Abstract
Apoptosis inducing factor (AIF) is a bifunctional mitochondrial flavoprotein critical for energy metabolism and induction of caspase-independent apoptosis, whose exact role in normal mitochondria remains unknown. Upon reduction with NADH, AIF undergoes dimerization and forms tight, long-lived FADH2-NAD charge-transfer complexes (CTC) proposed to be functionally important. To get a deeper insight into structure/function relations and redox mechanism of this vitally important protein, we determined the x-ray structures of oxidized and NADH-reduced forms of naturally folded recombinant murine AIF. Our structures reveal that CTC with the pyridine nucleotide is stabilized by (i) π-stacking interactions between coplanar nicotinamide, isoalloxazine and Phe309 rings, (ii) rearrangement of multiple aromatic residues in the C-terminal domain, likely serving as an electron delocalization site, and (iii) an extensive hydrogen-bonding network involving His453, a key residue undergoing a conformational switch to directly interact and orient the nicotinamide in position optimal for charge transfer. Via the His453-containing peptide, redox changes in the active site are transmitted to the surface, promoting AIF dimerization and restricting access to a primary nuclear localization signal through which the apoptogenic form is transported to the nucleus. Structural findings agree with the biochemical data and support the hypothesis that both normal and apoptogenic functions of AIF are controlled by NADH.
Keywords: apoptosis inducing factor, flavoprotein, mitochondria, crystal structure, charge-transfer complex, redox signaling
INTRODUCTION
Programmed cell death (PCD), also called apoptosis, is essential for early embryological development and tissue homeostasis. Dysregulation of apoptosis may lead to abnormal embryogenesis and a variety of human diseases, such as myocardial infarction, autoimmune syndromes, neurodegenerative disorders and cancer 1. Apoptosis can be initiated by the caspase family of cysteine proteases or occur independently of caspase activation 2; 3. One of the key caspase-independent death effectors is a phylogenetically old mitochondrial flavoprotein, apoptosis inducing factor (AIF) 4; 5; 6. An AIF precursor is encoded by a nuclear gene and posttranslationally imported into mitochondria. The processed 60 kDa mature protein (Δ1–53) is inserted into the inner mitochondrial membrane with the N-terminus facing the matrix and the C-terminal catalytic domain exposed into the intermembrane space 7. Upon an apoptotic insult, the membrane linker of AIF becomes proteolyzed and the detached Δ1–101 fragment is transported via two nuclear localization signals (NLS) into the nucleus, where it triggers chromatin condensation and large-scale DNA fragmentation 4, a hallmark of caspase-independent apoptosis.
AIF does not possess endonuclease activity and cooperates with other proteins, such as endonuclease G and cyclophilin A, to degrade DNA 8; 9; 10. The lymphoid protein TULA/Sts-2 is another enhancer of the AIF-mediated apoptogenic effect 11, wherein Hsp70 and X-linked inhibitor of apoptosis suppress the pro-apoptotic action of the flavoprotein by preventing its nuclear translocation and interaction with PCD-related partners 12; 13; 14;15. While the lethal function of AIF has been extensively investigated, the physiological role of the protein in normal mitochondria remains unclear. Based on properties of the refolded Δ1–120 fragment, AIF was proposed to function as a superoxide-generating NADH oxidase or electron transferase 16; 17 with unrelated redox and apoptogenic actions 4. In vivo studies, on the other hand, indicated that AIF may act as an antioxidant 18, assist biogenesis/maintenance of respiratory complexes I and III 19; 20; 21 and regulate mitochondrial morphology and cristae formation 22, possibly through interaction with the dynamin-related opa1 protein 21.
To date, only crystal structures of recombinant mouse and human AIFΔ1–120, expressed as apoproteins and refolded to incorporate FAD, have been determined 17; 23. The two structures are nearly identical and closely resemble that of BphA4, a glutathione reductase (GR)-like ferredoxin reductase component of biphenyl reductase from Pseudomonas sp. strain KKS102 24. The mouse and human AIF structures differ only in the number of monomers (two and one, respectively) and folding of a 540–559 peptide (disordered in the human model), most likely due to different crystal packing. The 540–559 peptide is part of the AIF-specific insertion (aa. 509–559) that contains two regulatory elements: the proline-rich motif (PPxxPxxPxxP), a potential recognition site for Src homology 3 (SH3) domains found in cytoskeletal and signaling proteins 25; 26, and a proline/glutamate/serine/threonine-rich (PEST) sequence, a putative proteolytic signal and a phosphorylation site 27. Since the 509–559 peptide is absent in the AIF homologues from lower eukaryotes, Mate et al. argued against its direct role in the apoptogenic and redox functions of the protein 17. Furthermore, observations that recombinant AIF becomes apoptogenic after refolding in the absence of the flavin cofactor and a naturally occurring FAD-free isoform (AIFsh, aa. 353–613) provokes the same apoptogenic effect as an intact flavoprotein, led to a conclusion that the oxidoreductase and apoptogenic activities of AIF can be separated 4; 28.
Our recent study challenged this concept and suggested that AIF may act as a redox signaling molecule whose normal and apoptotic functions are interrelated 29. First, we demonstrated that natural flavin incorporation is critical for preservation of the redox properties of AIF and, when properly folded, the protein has a 100-fold preference for NADH over NADPH, reacts with the reduced cofactor orders of magnitude slower than other flavoenzymes, and forms tight, dimeric and air-stable FADH2-NAD charge-transfer complexes (CTC) inefficient in electron transfer. Second, formation of CTC was found to induce conformational changes in the regulatory 509–559 peptide and affected susceptibility of the N-terminus to proteolysis and AIF-DNA interaction, the two events critical for initiation of the caspase-independent apoptosis. These results, as well as the finding that native AIF undergoes redox-dependent monomer-dimer transition and its release from mitochondria is inhibited by pyridine nucleotides, led us to hypothesize that both normal and apoptogenic functions of AIF are redox-controlled.
To further investigate the mechanism and structure/function relations in AIF and elucidate how reduction with NADH changes molecular properties of the protein, we determined the x-ray structures of oxidized and NADH-reduced forms of naturally folded recombinant murine AIF. The atomic models helped to identify aberrations caused by refolding and to understand how CTC with the pyridine nucleotide is formed and how AIF-NADH association/dissociation could affect normal and apoptogenic actions of the protein. In accord with the biochemical data, this study suggests that AIF may represent a novel redox sensor linking NAD(H)-dependent metabolic pathways to apoptosis.
