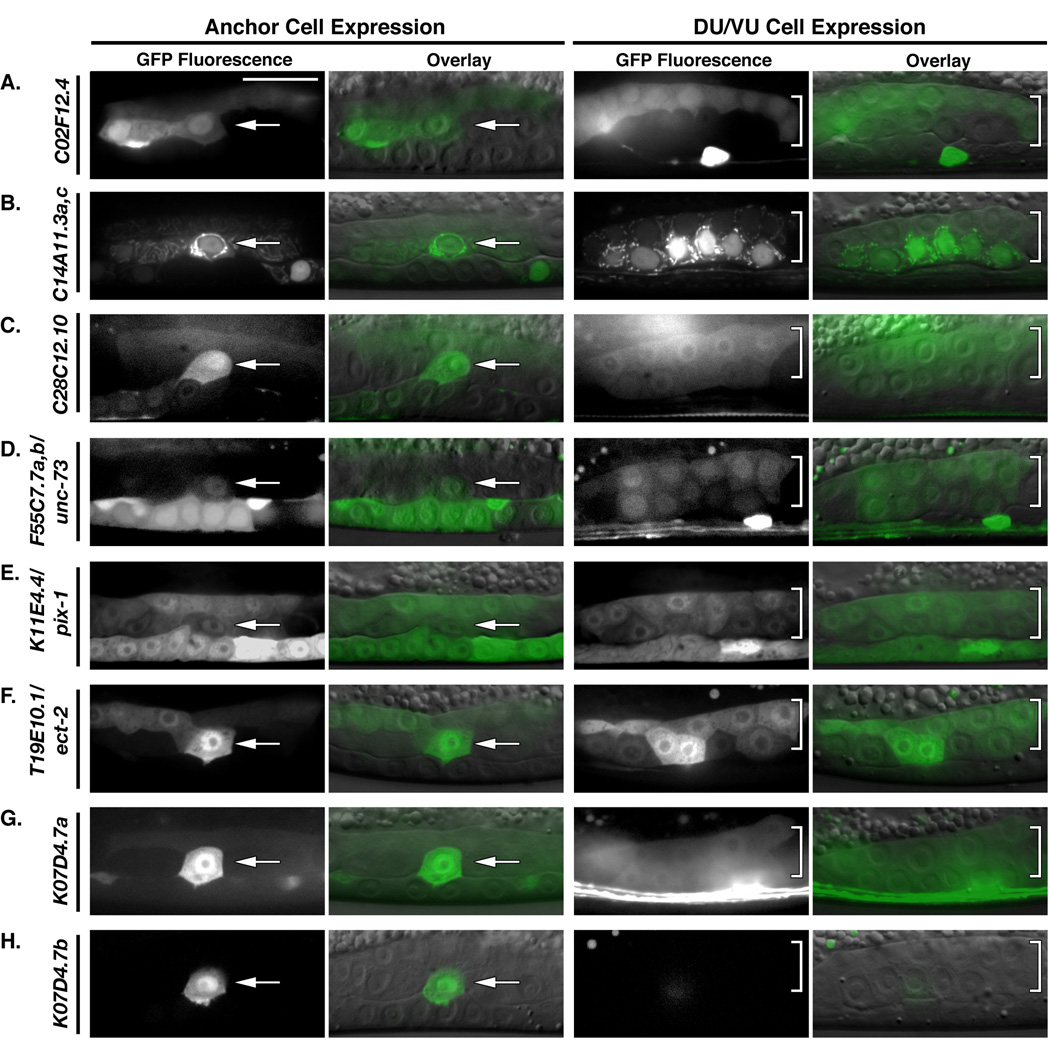
Figure 3.
Eight RhoGEF 5’CRE-reporter constructs were active around the time of AC invasion. (A) C02F12.4, (B) C14A11.3a,c, (C) C28C12.10, (D) F55C7.7a,b/unc-73 (E) K11E4.4/pix-1, (F) T19E10.1/ect-2 and (G) K07D4.7a were all active broadly within the central gonad at the mid-to-late L3 stage, being expressed in both the AC (arrows) and the VU/DU cells (brackets). (H) In contrast, 5’CREs from K07D4.7b were specifically active within the AC at the time of invasion and were not expressed within the VU/DU cells. Scale bar in (A) represents 20 microns.
