Abstract
During the tail end of the 20th century, a “golden period” in Alzheimer disease (AD) research, many of the pathogenic molecules of the autosomal dominant form of the disease were isolated. These molecular defects, however, do not exist in “sporadic” late-onset AD, the form of the disease that accounts for more than 95% of all cases. Pinpointing the pathogenic molecules of late-onset AD has, therefore, become an urgent goal, both for understanding disease mechanisms and for opening up novel therapeutic avenues. The retromer sorting pathway transports cargo along the endosome–trans-Golgi network, and retromer defects were first implicated in late-onset AD by a study that combined brain imaging with microarray. A range of studies have confirmed that defects in this pathway can play a pathogenic role in the disease. Herein, these findings will be reviewed, the details of the retromer sorting pathway will be discussed, and a biological model that can account for the disease’s regional selectivity will be elaborated.
Isolating the primary molecular defects of autosomal-dominant early-onset Alzheimer disease (AD) heralded a new era in AD research and served as the cornerstone on which biological insights into the disease have been made. In particular, expressing these molecules in cells and then in genetically engineered mice resolved many questions about the processing of the amyloid precursor protein (APP) and the neurotoxic effects of its cleaved product, the Aβ peptide.1 Nevertheless, the pathogenic molecules causing the early-onset form of the disease are not defective in sporadic late-onset AD, the dominant form of the disease accounting for most cases. Although a complex disorder—emerging from an interplay of genetic and epigenetic factors—isolating the pathogenic molecules of late-onset disease is acknowledged as the next important step in unraveling its causes and developing effective treatment.
As with all neurodegenerative diseases, a focus on different levels of analysis can provide clues about pathogenic molecules. Historically, a focus on histological abnormalities was the first level that offered early insight into the molecular biological features of AD. When, in 1984, the Aβ peptide was finally identified as the core of amyloid plaques (isolated from meningovascular tissue, not the brain),2 this led to the identification and cloning of its parent protein, APP.3 The cloning of APP was a required step for elucidating its metabolic pathway, serially cleaved by β-site APP-cleaving enzyme (BACE) and then by the γ-secretase, liberating Aβ and initiating the amyloid cascade.1 A focus on genetic mutations was a second level of investigation, and during the 1990s linkage analyses successfully isolated mutations in APP and the presenilins as pathogenic defects underlying early-onset AD.4 Linkage analysis, however, has proved less successful for pinpointing pathogenic molecules underlying complex disorders, including late-onset AD.
When gene expression techniques like microarray were introduced in the late 1990s,5 they provided an unprecedented opportunity to focus on brain tissue itself as a third level of analysis amenable to molecular discovery. In principle, because the expression profile of affected brain cells is a reflection of genetic and epigenetic factors, techniques like microarray are well suited for pinpointing molecules underlying complex disorders.6 In practice, however, microarray has a number of analytic challenges, such as poor signal-to-noise ratio and high false positivity, hampering its utility and dampening the overall enthusiasm for this approach.6
Early failures of microarray, however, have not impugned its technical validity but have simply emphasized the importance of using more sophisticated experimental designs to overcome its analytic challenges. With this in mind, an approach called “imaging-guided microarray,” specifically designed to address the analytic limitations inherent to microarray when applied to disorders of the brain, was recently introduced. A detailed description is provided elsewhere,6,7 but in general the approach relies on in vivo imaging to first construct a spatiotemporal model hypothesizing a priori how a pathogenic molecule should behave—anatomically and across age groups. Then, the spatiotemporal model is used as a guide in generating microarray data and in analyzing the gene expression data set. By converting a microarray experiment from one that is typically hypothesis free to one that is hypothesis driven, imaging-guided microarray naturally addresses many analytic challenges. As in any hypothesis-driven study, the results are only as good as the hypothesis and, therefore, as discussed later, any microarray finding needs to be independently confirmed and validated.