RESULTS AND DISCUSSION
Structural differences between naturally folded and refolded AIF
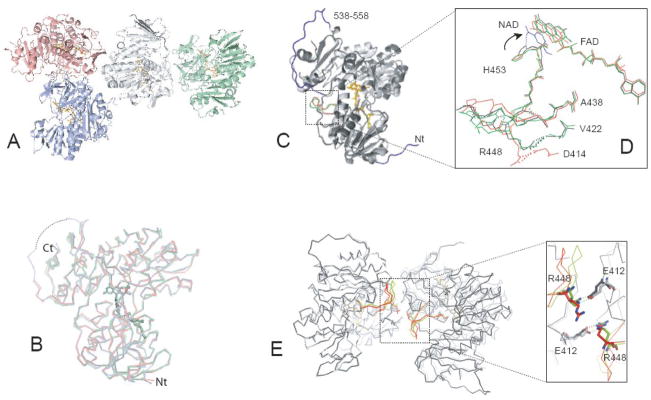
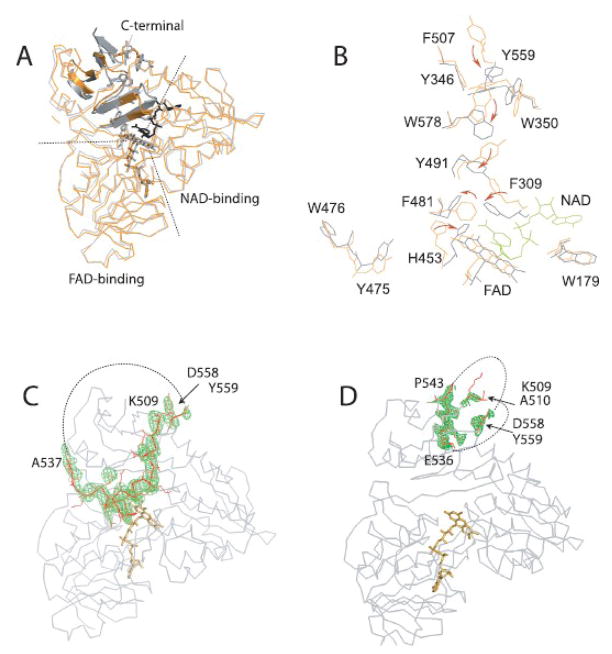
Given that only structural studies on refolded, ligand-free AIFΔ1–120 have been conducted 17; 23 and refolding significantly perturbs redox properties of the flavoprotein 29, structural information on both oxidized and reduced forms of naturally folded AIF was necessary to accurately identify redox-linked conformational changes. For this purpose, we first solved the 2.95 Å x-ray structure of oxidized, ligand-free AIFΔ1–77 (Table 1). The Δ1–77 protein lacks only the trans-membrane peptide, resembles mature AIFΔ1–53 in structure and function 29 and is more suitable for biochemical and structural studies because it is highly expressed in E. coli and less susceptible to the N-terminal proteolysis. The asymmetric unit of oxidized AIFΔ1–77 is comprised of four topologically similar monomers (molecules A, B, C and D, Fig. 1A). Molecules A and D are the best and the least defined, respectively. In all four monomers, the first fifty N-terminal and six C-terminal residues as well as parts of the 538–557 peptide are disordered and not visible in the structure. When the whole monomer structures are superposed (Fig. 1B), the average pair-wise root mean square deviation (rmsd) for their α-carbons is only 0.50–0.62 Å. Although oxidized AIF exists as a monomer in solution 29, the molecules in the crystal lattice are arranged in two dimers, highly similar to the crystallographic dimer of refolded AIFΔ1–120;17. Using molecules A from our and the latter structure for comparative analysis, we found that, in addition to subtle differences in the side chain and backbone conformations (the Cα rmsd of 0.56 Å), there was notable displacement of FAD in the refolded protein (0.5 and 0.7 Å for the adenosine and isoalloxazine moieties, respectively, Fig. 1D). Another important dissimilarity is around the 438–453 peptide, extending from the active site to the dimer interface. Part of this peptide (aa. 438–540) represents an elongated loop in refolded AIF and a β-turn in our protein (Fig. 1C, D). This conformational variation affects position of the Arg448 side chain, which forms a salt bridge with Asp414 in refolded AIF and an H-bond with the Val422 carbonyl oxygen in our structure (Fig. 1D). As a consequence, the crystallographic monomer-monomer interface in naturally folded AIF is more extensive and contains two Glu412-Arg448 inter-subunit salt bridges (Fig. 1E), playing a central role in stabilization of the NADH-reduced dimer (discussed below).
Table 1.
Data Collection, Refinement and Model Statistics
| AIFΔ1–77 oxidized | AIFΔ1–101 reduced, NAD-bound | |
|---|---|---|
| wavelength (Å) | 1.1 | 1.1 |
| oscillation range (deg.) | 0.5 | 1.0 |
| measurement angle (deg.) | 160 | 163 |
| Crystallographic Data | ||
| space group | P 212121 | P212121 |
| a (Å) | 75.1 | 60.3 |
| b (Å) | 80.3 | 120.4 |
| c (Å) | 419.6 | 178.2 |
| molecules per unit | 4 | 2 |
| resolution (Å) | 2.95 | 2.24 |
| total observations | 162,075 | 348,452 |
| unique reflections | 46,752 | 57,099 |
| I/σ(I) at dmin | 10.5 (2.0) | 32.9 (3.6) |
| Rsym (%) | 7.0 (49.2) | 3.6 (33.3) |
| Wilson B factor (Å2) | 92.3 | 33.9 |
| completeness | 85.2 (81.7) | 95.1 (85.3) |
| Refinement Statistics | ||
| Resolution range (Å) | 408.3-2.95 | 30.0-2.24 |
| R/Rfreea (%) | 25.4/29.7 | 18.6/22.2 |
| number of protein atoms | 14,480 | 6,806 |
| number of solvent atoms | 27 | 521 |
| average B factor (Å2) | 79.3 | 46.4 |
| rmsd bond lengths (Å) | 0.01 | 0.01 |
| rmsd angles (deg.) | 1.68 | 1.67 |
- Rfree is calculated from a subset of 5% of the data, which were excluded during refinement.
Figure 1.
Crystal structure of oxidized AIFΔ1–77. A, Molecules A, B, C, and D comprising the asymmetric unit are displayed in gray, blue, pink and green, respectively. Monomer A is the best defined in the crystal structure and monomer D the least. B, The least-squares backbone superposition of four crystallographically independent molecules. A disordered part of the 509–559 peptide is shown as a dotted line. C, Structural overlay of molecules A of AIFΔ1–77 (light gray) and refolded AIFΔ1–120 (dark gray, PDB code 1DV4). The N-terminal and 538–558 fragments (not seen in the AIFΔ1–77 structure) are in dark blue; differently folded 438–453 peptides are in green and red; FAD is in yellow and orange. D, Conformational differences in FAD and the 438–453 peptide in refolded (red), and naturally folded ligand-free AIF (green). The corresponding elements in the CTC structure are in black. Redox-induced movement of His453 toward the nicotinamide (shown in blue) is indicated by an arrow. E, The 438–453 peptide defines interactions at the subunit interface and allows formation of two inter-subunit Glu412-Arg448 salt-bridges in naturally folded (light gray and green) but not in refolded AIF (dark gray and red).