1.6 RhoGEF 5’CRE activity during Uterine, Spermathecal and Gonad Sheath development
The gonad sheath as well as the uterine and spermathecal epithelia all derive from the somatic gonad primordium (Kimble and Hirsh, 1979). During gonad arm elongation, sheath cells, which derive from the same precursor cells as some spermathecal cells, ultimately form a contractile apparatus around the gonad arms (Kimble and Hirsh, 1979). Also during the late L3 stage and continuing into the L4 stage, cells of the dorsal and ventral uterus form a mature, lumenalized epithelial tube from a previously uniform cluster of cells (Newman et al., 1996). Importantly, Rho GTPase signaling is critical for establishing the apical-basolateral polarity characteristic of cells within mature epithelia (Van Aelst and Symons, 2002). During the late L3 stage we detected 5’CRE activity from a large number of RhoGEF genes within this tissue. In total eight 5’CREs (C02F12.4, C14A11.3a,c, C28C12.10, C33D9.1/exc-5, F55C7.7ab/unc-73, K07D4.7a, K11E4.4/pix-1 and T19E10.1/ect-2) were active in the VU/DU cells prior to epithelialization during the L3 stage (Figure 1K; Figure 2). Some of these elements were also active in the spermathecal and sheath lineages (C14A11.3a,c, C33D9.1/exc-5, K11E4.4/pix-1 and T19E10.1/ect-2) at the L3 stage (Figure 2). 5’CREs from C11D9.1 and F22G12.5 were active in the spermathecal and sheath lineages but not within the DU/VU cells (Figure 1K; Figure 2).
5’CREs from C02F4.1/ced-5, F55C7.7d, K07D4.7b and F22G12.5 became active within the mature epithelium during the L4 stage (Figure 2). Most of these genes were also active within the spermathecal epithelia at the L4 stage (Figure 2). Notably, four RhoGEF 5’CREs (from C11D9.1, C35B8.2/vav-1, F55C7.7e/unc-73 and F13E6.6) were active only in the spermathecal epithelia during the L4 stage (Figure 1L; Figure 2).
1.7 Conclusion
During gonadogenesis, Rho family GTPases are known to regulate early gonad organization, DTC migration and AC invasion (Welchman et al., 2007; Ziel et al., 2009; Zipkin et al., 1997). Based on experimental results from other systems, they are also predicted to regulate AC fusion to its neighboring uterine cells as well as lumenalization of the uterine and spermathecal epithelia (Cote and Vuori, 2007; Van Aelst and Symons, 2002). Thus the Rho GTPases are expected to have multiple functions during gonad organogenesis. How these functions would be coordinated and segregated during development, however, remains unclear. In our current study we provide the first comprehensive examination of RhoGEF 5’CRE activity during a complex developmental process in C. elegans. Our data show that a large portion of the C. elegans RhoGEF complement is expressed during gonad formation. 5’CRE activity associated with these genes is largely overlapping, both temporally and spatially, and all cell types examined express multiple RhoGEFs. Together these results support the idea that distinct RhoGEFs may be used to activate Rho GTPases for specific functions during development. The fact that multiple RhoGEF genes are expressed in most gonadal cell types also raises the possibility of extensive genetic redundancy. Consistent with this notion, only five of these nineteen RhoGEF genes (C09D1.1/unc-89, C33D9.1/exc-5, F55C7.7/unc-73, T19E10.1/ect-2 and C02F4.1/ced-5) have been identified by traditional forward genetic screens. Given this possibility, the 5’CRE activity profiles described here, as well as the transgenic reporter lines developed during this study will be useful in designing genetic experiments to examine the combinatorial functions of RhoGEF proteins during gonadogenesis and other developmental processes in C. elegans.
2. Materials and Methods
2.1 Generation of C. elegans RhoGEF 5’CRE reporter transgenes
Some strains (indicated by the prefix sIs or sEx) were provided by the Caenorhabditis Genetics Center and were made available by the Genome B.C. C. elegans Gene Expression Consortium (Hunt-Newbury et al., 2007). 5’CRE-reporter transgenes were generated using a previously reported PCR fusion strategy (Hobert, 2002). Briefly, putative regulatory sequences upstream of each transcript were amplified from N2 genomic DNA using a specialized reverse primer containing sequence homologous to a small region of the coding sequences for GFP. In a second reaction, the resulting PCR product was combined with PCR-amplified coding sequences from pPD95.75, which contained synthetic introns for robust expression in C. elegans and a 3’ untranslated region from the unc-54 gene. This mixture was then amplified with a forward primer nested within the 5’CRE amplicon and a reverse primer nested within the GFP amplicon. The resulting PCR products were examined by gel electrophoresis and their concentration estimated. All PCR reactions were performed using the high-fidelity Expand Long Range PCR kit (Roche). Putative regulatory regions were typically either the intergenic region or approximately 2.5 kb of sequence upstream of each transcript.