THE RETROMER SORTING PATHWAY IMPLICATED BY IMAGING-GUIDED MICROARRAY
Alzheimer disease begins in the hippocampal formation before sweeping over the neocortex, ravaging the mind and causing dementia in its wake. The hippocampal formation itself, however, is a circuit made up of separate but interconnected subregions—the entorhinal cortex, the dentate gyrus, the CA3 and CA1 subfields, and the subiculum. Each hippocampal subregion expresses a unique molecular profile, accounting for why each subregion is differentially vulnerable to mechanisms of disease.8
During the past few years, variants of functional imaging have been used to investigate the hippocampus as a circuit—ie, simultaneously investigating multiple subregions—establishing a spatiotemporal profile of AD-related dysfunction.6 Agreeing with some, although not all, postmortem indicators of disease, the spatial pattern of dysfunction suggests that, early on, AD targets the entorhinal cortex with relative sparing of the dentate gyrus. In contrast to the spatial pattern, the temporal pattern of dysfunction uncovered by the imaging studies was unexpected and could not have been inferred from postmortem indicators alone. Specifically, entorhinal dysfunction detected in early AD was age invariant.6,7
This spatiotemporal profile was used to construct a model predicting how a pathogenic molecule related to AD should behave. Guided by the model, the entorhinal cortex and the dentate gyrus from postmortem brain specimens with and without AD were harvested, purposefully covering a broad age span, and microarray analysis was performed on each tissue sample. The final analysis revealed that, among a handful of hits, the expression level of vacuolar protein sorting 35 (VPS35) best conformed to the full spatiotemporal model of late-onset AD.9
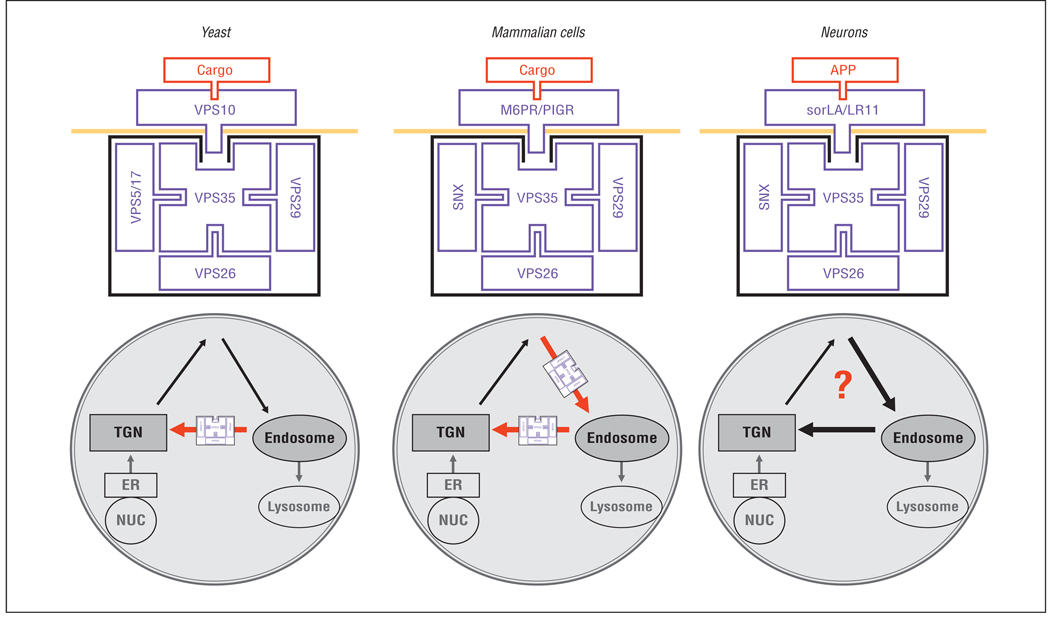
Vacuolar protein sorting 35 turns out to be the core component of the retromer sorting pathway. First described in yeast, the retromer sorting pathway consists of a multimeric retromer complex, comprising VPS35, VPS26, VPS29, VPS5, and VPS17. This complex acts as a “coat” that binds and transports the transmembrane receptor VPS10 from the endosome back to the trans-Golgi network10 (Figure 1). Except for VPS17, mammalian homologues of the retromer complex have been identified11 and are expressed in the brain and among other tissue types. Previous studies have shown that a primary reduction in any retromer element will lead to secondary degradation of other elements of the complex, causing general retromer dysfunction.12 Indeed, it was found that VPS35 and VPS26 proteins were differentially reduced in AD.9 To test whether this finding was potentially pathogenic, small interfering RNA was used to systematically decrease retromer elements in cell culture, showing that this reduction led to increased concentrations of Aβ, while overexpressing retromer elements decreased Aβ levels.9
Figure 1.