Crystallization and overall structure of reduced, NAD-bound AIF
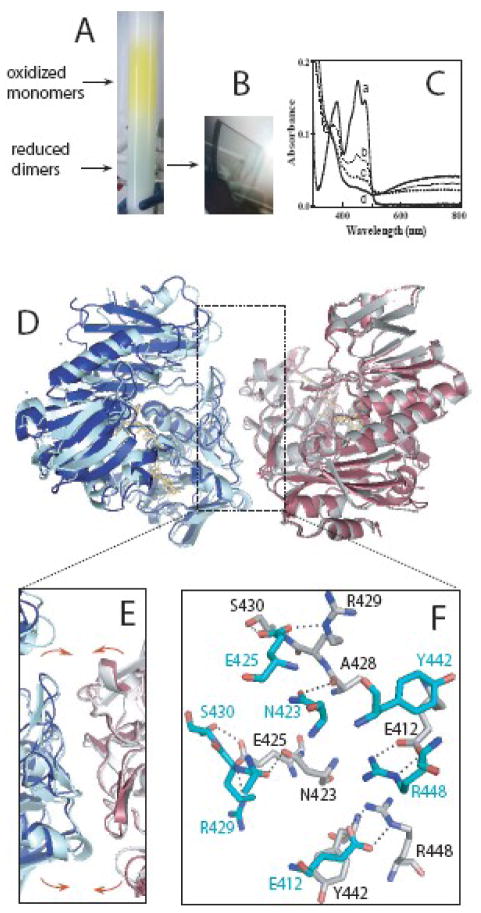
Soaking crystals of oxidized AIF with NADH led to crystal cracking, likely due to structural reorganization caused by cofactor binding and FAD reduction. Attempts to crystallize AIF freshly reduced with NADH were also unsuccessful. The reduced, dimeric fraction of AIFΔ1–101 purified from E. coli, however, easily produced well diffracting crystals suitable for x-ray analysis (Fig. 2A, B). One possible reason for this dissimilarity could be conformational heterogeneity of the freshly reduced AIF which, as we showed previously, is also dimeric in solution 29. Taking into account thermodynamic instability of the FAD semiquinone, that does not accumulate during AIF oxidoreduction 29, and the optical spectrum of the crystalline protein, typical for the FADH2-NAD charge-transfer complex (Fig. 2C), we conclude that FAD in the crystals is in the 2-electron reduced state.
Figure 2.
Crystal structure of the dimeric charge-transfer complex. A and B, Separation during gel filtration and crystals of reduced, NAD-bound AIFΔ1–101. C, Absorbance spectra of monomeric and dimeric fractions (a and b, respectively), crystalline protein (c, recorded in a suspension of crashed crystals) and freshly NADH-reduced AIF (d). Resemblance between the spectra c and d and inability of AIF to stabilize FAD semiquinone 29 allow to conclude that the cofactor in the crystals is in the 2-electron reduced form. D, Superposition of the crystallographic dimer of ligand-free AIFΔ1–77 (molecules A and D, shown in red and blue, respectively) and the biological dimer of reduced, NAD-bound AIFΔ1–101 (gray and cyan). E and F, A magnified view and interactions at the dimer interface in the latter structure, respectively. Although the manner of monomer-monomer interaction in the oxidized and reduced structures is highly similar, the CTC subunits are positioned closer (shown by arrows in panel E) and establish a more extensive H-bonding network.
The structure of the AIF-pyridine nucleotide complex was solved to 2.24 Å resolution (Table 1) and consists of one biological dimer, topologically similar to the crystallographic dimers of ligand-free AIF (Fig. 2D). The CTC monomers are positioned slightly closer and establish a more intricate network of hydrophobic and polar interactions, central of which are two inter-subunit Glu412-Arg448 salt bridges (Fig. 2E, F). Since in refolded AIF, incapable of forming CTC with equimolar NAD(P)H 16, Arg448 is locked in an alternative conformation inaccessible to Glu412 (Fig. 1D), it is plausible to conclude that electrostatic interactions between Arg448 and Glu412 assist dimerization and/or stabilization of the cofactor-bound intermediate.
Pyridine nucleotide-induced changes in the active site
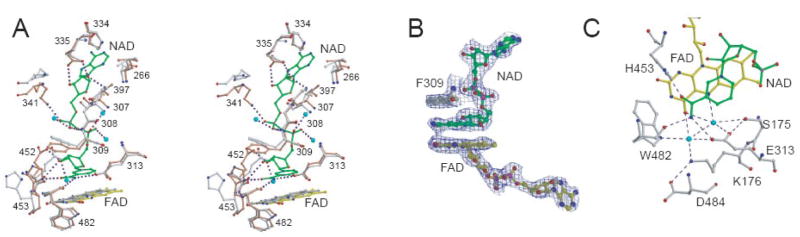
Flavin reduction and establishing of a FADH2-NAD CTC lead to significant changes in the pyridine nucleotide binding channel and the active site (Fig. 3A). In both AIF molecules, NAD is well defined and bound in an extended conformation with the nicotinamide group parallel stacked between the isoalloxazine and Phe309 rings (Fig. 3B). The flavin nucleotide portion of FAD shifts by ca. 1.2 Å to optimize π-π charge-transfer interactions with the nicotinamide. Two active site water molecules 17 are displaced by NAD and move aside, shifting the Lys176 side chain and forming an extensive hydrogen bonding network with the cofactors (Fig. 3C). Conserved in AIFs Lys176 and Glu313 provide a majority of interactions that position the solvent molecules, most likely proton donors, within an H-bond distance from FAD N5 and O4. This arrangement could explain why the K176A and E313A mutants of AIF have considerably lower KM for NADH 17: elimination of bulky lysine and glutamate side chains may create space for displaced waters and facilitate ligand binding.
Figure 3.
A, Stereodiagram of the superimposed pyridine nucleotide binding channels in ligand-free (gray) and NAD-bound AIF (light brown). Dotted lines represent hydrogen bonds; blue spheres are water molecules. B, The 2Fo-Fc electron density map at the active site contoured at 1σ shows that the nicotinamide of NAD is parallel stacked between the isoalloxazine and Phe309 rings. C, A hydrogen-bonding network in the active site involves two water molecules and optimally orients the nicotinamide for charge transfer.
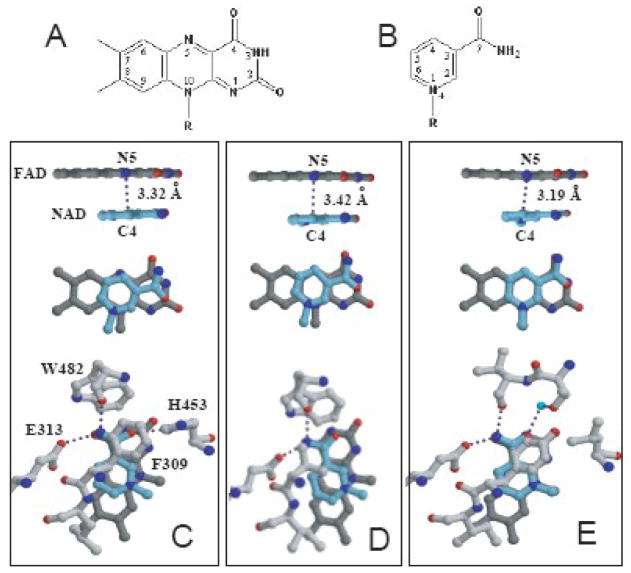
The relative orientation and H-bonding network between the flavin and pyridine nucleotide in our structure resemble those observed in the x-ray models of homologous proteins complexed with NAD(P)H (Fig. 4) 24; 30. A distinctive feature of AIF is the active site His453 (Leu or Ile in other GR-like enzymes), which undergoes a 3.5 Å shift to establish a hydrogen bond with NAD O7 and optimally orient the nicotinamide for charge transfer (Fig. 1D and 3C). This interaction is critical for several reasons. First of all, redox-induced movement of His453 affects positioning of the 438–453 peptide (Fig. 1D) and, as a result, changes the subunit interface and facilitates AIF dimerization. Secondly, the H453L mutation drastically lowers affinity of AIF for NADH and prevents CTC formation 29. Since the active site architecture in the GR family of enzymes is highly similar (Fig. 6 in 29) and neither Leu nor Ile at position 453 (AIF numbering) preclude FADH2-NAD(P) charge-transfer interactions in other flavoproteins, we postulate that formation of long-lived CTC and, possibly, AIF dimerization are assisted by conformational changes not only in the active site but also in the adjacent areas. One such region may be the C-terminal domain, as discussed in the next section.