Extrachromosomal arrays carrying each 5’CRE-reporter transgene were generated by germ-line transformation of unc-119(ed4) mutants. A standard injection mix containing 1–10 ng/ul 5’CRE-reporter DNA, 75 ng/ul pBSSK-, and 50 ng/ul unc-119-rescuing sequence (pDP#MM016B) was used. Transformants were distinguished by their non-Unc phenotype and transgenic lines were established from single non-Unc F2 progeny. These strains were propagated using standard techniques (Brenner, 1974). A complete list of primers, 5’CRE sizes, and resulting extrachromosomal arrays is presented in Table S1. Schematic representations of each 5’CRE-reporter construct in the context of the endogenous locus are presented in Supplemental Figure 1.
2.2 Phylogenetic analysis of predicted RhoGEF protiens
An amino acid alignment of the ~200–amino acid (AA) DH domains from the C. elegans DH-domain containing RhoGEF proteins was generated using MUSCLE v.3.6 (Edgar, 2004). Bayesian phylogenetic analyses were conducted using MrBayes 3.1.2 via a web portal at the CBSU Web Computing Resources Microsoft High-Performance Computing Institute (http://cbsuapps.tc.cornell.edu/index.aspx) (Ronquist and Huelsenbeck, 2003). The Bayesian analysis was conducted using a mixed amino acid model with gamma which selected the WAG model of amino acid evolution with a 100% posterior probability with 2,000,000 generations sampled every 100 generations with four chains over four independent runs. A summary tree was produced from the final 18,000 trees representing 1,800,000 stationary generations per run, and 72,000 trees representing 7,200,000 stationary generations for the consensus tree. In addition, neighbor joining (NJ) (using mean AA distances) was performed using PAUP* 4.0b10 (Swofford, 2000). ProtTest selected the LG+I+G model of amino acid evolution, which was used for a maximum likelihood (ML) bootstrap analysis using RaXML v7.0.4 (Stamatakis, 2006; Abascal et al., 2005; Le and Gascuel, 2008) performed on the Duke Shared Cluster Resource. The LG model was provided at http://www.kramer.in.tum.de/exelixis/software.html. Domain architecture of the C. elegans RhoGEF proteins was determined using the SMART database or the NCBI Conserved Domain Database (Marchler-Bauer et al., 2009).
2.3 Scoring of RhoGEF 5’CRE activity during gonadogenesis
Living worms were examined and imaged using a Zeiss AxioImager A1 microscope with a 100 plan-apochromat objective and a Zeiss AxioCam MRm CCD camera, controlled by Zeiss Axiovision software (Zeiss Microimaging), or using a Yokogawa spinning disk confocal mounted on a Zeiss AxioImager A1 microscope controlled by the iVision software package (Biovision Technologies). Images were processed and overlaid using Photoshop 8.0 (Adobe Systems).
RhoGEF 5’CRE-reporter transgenes were first screened for general gonadal expression by examining at least thirty transgenic worms across the four larval stages. 5’CRE reporter transgenes that were active during any stage and in any gonadal cell-type were subjected to a more detailed screening process at the discrete developmental stages described in Figure 1. Because extrachromasomal arrays are unstable and can be lost during mitosis, at least ten animals were examined at each specific stage. Raw scoring data for each 5’CRE-reporter construct is provided in Supplemental Table 2.
Supplementary Material
Acknowledgements
We thank the Caenorhabditis Genetics Center for providing strains and E. Hagedorn for helpful comments during preparation of the manuscript. This work was supported by a Basil O’Connor Award, Pew Scholars Award, Howard Temin K01 Award CA098316-01, and NIH Grant GM079320 to D.R.S. D.Q.M. is a Robert Black Fellow of the Damon Runyon Cancer Research Foundation (DRG-1949-07).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abascal F, et al. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Benian GM, et al. The Caenorhabditis elegans gene unc-89, required fpr muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J Cell Biol. 1996;132:835–848. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch R, et al. Control of cell migration during Caenorhabditis elegans development. Curr Opin Cell Biol. 1999;11:608–613. doi: 10.1016/s0955-0674(99)00028-9. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote JF, Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 2007;17:383–393. doi: 10.1016/j.tcb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cram EJ, et al. A systematic RNA interference screen reveals a cell migration gene network in C. elegans. J Cell Sci. 2006;119:4811–4818. doi: 10.1242/jcs.03274. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock EM, et al. Genetics of cell and axon migrations in Caenorhabditis elegans. Development. 1987;100:365–382. doi: 10.1242/dev.100.3.365. [DOI] [PubMed] [Google Scholar]