The retromer sorting pathway is made up of a coat complex that binds and transports transmembrane receptors between the endosome and the trans-Golgi network (TGN). In yeast, the coat complex, made up of vacuolar protein sorting (VPS) 35, VPS26, VPS29, VPS5, and VPS17, binds and transports VPS10 from the endosome back to the TGN (red arrow). In nonneuronal mammalian cells, the coat complex, made up of VPS35, VPS26, VPS29, VPS5, and VPS17, binds and transports the mannose-6-phosphate (M6PR) and the polymeric immunoglobulin (PIGR) receptors from the cell surface to endosome and from endosome back to the TGN (red arrows). In neurons, the coat complex binds and transports the family of VPS10-containing receptors, including sorLA/LR11. sorLA/LR11 binds the amyloid precursor protein (APP). The exact transport itinerary of the neuronal retromer sorting pathway remains undetermined (?). ER indicates endoplasmic reticulum, NUC, nucleus; and SNX, sorting nexin.
THE NEURONAL RETROMER AND ITS RELATION TO APP PROCESSING
Why would retromer dysfunction cause an increase in Aβ levels? Identifying the type 1 transmembrane receptor sorted by the neuronal retromer might offer clues. In contrast to nonneuronal mammalian cells,10 the receptor of the neuronal retromer had not been elucidated, although retromer-related molecules are highly expressed in the brain. In an attempt to identify candidate receptors of the neuronal retromer, an analytic approach was applied to the microarray data set; this approach has been used in prior gene expression studies to search for potentially interacting molecules. Underlying this approach is the assumption that molecules that interact with each other are more likely to have expression levels that cross correlate.13 Because VPS35 serves as the key retromer element that directly binds the retromer receptor, the microarray data set was searched for correlations between the expression levels of VPS35 and type I transmembrane molecules, a search that identified, among other molecules, sorLA as a candidate receptor of the neuronal retromer.14,15
sorLA is a complex molecule with multiple domains, including a VPS10 domain and low-density lipoprotein receptor domains. It is this complexity that accounts for its numerous names (eg, sorLA, sorl1, and LR11) and for why this molecule has been grouped together with different families of proteins. Vacuolar protein sorting 10 is the receptor of the yeast retromer, the species in which the retromer was first described,10 and so it was sorLA’s VPS10 domain that seemed most intriguing.12,13 Mammals express a family of 5 VPS10-containing proteins that, together with sorLA, include sortilin, sorCS1, sorCS2, and sorCS3.16 Because all members of the family are type I transmembrane receptors and are highly expressed in the brain, it was proposed that, in contrast to nonneuronal mammalian cells,10 the VPS10 family of proteins might function as receptors of the neuronal retromer9 (Figure 1).
More important, work by Scherzer et al17 had previously shown that sorLA is down-regulated in late-onset AD (in their article, they focused on the low-density lipoprotein receptor domain of the molecule, using the name LR11). Shortly thereafter, a collaborative series of studies by the laboratories of Andersen et al18 reported on the cell’s biological properties of sorLA, focusing more on its possible role in sorting APP. Since then, all of these groups have extended their work, suggesting that sorLA might interact with APP or with BACE. Put into the context of the retromer, the neuronal retromer might be involved, directly or indirectly, via sorLA or other VPS10-containing proteins, in sorting APP and/or BACE along the endosome–trans-Golgi network trafficking pathway.9
Taken together, it has been proposed that retromer dysfunction would increase the resident time of APP and its cleaving enzymes in the same organelle, accelerating APP processing and accounting for the Aβ elevation observed in retromer-deficient states.9,19
CONFIRMING THE PATHOGENICITY OF RETROMER SORTING
Although a priori modeling and sophisticated statistics can increase the odds that a given microarray finding is relevant to a disease process, microarray findings by themselves do not inform about pathogenicity. As previously described, because tissue samples are harvested years after the disease has begun, it is impossible to know whether the retromer defects observed in AD brain specimens are truly pathogenic or whether the finding simply reflects a secondary response to a sick and dying cell. As with all microarray findings, 3 types of studies can be used to potentially confirm the pathogenicity of retromer sorting in AD.6 First, cell culture studies can test whether manipulating retromer-related molecules affects Aβ production. Second, genetically engineered mice studies can test whether retromer deficiency affects Aβ production in the brain and causes hippocampal dysfunction. Third, genetic studies can test whether polymorphisms in retromer-related molecules increase the risk for late-onset disease.