Figure 4.
Structures (A and B) and relative orientation (C–E) of the isoalloxazine and nicotinamide groups (rendered in dark gray and blue, respectively) in the x-ray models of AIF (C), BphA4 (D, PDB code 1FP3), and human GR (E, PDB code 1GRB). AIF-specific His453, critical for NADH binding and CTC formation, undergoes a 3.5 Å shift to establish an H-bond with the nicotinamide.
Redox reorganization in the C-terminal domain
The C-terminal domain undergoes the most dramatic redox changes. In particular, the reorganization involves Phe481, Phe309, Tyr491, Trp578, Trp350, Tyr346, Phe507, and Tyr559, that rearrange and align in an aromatic tunnel stretching from the active site to the surface (Fig. 5A, B). Displacement of Phe481 and π-stacking between Phe309 and the nicotinamide could initiate this rearrangement, whereas a 9.4 Å shift of surface Tyr559 may be the last step bringing the tyrosine ring, a “lid” of the tunnel, in the vicinity of the Tyr346-Trp350-Phe507 cluster. With the distances between the aromatic rings < 4.0 Å, the tunnel is perfectly suited for transfer and delocalization of electrons, which could be one of the factors prolonging the CTC lifetime. Another important outcome of structural reorganization in the C-terminal domain and active site is dislocation of the entire 509–558 peptide, whose α-helical portion (aa. 517–523) unwinds and the 536–543 stretch transforms into the 6th strand of the C-terminal β-sheet, while residues 511–535 and 544–557 become/remain fully disordered (Fig. 5C, D). The NADH-triggered unwinding of the 517–523 helix explains why trypsinolysis of the Lys517-Ser518 bond is redox-controlled 29. In contrast to drastic structural changes seen in the crystalline BphA4-NADH complex 31, no notable domain movement was observed in AIF upon reduction.
Figure 5.
NADH-induced reorganization in the C-terminal domain. A, Structural overlay of oxidized (orange) and reduced AIF (gray). Dotted lines represent borders between the FAD-binding, NAD-binding and C-terminal domains. FAD and NAD (shown in black) are in stick representation. The most notable changes are observed in the 480–506 and 563–574 peptides (in cartoon representation). Only the side chains of residues comprising an aromatic tunnel, that extends from the active site to the surface, are displayed. B, Redox-induced rearrangement of phenylalanines, tyrosines and tryptophans, leading to formation of an aromatic tunnel, a potential electron delocalization site, is indicated by arrows. Tyr559 shifts by 9.4 Å to become a “lid” of a tunnel, which may assist dislocation of the preceding 509–558 peptide. C and D, SA-omit maps for the 509–559 peptide in oxidized and reduced AIF, respectively. Missing parts are shown as dotted lines.
Changes in the Trp195-containing peptide
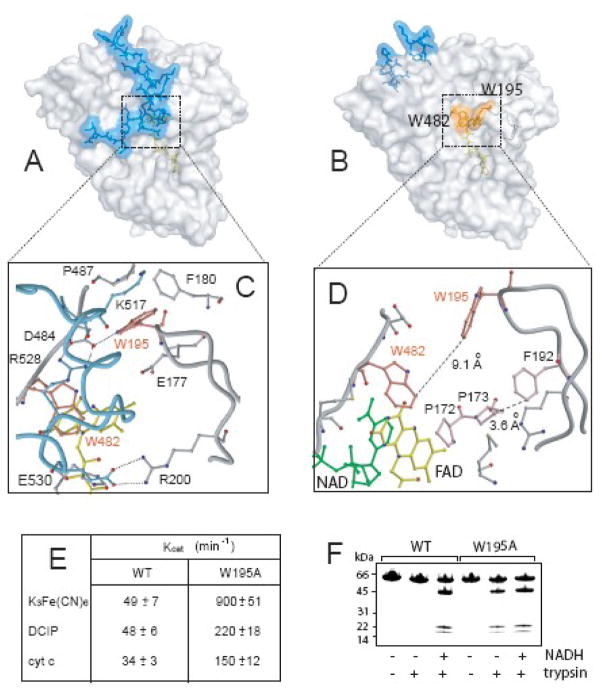
Redox-dependent transformations in the 509–559 peptide have far-reaching consequences. First of all, they change position of the 190–202 fragment, another AIF-specific insertion folded as a β-hairpin with five residues per strand and a two-residue turn. In oxidized AIF, residing at the tip Trp195 is buried and provides both hydrophobic and H-bonding interactions that retain the hairpin next to the 517–523 α-helix (Fig. 6A, C). Upon FAD reduction, this helix unwinds and the hairpin is released into the solvent (Fig. 6B, D). Since solvent exposed tryptophans are rare and usually functionally important, we examined the role of Trp195 by replacing it with alanine. The mutation dramatically changed properties of AIF. Compared to wild type, AIF W195A has a 70 mV higher redox potential, reacts with NADH two orders of magnitude faster (kET of 0.3 versus 26.3 sec−1 at 20°C), forms two-fold longer lived CTC, possesses several-fold higher electron transferring ability, especially toward one-electron acceptors (Fig. 6E), and its trypsin digest pattern is redox independent (Fig. 6F). The latter observation means that the Lys517-Ser518 bond is accessible due to unfolding of the 509–538 peptide 29 in both oxidized and reduced forms of the mutant. The longer-lived CTC, in turn, suggests that Trp195 may serve as an electron leakage/exit site. According to the structure, reducing equivalents could reach Trp195 via Pro172 → Pro173 → Phe192 and a β-hairpin wire (Fig. 6D). If this assumption is correct, the flexible (high B-factors) and proximal to the membrane hairpin could mediate interaction and slow electron/radical exchange with a membrane-associated partner (Fig. 7). Moreover, preferable incorporation of the NADH-reduced mature flavoprotein into liposomes in inward orientation (our unpublished data) allows us to speculate that the redox-controlled monomer-dimer transition in AIF can modulate the curvature of the inner mitochondrial membrane directly or indirectly via interaction with other proteins maintaining cristae morphology, such as opa1 21. The redox-dependent changes in protein-protein and protein-membrane interactions could explain the ability of AIF to assist radical scavenging and assembly/maintenance of the respiratory complexes 19; 20; 32; 33; 34.
Figure 6.
A and B, Molecular surface of oxidized and reduced AIF, respectively. The 509–559 peptide (blue) occludes the active site in the oxidized protein but translocates upon FAD reduction, releasing the 190–202 hairpin into the solvent. As a result, flanking FAD Trp482 and residing at the tip of the hairpin Trp195, buried in ligand-free AIF, become solvent accessible (shown in orange). C and D, Positioning of the 190–202 hairpin in oxidized and reduced AIF, respectively. E, The W195A mutation significantly elevates the NADH-dependent redox activity of AIF by affecting the FAD redox potential and hydride transfer rate. F, Tryptic digests of wild type AIF and the W195A mutant. In the wild type, the Lys517-Ser518 bond becomes accessible to trypsin only upon reduction with NAD(P)H 29. In contrast, the digest pattern of the W195A mutant is redox-independent.