- Hikita T, et al. Identification of a novel Cdc42 GEF that is localized to the PAT-3-mediated adhesive structure. Biochem Biophys Res Commun. 2005;335:139–145. doi: 10.1016/j.bbrc.2005.07.068. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hubbard EJ, Greenstein D. The Caenorhabditis elegans gonad: a test tube for cell and developmental biology. Dev Dyn. 2000;218:2–22. doi: 10.1002/(SICI)1097-0177(200005)218:1<2::AID-DVDY2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Hunt-Newbury R, et al. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Letunic I, et al. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucanic M, Cheng HJ. A RAC/CDC-42-independent GIT/PIX/PAK signaling pathway mediates cell migration in C. elegans. PLoS Genet. 2008;4:e1000269. doi: 10.1371/journal.pgen.1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist EA. Small GTPases. WormBook. 2006:1–18. doi: 10.1895/wormbook.1.67.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;32:D205. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, et al. The Caenorhabditis elegans ect-2 RhoGEF gene regulates cytokinesis and migration of epidermal P cells. EMBO Rep. 2005;6:1163–1168. doi: 10.1038/sj.embor.7400533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narumiya S. The small GTPase Rho: cellular functions and signal transduction. J Biochem. 1996;120:215–228. doi: 10.1093/oxfordjournals.jbchem.a021401. [DOI] [PubMed] [Google Scholar]
- Newman AP, Sternberg PW. Coordinated morphogenesis of epithelia during development of the Caenorhabditis elegans uterine-vulval connection. Proc Natl Acad Sci U S A. 1996;93:9329–9333. doi: 10.1073/pnas.93.18.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, et al. The Caenorhabditis elegans lin-12 gene mediates induction of ventral uterine specialization by the anchor cell. Development. 1995;121:263–271. doi: 10.1242/dev.121.2.263. [DOI] [PubMed] [Google Scholar]
- Newman AP, et al. Morphogenesis of the C. elegans hermaphrodite uterus. Development. 1996;122:3617–3626. doi: 10.1242/dev.122.11.3617. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat Cell Biol. 2000;2:131–136. doi: 10.1038/35004000. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schiller MR. Coupling receptor tyrosine kinases to Rho GTPases--GEFs what's the link. Cell Signal. 2006;18:1834–1843. doi: 10.1016/j.cellsig.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- Sharma-Kishore R, et al. Formation of the vulva in Caenorhabditis elegans: a paradigm for organogenesis. Development. 1999;126:691–699. doi: 10.1242/dev.126.4.691. [DOI] [PubMed] [Google Scholar]
- Sherwood DR, et al. FOS-1 promotes basement-membrane removal during anchor-cell invasion in C. elegans. Cell. 2005;121:951–962. doi: 10.1016/j.cell.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Sherwood DR, Sternberg PW. Anchor Cell Invasion into the Vulval Epithelium in C. elegans. Developmental Cell. 2003;5:21–31. doi: 10.1016/s1534-5807(03)00168-0. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Steven R, et al. UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell. 1998;92:785–795. doi: 10.1016/s0092-8674(00)81406-3. [DOI] [PubMed] [Google Scholar]
- Steven R, et al. The UNC-73/Trio RhoGEF-2 domain is required in separate isoforms for the regulation of pharynx pumping and normal neurotransmission in C. elegans. Genes Dev. 2005;19:2016–2029. doi: 10.1101/gad.1319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, et al. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Sundaram MV. Vulval Development: The Battle between Ras and Notch. Current Biology. 2004;14:R311. doi: 10.1016/j.cub.2004.03.052. [DOI] [PubMed] [Google Scholar]
- Swofford DL PAUP*. Phylogenetic Analysis Using Parsimony *and Other Methods. Sunderland, Massachusetts: Sinauer Associates; 2000. [Google Scholar]
- Van Aelst L, Symons M. Role of Rho family GTPases in epithelial morphogenesis. Genes Dev. 2002;16:1032–1054. doi: 10.1101/gad.978802. [DOI] [PubMed] [Google Scholar]
- Welchman DP, et al. Similar requirements for CDC-42 and the PAR-3/PAR 6/PKC-3 complex in diverse cell types. Dev Biol. 2007;305:347–357. doi: 10.1016/j.ydbio.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YC, Horvitz HR. C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature. 1998;392:501–504. doi: 10.1038/33163. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Greenwald I. LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans. Science. 2005;310:1330–1333. doi: 10.1126/science.1119481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziel JW, et al. UNC-6 (netrin) orients the invasive membrane of the anchor cell in C. elegans. Nat Cell Biol. 2009;11:183–189. doi: 10.1038/ncb1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipkin ID, et al. Role of a new Rho family member in cell migration and axon guidance in C. elegans. Cell. 1997;90:883–894. doi: 10.1016/s0092-8674(00)80353-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.