As mentioned, the first confirmatory studies were reported in a previous retromer study,9 and showed that manipulating retromer-related molecules in cell culture had a commensurate effect on Aβ levels. The second confirmatory studies using genetically engineered mice are under way. A colony of retromer-deficient mice has been recently bred, and the process of establishing their behavioral, electrophysiological, and biochemical phenotypes is under way. These mice have partial reductions in VPS26 and VPS35, thereby modeling the molecular defects found in AD brain specimens. Although still a work in progress, the preliminary results are encouraging.20
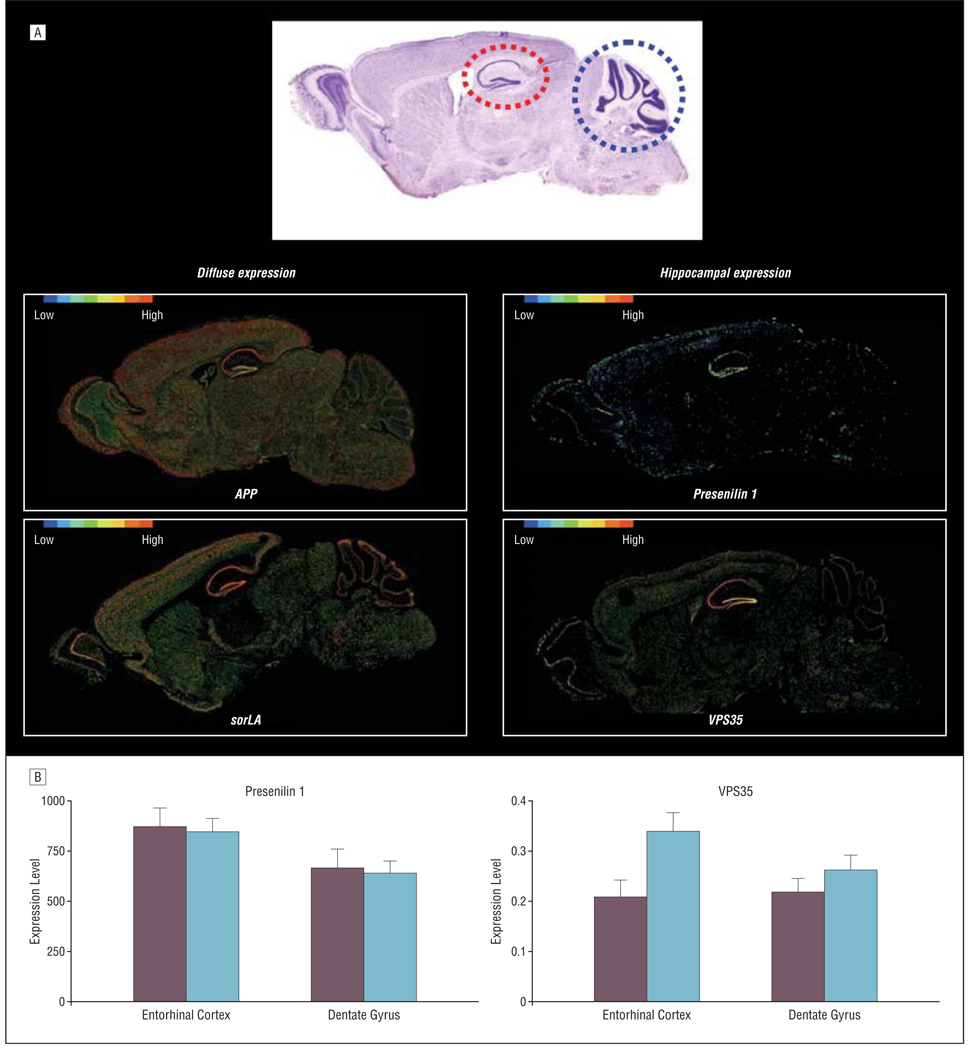
Mice can also be used to establish the normal anatomical expression pattern of retromer-related molecules in the brain. With this question in mind, I recently explored the Allen Brain Atlas,21 which has made available the expression maps of most of the murine genome. Unexpectedly, VPS35 and VPS26 are expressed with highest levels in the pyramidal cells of the hippocampus, more so than in other regions of the brain (Figure 2). This contrasts with the diffuse expression pattern observed for APP, BACE, and sorLA. Interestingly, although presenilin 1 is expressed in many regions of the brain, compared with APP or BACE, presenilin 1 does show some degree of differential expression in the hippocampal formation (Figure 2A). Furthermore, examining the microarray data set of human hippocampal tissue shows that presenlin 1 has higher expression levels in the entorhinal cortex compared with the dentate gyrus (P<.04) (Figure 2B). Of course only suggestive, the fact that retromer-related molecules track the anatomical pattern of AD provides indirect, but intriguing, support for a role in the disease.