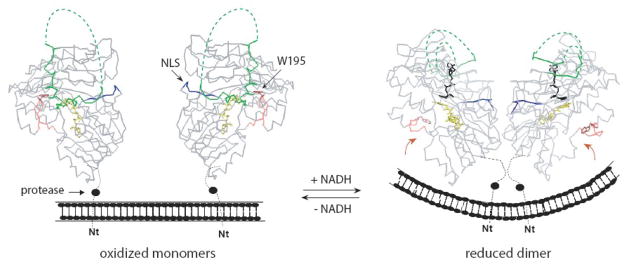
Figure 7.
Formation of long-lived dimeric complexes with NADH may affect normal and apoptogenic functions of AIF and AIF-mediated cell signaling. The redox-linked monomer-dimer transition in AIF could modulate the curvature of the inner mitochondrial membrane and change accessibility of the nuclear leading signal (NLS, shown in blue) and N-terminal proteolysis site (filled circle). Through reorganization in the 509–559 peptide (green; disordered parts depicted as dotted lines), the AIF-NADH association/dissociation may regulate the lifetime, posttranslational modification and interaction of AIF with partnering proteins and DNA. Redox perturbations in the 190–202 hairpin (pink), in turn, change solvent accessibility of Trp195, a potential site for interaction and slow electron/radical exchange with a membrane-bound partner (shown by arrows).
Redox control of the apoptogenic action of AIF
Owing to structural disorder of the N-termini in both oxidized and reduced AIF, it was not possible to determine how the redox state affects susceptibility of the N-terminal linker to proteolysis 29, one of the initial steps in the AIF-mediated apoptogenic pathway 7. Nevertheless, the available structural data provide evidence that association/dissociation of NADH may affect the apoptogenic function of AIF. First of all, this follows from transformations in the 509–559 insertion. This regulatory peptide contains several functionally important elements, such as lysines 509 and 517 (510 and 518 in human) critical for AIF-DNA interaction and induction of apoptosis 23, PEST sequence, and proline-rich motif. Since the DNA binding ability of AIF is redox-controlled and decreases upon FAD reduction 29, it is possible that the AIF-DNA complex affinity is modulated by the folding of the 509–517 fragment and spatial positioning of Lys509 and 517. The lysines may serve as ubiquitin attachment sites as well; AIF is known to undergo ubiquitinization 11; 15 but the residues modified by ubiquitin have not been identified. The PEST sequence is a putative proteolytic signal and a phosphorylation site 27; 35; 36, via posttranslational modification of which AIF destruction and AIF-mediated signaling pathways could be regulated. The proline-rich motif, on the other hand, is a potential recognition/interaction site for SH3 domains, present in many proteins involved in intracellular signaling, cytoskeletal rearrangements, cell growth and differentiation, protein trafficking and immune response 37, and executing specific functions through highly regulated assembly of multiprotein complexes 38. In addition to the SH3-domain containing partners that yet to be identified, the prolin-rich fragment may be involved in binding of cyclophilin A, a prolyl isomerase regulating nuclear translocation of AIF and apoptosis-associated chromatinolysis 10; 39. Thus, through conformational changes in the 509–559 peptide, the FAD oxidoreduction could control the lifetime, posttranslational modification, nuclear translocation and interaction of AIF with partnering proteins and DNA.
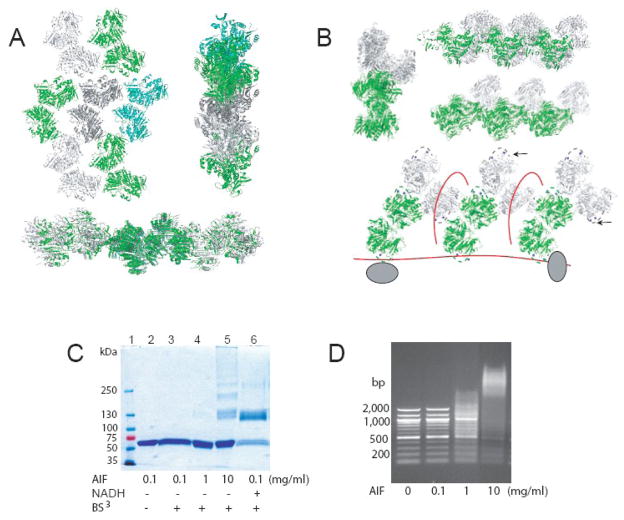
The structural data suggest also that transport of AIF to the nucleus and PCD induction can be regulated via the NADH-induced monomer-dimer transition. In CTC, the nuclear leading sequence (aa. 445–451), through which apoptogenic AIF is predominantly transported from the cytosol to the nucleus 13, becomes part of the dimer interface (Fig. 7). Having inaccessible NLS, CTC would possess a lesser ability, if any, to translocate to the nucleus and induce apoptosis. Other supporting evidence comes from examination of crystal packing. In the crystal lattice, oxidized but not reduced AIF forms fibril-like polymers perfectly suited for DNA binding (Fig. 8A, B). Since the N-terminal tail of crystalline AIF does not participate in monomer-monomer interaction and molecular properties of the full-length and apoptogenic flavoproteins are highly similar 29, the latter form is expected to produce oligomers analogous to those seen in the crystal structure.
Figure 8.
Packing of reduced (A) and oxidized (B) AIF in the crystal lattice. Reduced dimers have fewer intermolecular contacts and form a zig-zag type network, whereas molecules of oxidized AIF are aligned into fibril-like bladed polymers. Solvent exposed fragments of the regulatory peptide (dashed lines) and critical for DNA binding lysines 509 and 517 (blue spheres) are situated at the bottom of the crevices and on the edges of the blades (indicated by arrows), which makes AIF fibrils perfectly suited for binding of DNA (red lines) and recruitment of DNA degrading proteins (gray ovals). C, Oxidized apoptogenic AIF forms high-molecular weight oligomers in solution when protein concentration is high (row 5). Polymer crosslinking was achieved with BS3, a primary amine-reactive reagent specifically conjugating reduced dimers but not oxidized monomers of AIF when protein concentration is low (rows 3 and 6). Reactions and sample separation were conducted as described in Materials and Methods. D, Significant perturbation of electrophoretic mobility of linearized DNA and formation of high-molecular weight AIF-DNA complexes are observed at high protein concentration and could be triggered by AIF oligomerization. Different amounts of AIFΔ1–101 were incubated with 250 ng of 100-bp DNA ladder (New England Biolabs) for 15 min, separated on a 2% agarose gel and visualized with ethidium bromide.
To demonstrate the ability of oxidized AIFΔ1–101 to polymerize in solution, we performed crosslinking experiments with bis(sulfosuccinimidyl)suberate (BS3), a primary amine-reactive reagent. As seen from Figure 8C, BS3 specifically conjugates NADH-reduced dimers but not ligand-free monomers of AIF when the protein concentration is low (compare rows 3 and 6). However, at high concentration of oxidized AIF (10 mg/ml), accumulation of BS3-linked oligomers of various lengths is observed (row 5). These protein levels are also required for crystallization and, most importantly, for formation of high-molecular weight AIF-DNA complexes (Fig. 8D). Taking into account these results and the facts that (i) AIF binds DNA and RNA in a sequence-independent manner 40 via positively charged surface residues 23, and (ii) AIF-DNA interaction is cooperative, accompanied by DNA condensation 40, assists recruitment of DNA degrading proteins 41 and is redox dependent 29, it is plausible to propose that an increase in nuclear concentrations of oxidized apoptogenic AIF could trigger its oligomerization and, consequently, facilitate DNA binding and recruit DNA degrading partners, leading to formation of degradosome 41.
Differences between AIFs from higher and lower eukaryotes
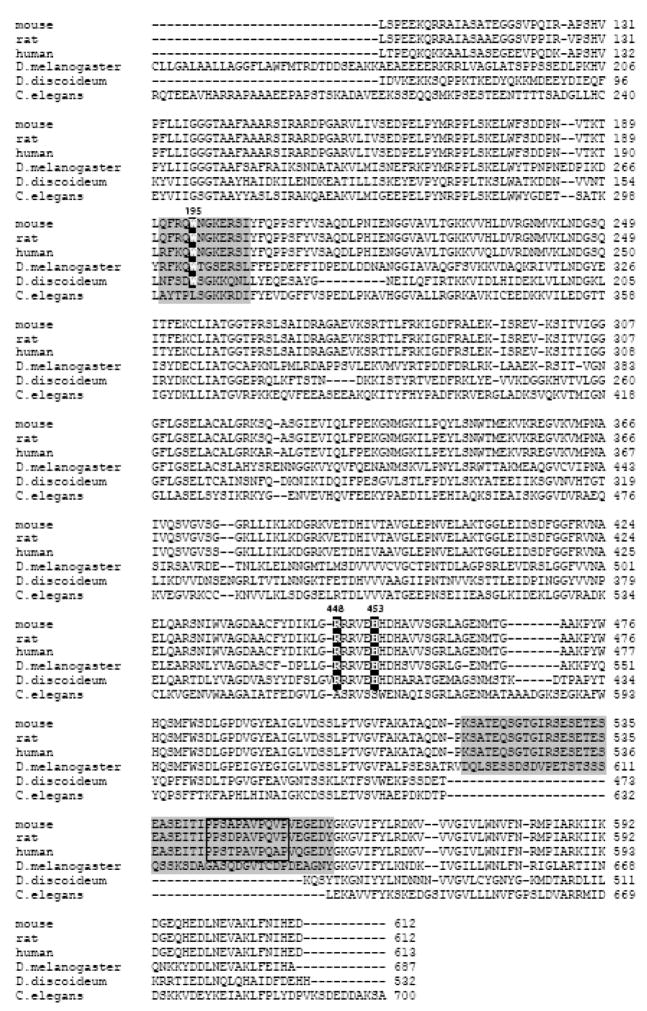
Analysis of sequence conservation reveals that all redox-sensitive and functionally important structural elements (190–202 and 509–559 peptides, Arg448 and His453) are present only in AIFs from higher eukaryotes (Fig. 9). The protein from D. melanogaster contains the key features but its regulatory insertion (aa. 585–636) has a distinct sequence lacking the proline-rich motif. This suggests that the redox properties of fly and mammalian AIF likely to be similar but the AIF-mediated signal transduction pathways may differ. AIF homologues from lower eukaryotes, on the other hand, are missing some of the aforementioned elements. In particular, the protein from C. elegans contains only the 190–202 peptide lacking Trp195. Thus, this organism, widely utilized as a model system to address fundamental questions in developmental biology, may not be suitable for investigation of cellular processes involving AIF.
Figure 9.
Sequence alignment of the catalytic domains of AIFs from different species, performed with the ClustalW2 program, shows that redox-sensitive and functionally important elements, such as Trp195, Arg448, His453, the 190–202 and 509–559 peptides (highlighted in gray) and prolin-rich motif (boxed), are conserved only in AIF from higher eukaryotes.
In conclusion, determination and comparison of the crystal structures of the oxidized and reduced forms of naturally folded AIF helped us to identify aberrations caused by refolding, pinpoint unique elements involved in redox-linked conformational reorganization and explain previously reported experimental results. The structural data suggest that redox-induced conformational changes in the active site are transmitted to the dimer interface through the 438–453 peptide, promoting interaction between the subunits. This and reorganization in the C-terminal domain lead to AIF dimerization, formation of a long-lived charge-transfer complex, and conformational changes in the 190–202 and 509–559 peptides, which could regulate AIF-mediated protein-protein interactions in normal and dying cell. The structural findings suggest that AIF has evolved to form long-lived, functionally important complexes with NADH and may operate as a redox-signaling molecule linking NAD(H)-dependent metabolic pathways to apoptosis.
MATERIALS AND METHODS
Expression and purification of recombinant mouse AIF were carried out as described previously 29. Crystals of oxidized ligand-free AIFΔ1–77 were grown at room temperature by the microbatch method under oil. Five microliters of 170 μM AIF in 70 mM HEPES, pH 7.0, 4 mM CHAPS were mixed with 5 μl of 16% polyethylene glycol 4000, 0.35 M KNO3, 20% 1,2 butanediol and covered with paraffin oil. Crystals were harvested 3–4 days later and belonged to the P212121 space group with unit cell dimensions of 75.1 Å × 80.3 Å × 419.6 Å. Reduced and NAD-bound dimers of six-histidine tagged AIFΔ1–101 were purified from E. coli by Ni2+-affinity chromatography and separated from the monomeric oxidized fraction by gel filtration on Sephacryl S100 HR (Sigma, USA). The charge-transfer complex was crystallized overnight under aerobic conditions at room temperature by the hanging drop vapor diffusion method. One microliter of 200 μM protein in 100 mM bis-tris propane, pH 7.4, was mixed with 2 μl of 10% polyethylene glycol 4000, 10% isopropanol and 0.1 M lithium citrate, pH 6.0. Blue-colored crystals belonged to the P212121 space group with unit cell dimensions of 60.3 Å × 120.4 Å × 178.2 Å. For both types of crystals, 30% glycerol was used as a cryoprotectant.
Diffraction data were collected at the Stanford Synchrotron Research Laboratories and processed with HKL2000 and D*TREK 42; 43. Both oxidized and reduced structures were solved by molecular replacement with PHASER 44 using molecule A of mouse AIFΔ1–120 (PDB code 1GV4) as a search model. The initial models were rebuilt and refined with O 45 and CNS 46 and at the final stages subjected to TLS and restrained refinement with REFMAC 44. Water molecules were picked automatically with CNS and inspected visually in O. The N- and C-termini as well as parts of the 538–559 peptide are missing in both structures due to disorder. In the oxidized AIFΔ1–77 model, there are two stretches of 8 and 9 residues each near the 501–508 β-strand of molecule A that may belong to the missing N-termini of the neighboring molecules. These peptides could not be identified due to insufficient resolution and were modeled as poly-alanines. In the reduced AIFΔ1–101 structure, electron density for residues 536–543 was clearly seen and the fragment could be modeled only in molecule A. Data collection and refinement statistics are summarized in Table 1. Wilson B-factors were included to show that the high average B-factor value for the oxidized structure reflects the nature of the crystal. The least-squares superposition and rmsd calculations were performed using the program LSQMAN 44.
Redox potential and electron transferring activity measurements as well as tryptic digests were conducted as described previously 29. Crosslinking with BS3 (Pierce) was performed in 20 mM phosphate buffer, pH 7.0. Different concentrations of AIF were mixed with a 100-fold excess of BS3 in the absence or presence of NADH and incubated for 30 min at room temperature. Reactions were stopped by adding an aliquot of 1 M Tris (10 mM final concentration). Six micrograms of AIF from each sample were subjected to 3–8% SDS-PAGE and visualized with Coomassie staining after separation.