Figure 2.
Retromer and presenilin 1 are differentially expressed in the entorhinal cortex. A, Compared with the diffuse expression pattern of amyloid precursor protein (APP) and sorLA, presenilin 1, vacuolar protein sorting (VPS) 35, and VPS26 are differentially expressed in the hippocampal formation compared with the cerebellum. The mouse hippocampal formation (red stippled circle) and the cerebellum (blue stippled circle) are shown in a sagittal histological slice (upper panel). Gene expression maps (generated by the Allen Brain Atlas,21 lower panels) are color coded such that warmer colors reflect more expression of a particular gene, while cooler colors reflect less expression. B, Within the human hippocampal formation, presenilin 1 and VPS35 are differentially expressed in the entorhinal cortex compared with the dentate gyrus. Left panel shows messenger RNA expression levels of presenilin 1, as measured by Affymetrix HG-U133A GeneChip (Affymetrix, Santa Clara, California). Right panel shows protein expression levels of VPS35, as measured by quantitative Western blotting normalized to actin. For the entorhinal cortex, red bars indicate Alzheimer disease; orange bars, controls. For the dentate gyrus, light blue bars indicate Alzheimer disease; dark blue bars, controls.
Recently, a genetic study was reported by Rogaeva et al.22 Investigating multiple cohorts with late-onset AD, they genotyped VPS35, VPS26, and the family of VPS10-containing molecules. Remarkably, genetic variants in sorLA were associated with late-onset AD. The researchers interpret their results in the context of the retromer sorting pathway and, indeed, provide direct evidence that VPS35 binds sorLA and that knocking down VPS26 in cell culture increases Aβ production.
OVERLAPPING FUNCTIONS OF RETROMER AND PRESENILIN SUGGEST UNIFYING MECHANISMS OF DISEASE PATHOGENESIS?
“Localizing the lesion” is a basic tenet in all neurology. Not only does pinpointing a targeted neuronal population promise to improve diagnostic precision, but more important, this anatomical information provides clues into a disease’s primary pathophysiological features. As discussed, in contrast to APP, BACE, retromer, and presenilin are differentially expressed in the pyramidal neurons of the hippocampal formation (Figure 2). Thus, identifying cellular mechanisms in which retromer and presenilin play a shared role might expand our understanding of the disease process.
Besides Aβ production, to date, there are 2 additional cellular mechanisms in which retromer and presenilin apear to play an active role. First, like the retromer, a growing number of studies have established that the presenilins play a general role in sorting type I transmembrane proteins, and that disease-causing mutations in presenilin cause protein missorting (as reported by Small and Gandy19). Second, and perhaps more interesting, retromer23 and presenilin24 play critical roles in the Wnt signaling pathway.
Which of these 3 overlapping functions—Aβ production, transmembrane protein sorting, and Wnt signaling—might account for the differential expression pattern of presenilin and retromer in the entorhinal cortext? Aβ production is an unlikely candidate because within unaffected specimens, Aβ levels are not higher in the entorhinal cortex compared with other brain regions.25 The second function, protein sorting, is a better candidate. Sorting type I transmembrane proteins is important for synaptogenesis, because these proteins are the dominant players in the synaptogenic process.26 As the main gateway into the hippocampus, the entorhinal cortex receives constant input from the whole neocortical mantle, and integrating this information requires highly active dendritic remodeling and extremely high metabolic activity.27
The overlap of retromer and presenilin in the function of the Wnt signaling pathway is perhaps the best candidate for why they are differentially expressed in the entorhinal cortex. Morphologically, the entorhinal cortex exhibits 2 distinct features. First, at the single-cell level, entorhinal cortex neurons exhibit a very complex dendritic organization. Indeed, it is for this reason that many entorhinal cortex neurons are called “stellate” cells, in contrast to morphologically simpler “pyramidal” or “granule” cells found in other hippocampal subregions.28 Second, an even more unique morphological feature is that stellate neurons are organized as clusters of “islands” extending throughout the tangential axis of the entorhinal cortex.29 The Wnt signaling pathway turns out to play an important role in establishing complex cellular30 and anatomical morphology,31 which can account for why retromer and presenilin are differentially needed in the entorhinal cortex.