Protein Data Bank accession codes
Coordinates for the oxidized and reduced AIF structures have been deposited into the RCSB Protein Data Bank with PDB ID codes 3GD3 and 3GD4, respectively.
Acknowledgments
This work was supported by the National Institutes of Health grant GM67637 and based on research conducted at the Stanford Synchrotron Radiation Laboratory, which is funded by the Department of Energy, Office of Basic Energy Sciences, and NIH. The author thanks H. Li, L. Lad and B. Bhaskar for help with synchrotron data collection and processing, I. Churbanova for technical assistance with protein purification and crosslinking experiments, and T. L. Poulos for critical reading of the manuscript.
Abbreviations
- AIF
apoptosis inducing factor
- PCD
programmed cell death
- NLS
nuclear leading signal
- CTC
FADH2-NAD charge-transfer complex
- GR
glutathione reductase
- PEST
proline/glutamate/serine/threonine-rich sequence
- BphA4
a ferredoxin reductase component of biphenyl reductase from Pseudomonas sp. strain KKS102
- BS3
bis(sulfosuccinimidyl)suberate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–62. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 2.Samali A, Zhivotovsky B, Jones D, Nagata S, Orrenius S. Apoptosis: cell death defined by caspase activation. Cell Death Differ. 1999;6:495–6. doi: 10.1038/sj.cdd.4400520. [DOI] [PubMed] [Google Scholar]
- 3.Kroemer G, Martin SJ. Caspase-independent cell death. Nat Med. 2005;11:725–30. doi: 10.1038/nm1263. [DOI] [PubMed] [Google Scholar]
- 4.Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Goodlett DR, Aebersold R, Siderovski DP, Penninger JM, Kroemer G. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–6. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 5.Joza N, Susin SA, Daugas E, Stanford WL, Cho SK, Li CYJ, Sasaki T, Elia AJ, Cheng HYM, Ravagnan L, Ferri KF, Zamzami N, Wakeham A, Hakem R, Yoshida H, Kong YY, Mak TW, Zuniga-Pflucker JC, Kroemer G, Penninger JM. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature. 2001;410:549–554. doi: 10.1038/35069004. [DOI] [PubMed] [Google Scholar]
- 6.Lorenzo HK, Susin SA, Penninger J, Kroemer G. Apoptosis inducing factor (AIF): a phylogenetically old, caspase-independent effector of cell death. Cell Death Differ. 1999;6:516–24. doi: 10.1038/sj.cdd.4400527. [DOI] [PubMed] [Google Scholar]
- 7.Otera H, Ohsakaya S, Nagaura ZI, Ishihara N, Mihara K. Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J. 2005;17:1–12. doi: 10.1038/sj.emboj.7600614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Yang C, Chai J, Shi Y, Xue D. Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. Science. 2002;298:1587–92. doi: 10.1126/science.1076194. [DOI] [PubMed] [Google Scholar]
- 9.Niikura Y, Dixit A, Scott R, Perkins G, Kitagawa K. BUB1 mediation of caspase-independent mitotic death determines cell fate. J Cell Biol. 2007;178:283–96. doi: 10.1083/jcb.200702134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cande C, Vahsen N, Kouranti I, Schmitt E, Daugas E, Spahr C, Luban J, Kroemer RT, Giordanetto F, Garrido C, Penninger JM, Kroemer G. AIF and cyclophilin A cooperate in apoptosis-associated chromatinolysis. Oncogene. 2004;23:1514–21. doi: 10.1038/sj.onc.1207279. [DOI] [PubMed] [Google Scholar]
- 11.Collingwood TS, Smirnova EV, Bogush M, Carpino N, Annan RS, Tsygankov AY. T-cell ubiquitin ligand affects cell death through a functional interaction with apoptosis-inducing factor, a key factor of caspase-independent apoptosis. J Biol Chem. 2007;282:30920–8. doi: 10.1074/jbc.M706870200. [DOI] [PubMed] [Google Scholar]
- 12.Ravagnan L, Gurbuxani S, Susin SA, Maisse C, Daugas E, Zamzami N, Mak T, Jaattela M, Penninger JM, Garrido C, Kroemer G. Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nat Cell Biol. 2001;3:839–43. doi: 10.1038/ncb0901-839. [DOI] [PubMed] [Google Scholar]
- 13.Gurbuxani S, Schmitt E, Cande C, Parcellier A, Hammann A, Daugas E, Kouranti I, Spahr C, Pance A, Kroemer G, Garrido C. Heat shock protein 70 binding inhibits the nuclear import of apoptosis-inducing factor. Oncogene. 2003;22:6669–78. doi: 10.1038/sj.onc.1206794. [DOI] [PubMed] [Google Scholar]
- 14.Ruchalski K, Mao H, Li Z, Wang Z, Gillers S, Wang Y, Mosser DD, Gabai V, Schwartz JH, Borkan SC. Distinct hsp70 domains mediate apoptosis-inducing factor release and nuclear accumulation. J Biol Chem. 2006;281:7873–80. doi: 10.1074/jbc.M513728200. [DOI] [PubMed] [Google Scholar]
- 15.Wilkinson JC, Wilkinson AS, Galban S, Csomos R, Duckett CS. Apoptosis-inducing factor is a target for ubiquitination through interaction with XIAP. Mol Cell Biol. 2008;28:237–47. doi: 10.1128/MCB.01065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miramar MD, Costantini P, Ravagnan L, Saraiva LM, Haouzi D, Brothers G, Penninger JM, Peleato ML, Kroemer G, Susin SA. NADH oxidase activity of mitochondrial apoptosis-inducing factor. J Biol Chem. 2001;276:16391–8. doi: 10.1074/jbc.M010498200. [DOI] [PubMed] [Google Scholar]
- 17.Mate MJ, Ortiz-Lombardia M, Boitel B, Haouz A, Tello D, Susin SA, Penninger J, Kroemer G, Alzari PM. The crystal structure of the mouse apoptosis-inducing factor AIF. Nat Struc Biol. 2002;9:442–6. doi: 10.1038/nsb793. [DOI] [PubMed] [Google Scholar]
- 18.Klein JA, Longo-Guess CM, Rossmann MP, Seburn KL, Hurd RE, Frankel WN, Bronson RT, Ackerman SL. The harlequin mouse mutation down-regulates apoptosis-inducing factor. Nature. 2002;419:367–74. doi: 10.1038/nature01034. [DOI] [PubMed] [Google Scholar]
- 19.Vahsen N, Cande C, Briere JJ, Benit P, Joza N, Larochette N, Mastroberardino PG, Pequignot MO, Casares N, Lazar V, Feraud O, Debili N, Wissing S, Engelhardt S, Madeo F, Piacentini M, Penninger JM, Schagger H, Rustin P, Kroemer G. AIF deficiency compromises oxidative phosphorylation. EMBO J. 2004;23:4679–89. doi: 10.1038/sj.emboj.7600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apostolova N, Cervera AM, Victor VM, Cadenas S, Sanjuan-Pla A, Alvarez-Barrientos A, Esplugues JV, McCreath KJ. Loss of apoptosis-inducing factor leads to an increase in reactive oxygen species, and an impairment of respiration that can be reversed by antioxidants. Cell Death Differ. 2005;82:354–7. doi: 10.1038/sj.cdd.4401776. [DOI] [PubMed] [Google Scholar]
- 21.Zanna C, Ghelli A, Porcelli AM, Karbowski M, Youle RJ, Schimpf S, Wissinger B, Pinti M, Cossarizza A, Vidoni S, Valentino ML, Rugolo M, Carelli V. OPA1 mutations associated with dominant optic atrophy impair oxidative phosphorylation and mitochondrial fusion. Brain. 2008;131:352–67. doi: 10.1093/brain/awm335. [DOI] [PubMed] [Google Scholar]
- 22.Cheung EC, Joza N, Steenaart NA, McClellan KA, Neuspiel M, McNamara S, Maclaurin JG, Rippstein P, Park DS, Shore GC, McBride HM, Penninger JM, Slack RS. Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. EMBO J. 2006;25:4061–73. doi: 10.1038/sj.emboj.7601276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye H, Cande C, Stephanoou NC, Jiang S, Gurbuxani S, Larochette N, Dauglas E, Garrido C, Kroemer G, Wu H. DNA binding is required for the apoptogenic action of apoptosis inducing factor. Nature Struc Biol. 2002;9:680–4. doi: 10.1038/nsb836. [DOI] [PubMed] [Google Scholar]
- 24.Senda T, Yamada T, Sakurai N, Kubota M, Nashizaki T, Masai T, Fukuda M, Mitsui Y. Crystal structure of NADH-dependent ferredoxin reductase component in biphenyl dioxygenase. J Mol Biol. 2000;304:397–410. doi: 10.1006/jmbi.2000.4200. [DOI] [PubMed] [Google Scholar]
- 25.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–41. [PubMed] [Google Scholar]
- 26.Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–63. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 27.Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–71. [PubMed] [Google Scholar]
- 28.Delettre C, Yuste VJ, Moubarak RS, Bras M, Lesbordes-Brion JC, Petres S, Bellalou J, Susin SA. AIFsh, a novel apoptosis-inducing factor (AIF) pro-apoptotic isoform with potential pathological relevance in human cancer. J Biol Chem. 2006;281:6413–27. doi: 10.1074/jbc.M509884200. [DOI] [PubMed] [Google Scholar]
- 29.Churbanova IY, Sevrioukova IF. Redox-dependent changes in molecular properties of mitochondrial apoptosis inducing factor. J Biol Chem. 2008;283:5622–31. doi: 10.1074/jbc.M709147200. [DOI] [PubMed] [Google Scholar]
- 30.Karplus PA, Schulz GE. Substrate binding and catalysis by glutathione reductase as derived from refined enzyme: substrate crystal structures at 2 Å resolution. J Mol Biol. 1989;210:163–80. doi: 10.1016/0022-2836(89)90298-2. [DOI] [PubMed] [Google Scholar]
- 31.Senda M, Kishigami S, Kimura S, Fukuda M, Ishida T, Senda T. Molecular mechanism of the redox-dependent interaction between NADH-dependent ferredoxin reductase and Rieske-type [2Fe-2S] ferredoxin. J Mol Biol. 2007;373:382–400. doi: 10.1016/j.jmb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 32.van Empel VP, Bertrand AT, van der Nagel R, Kostin S, Doevendans PA, Crijns HJ, de Wit E, Sluiter W, Ackerman SL, De Windt LJ. Downregulation of apoptosis-inducing factor in harlequin mutant mice sensitizes the myocardium to oxidative stress-related cell death and pressure overload-induced decompensation. Circ Res. 2005;96:e92–e101. doi: 10.1161/01.RES.0000172081.30327.28. [DOI] [PubMed] [Google Scholar]
- 33.van Empel VP, Bertrand AT, van Oort RJ, van der Nagel R, Engelen M, van Rijen HV, Doevendans PA, Crijns HJ, Ackerman SL, Sluiter W, De Windt LJ. EUK-8, a superoxide dismutase and catalase mimetic, reduces cardiac oxidative stress and ameliorates pressure overload-induced heart failure in the harlequin mouse mutant. J Am Coll Cardiol. 2006;48:824–32. doi: 10.1016/j.jacc.2006.02.075. [DOI] [PubMed] [Google Scholar]
- 34.Joza N, Oudit GY, Brown D, Benit P, Kassiri Z, Vahsen N, Benoit L, Patel MM, Nowikovsky K, Vassault A, Backx PH, Wada T, Kroemer G, Rustin P, Penninger JM. Muscle-specific loss of apoptosis-inducing factor leads to mitochondrial dysfunction, skeletal muscle atrophy, and dilated cardiomyopathy. Mol Cell Biol. 2005;25:10261–72. doi: 10.1128/MCB.25.23.10261-10272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez LO, Agerholm-Larsen B, Wang N, Chen W, Tall AR. Phosphorylation of a pest sequence in ABCA1 promotes calpain degradation and is reversed by ApoA-I. J Biol Chem. 2003;278:37368–74. doi: 10.1074/jbc.M307161200. [DOI] [PubMed] [Google Scholar]
- 36.Belizario JE, Alves J, Garay-Malpartida M, Occhiucci JM. Coupling caspase cleavage and proteasomal degradation of proteins carrying PEST motif. Curr Protein Pept Sci. 2008;9:210–20. doi: 10.2174/138920308784534023. [DOI] [PubMed] [Google Scholar]
- 37.Karkkainen S, Hiipakka M, Wang JH, Kleino I, Vaha-Jaakkola M, Renkema GH, Liss M, Wagner R, Saksela K. Identification of preferred protein interactions by phage-display of the human Src homology-3 proteome. EMBO Rep. 2006;7:186–91. doi: 10.1038/sj.embor.7400596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaneko T, Li L, Li SS. The SH3 domain--a family of versatile peptide- and protein-recognition module. Front Biosci. 2008;13:4938–52. doi: 10.2741/3053. [DOI] [PubMed] [Google Scholar]
- 39.Zhu C, Wang X, Deinum J, Huang Z, Gao J, Modjtahedi N, Neagu MR, Nilsson M, Eriksson PS, Hagberg H, Luban J, Kroemer G, Blomgren K. Cyclophilin A participates in the nuclear translocation of apoptosis-inducing factor in neurons after cerebral hypoxia-ischemia. J Exp Med. 2007;204:1741–8. doi: 10.1084/jem.20070193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vahsen N, Cande C, Dupaigne P, Giordanetto F, Kroemer RT, Herker E, Scholz S, Modjtahedi N, Madeo F, Le Cam E, Kroemer G. Physical interaction of apoptosis-inducing factor with DNA and RNA. Oncogene. 2005;25:1763–74. doi: 10.1038/sj.onc.1209206. [DOI] [PubMed] [Google Scholar]
- 41.Modjtahedi N, Giordanetto F, Madeo F, Kroemer G. Apoptosis-inducing factor: vital and lethal. Trends Cell Biol. 2006;16:264–72. doi: 10.1016/j.tcb.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–26. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr sect D. 1999;55:1718–25. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 44.CCP4. Collaborative Computational Project Number 4 The CCP4 suite programs for protein crystallography. Acta Crystallogr sect D. 1994;50:760–63. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 45.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr sect A. 1991;47:110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 46.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, GrosseKunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography + NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr sect D. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]