Thus, entorhinal cortex neurons might differentially express retromer and presenilin not because these neurons require high Aβ levels in their normal states but rather to support their unique metabolic and morphological characteristics. However, once presenilin and retromer are rendered dysfunctional, by genetic or epigenetic factors, these neurons are expected to differentially overproduce Aβ because presenilin and retromer can also affect APP processing. Future studies are required to test this proposed hypothesis, but if confirmed it would open up novel therapeutic avenues for treating this devastating and common disease.
Acknowledgments
Funding/Support: This study was supported in part by federal grant AG025161 from the National Institutes of Health; the McKnight Neuroscience of Brain Disorders Award; and the James S. McDonnell Foundation.
Footnotes
Financial Disclosure: None reported.
REFERENCES
- 1.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Glenner GG, Wong CW. Alzheimer’s disease and Down’s syndrome: sharing of a unique cerebrovascular amyloid fibril protein. Biochem Biophys Res Commun. 1984;122(3):1131–1135. doi: 10.1016/0006-291x(84)91209-9. [DOI] [PubMed] [Google Scholar]
- 3.Tanzi RE, Gusella JF, Watkins PC, et al. Amyloid beta protein gene: cDNA, mRNA distribution, and genetic linkage near the Alzheimer locus. Science. 1987;235(4791):880–884. doi: 10.1126/science.2949367. [DOI] [PubMed] [Google Scholar]
- 4.Price DL, Tanzi RE, Borchelt DR, Sisodia SS. Alzheimer’s disease: genetic studies and transgenic models. Annu Rev Genet. 1998;32:461–493. doi: 10.1146/annurev.genet.32.1.461. [DOI] [PubMed] [Google Scholar]
- 5.Lockhart DJ, Dong H, Byrne MC, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14(13):1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 6.Lewandowski NM, Small SA. Brain microarray: finding needles in molecular haystacks. J Neurosci. 2005;25(45):10341–10346. doi: 10.1523/JNEUROSCI.4006-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pierce A, Small S. Combining brain imaging with microarray: isolating molecules underlying physiologic disorders of the brain. Neurochem Res. 2004;29(6):1145–1152. doi: 10.1023/b:nere.0000023601.50101.7f. [DOI] [PubMed] [Google Scholar]
- 8.Small SA. Age-related memory decline: current concepts and future directions. Arch Neurol. 2001;58(3):360–364. doi: 10.1001/archneur.58.3.360. [DOI] [PubMed] [Google Scholar]
- 9.Small SA, Kent K, Pierce A, et al. Model-guided microarray implicates the retromer complex in Alzheimer’s disease. Ann Neurol. 2005;58(6):909–919. doi: 10.1002/ana.20667. [DOI] [PubMed] [Google Scholar]
- 10.Seaman MN. Recycle your receptors with retromer. Trends Cell Biol. 2005;15(2):68–75. doi: 10.1016/j.tcb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Haft CR, de la Luz Sierra M, Bafford R, Lesniak MA, Barr VA, Taylor SI. Human orthologs of yeast vacuolar protein sorting proteins Vps26, 29, and 35: assembly into multimeric complexes. Mol Biol Cell. 2000;11(12):4105–4116. doi: 10.1091/mbc.11.12.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arighi CN, Hartnell LM, Aguilar RC, Haft CR, Bonifacino JS. Role of the mammalian retromer in sorting of the caption-independent mannose 6-phosphate receptor. J Cell Biol. 2004;165(1):123–133. doi: 10.1083/jcb.200312055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhardwaj N, Lu H. Correlation between gene expression profiles and protein-protein interactions within and across genomes. Bioinformatics. 2005;21(11):2730–2738. doi: 10.1093/bioinformatics/bti398. [DOI] [PubMed] [Google Scholar]
- 14.Small S, Kim T. inventors; Trustees of Columbia, assignee.VPS35-based assays and methods for treating Alzheimer’s disease. US patent 60/518. 2003 November 3;
- 15.Small SA, Peirce AL, Kent K, Leung C, Honig L, Vonsattel JP. Combining functional imaging with microarray: identifying an unexplored cellular pathway implicated in sporadic Alzheimer’s disease. Paper presented at: Annual Meeting of the Society for Neuroscience; New Orleans, LA. 2003. [Google Scholar]
- 16.Hampe W, Rezgaoui M, Hermans-Borgmeyer I, Schaller HC. The genes for the human VPS10 domain-containing receptors are large and contain many small exons. Hum Genet. 2001;108(6):529–536. doi: 10.1007/s004390100504. [DOI] [PubMed] [Google Scholar]
- 17.Scherzer CR, Offe K, Gearing M, et al. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch Neurol. 2004;61(8):1200–1205. doi: 10.1001/archneur.61.8.1200. [DOI] [PubMed] [Google Scholar]
- 18.Andersen OM, Reiche J, Schmidt V, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2005;102(38):13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Small SA, Gandy S. Sorting through the cell biology of Alzheimer’s disease: intracellular pathways to pathogenesis. Neuron. 2006;52(1):15–31. doi: 10.1016/j.neuron.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flores I, Yu R, Kent K, et al. The retromer and Alzheimer’s disease: cellular mechanisms for elevated Aβ in late-onset disease. Paper presented at: 36th Annual Meeting of the Society for Neuroscience; Washington, DC. 2005. [Google Scholar]
- 21.Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 22.Rogaeva E, Meng Y, Lee JH, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39(2):168–177. doi: 10.1038/ng1943. published online ahead of print January 14, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coudreuse DY, Roel G, Betist MC, Destree O, Korswagen HC. Wnt gradient formation requires retromer function in Wnt-producing cells. Science. 2006;312(5775):921–924. doi: 10.1126/science.1124856. [DOI] [PubMed] [Google Scholar]
- 24.Soriano S, Kang DE, Fu M, et al. Presenilin 1 negatively regulates β-catenin/T-cell factor/lymphoid enhancer factor-1 signaling independently of β-amyloid precursor protein and notch processing. J Cell Biol. 2001;152(4):785–794. doi: 10.1083/jcb.152.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naslund J, Haroutunian V, Hohs R, et al. Correlation between elevated levels of amyloid beta-peptide in the brain and cognitive decline. JAMA. 2000;283(12):1571–1577. doi: 10.1001/jama.283.12.1571. [DOI] [PubMed] [Google Scholar]
- 26.Garner CC, Zhai RG, Gundelfinger ED, Ziv NE. Molecular mechanisms of CNS synaptogenesis. Trends Neurosci. 2002;25(5):243–251. doi: 10.1016/s0166-2236(02)02152-5. [DOI] [PubMed] [Google Scholar]
- 27.Hevner RF, Wong-Riley MT. Entorhinal cortex of the human, monkey, and rat: metabolic map as revealed by cytochrome oxidase. J Comp Neurol. 1992;326(3):451–469. doi: 10.1002/cne.903260310. [DOI] [PubMed] [Google Scholar]
- 28.Mikkonen M, Pitkanen A, Soininen H, Alafuzoff I, Miettinen R. Morphology of spiny neurons in the human entorhinal cortex: intracellular filling with lucifer yellow. Neuroscience. 2000;96(3):515–522. doi: 10.1016/s0306-4522(99)00592-8. [DOI] [PubMed] [Google Scholar]
- 29.Solodkin A, Van Hoesen GW. Entorhinal cortex modules of the human brain. J Comp Neurol. 1996;365(4):610–617. doi: 10.1002/(SICI)1096-9861(19960219)365:4<610::AID-CNE8>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 30.Yu X, Malenka RC. β-catenin is critical for dendritic morphogenesis. Nat Neurosci. 2003;6(11):1169–1177. doi: 10.1038/nn1132. [DOI] [PubMed] [Google Scholar]
- 31.Seto ES, Bellen HJ. The ins and outs of Wingless signalling. Trends Cell Biol. 2004;14(1):45–53. doi: 10.1016/j.tcb.2003.11.004. [DOI] [PubMed] [Google Scholar]