Summary
Background
It is unknown how oscillations in Cdk1 activity drive the dramatic changes in chromosome and spindle dynamics that occur at the metaphase/anaphase transition.
Results
We show that the S. pombe monopolin complex has distinct functions in metaphase and anaphase that are determined by the phosphorylation state of its Mde4 subunit. When Cdk1 activity is high in metaphase, Mde4 is hyperphosphorylated on Cdk1 phosphorylation sites, and localizes to kinetochores. A non-phosphorylatable mutant of Mde4 does not localize to kinetochores, appears prematurely on the metaphase spindle, and interferes with spindle dynamics and chromosome segregation, illustrating the importance of Cdk1 phosphorylation in regulating metaphase monopolin activity. When Cdk1 activity drops in anaphase, dephosphorylation of Mde4 triggers monopolin localization to the mitotic spindle, where it promotes spindle elongation and integrity, coupling the late mitotic loss of Cdk1 activity to anaphase spindle dynamics.
Conclusions
Together these findings illustrate how the sequential phosphorylation and dephosphorylation of monopolin helps ensure the orderly execution of discrete steps in mitosis.
Introduction
For equal segregation of chromosomes to each daughter cell, sister kinetochores must attach to microtubules emanating from opposite poles at metaphase in a bioriented fashion. Most defects in attachment of chromosomes to spindle microtubules trigger the spindle assembly checkpoint, which halts cell cycle progression and promotes correction of attachment defects [1]. However merotelic attachments, in which a single kinetochore binds to microtubules from both poles, cannot be sensed by the spindle assembly checkpoint since the requirements for both attachment and tension at kinetochores are satisfied [2]. Thus, cells with merotelically attached chromosomes proceed into anaphase but develop lagging chromosomes, which often do not segregate properly. Merotelically attached lagging chromosomes are the most common cause of aneupoloidy in cultured mammalian cells [3].
Unlike the budding yeast Saccharomyces cerevisiae [4], fission yeast and mammalian kinetochores each bind multiple microtubules (2–4 microtubules in the case of fission yeast)[5]. Since fission yeast kinetochores can bind multiple microtubules, mechanisms are required to prevent merotelic attachments. A recent study showed that both centromeric heterochromatin and a protein complex called monopolin are required to prevent merotelic attachments in the fission yeast S. pombe [6]. The monopolin complex was first identified in S. cerevisiae, where it is required during meiosis I to orient duplicated sister kinetochores to the same pole [7–9]. The S. pombe monopolin complex consists of the Pcs1 and Mde4 proteins, which localize to the central core of centromeres [6]. Although the fission yeast monopolin complex is not required for mono-orientation of sister chromatids during meiosis I, it is required during mitosis and meiosis II to prevent lagging chromosomes caused by merotelic attachments [6]. It has been proposed that monopolin prevents merotelic attachments by acting as a clamp to align microtubule binding sites together at kinetochores [6, 7, 10]. How monopolin function is regulated during mitosis in S. pombe remains unclear.
The evolutionarily conserved Cdc14 phosphatase is known to dephosphorylate Cdk1 substrates. The S. pombe Cdc14-like phosphatase Clp1/Flp1 (hereafter referred to as Clp1) carries out multiple functions in mitosis [11–14]. We showed previously that Clp1 localizes to kinetochores at metaphase and plays a role in chromosome segregation[15], however substrates of Clp1 at kinetochores have not been identified. Here we report that the monopolin subunit Mde4 is a substrate of Clp1. We show that phosphorylation of Mde4 on Cdk1 sites keeps monopolin on kinetochores by preventing its premature localization to the mitotic spindle. Dephosphorylation of Mde4 in anaphase is required for monopolin localization to the mitotic spindle, where it promotes spindle stability and normal spindle elongation. Thus both phosphorylation and dephosphorylation of Mde4 are important for proper chromosome segregation.
Results
Mde4 is phosphorylated during mitosis and interacts with the Cdc14 like phosphatase, Clp1
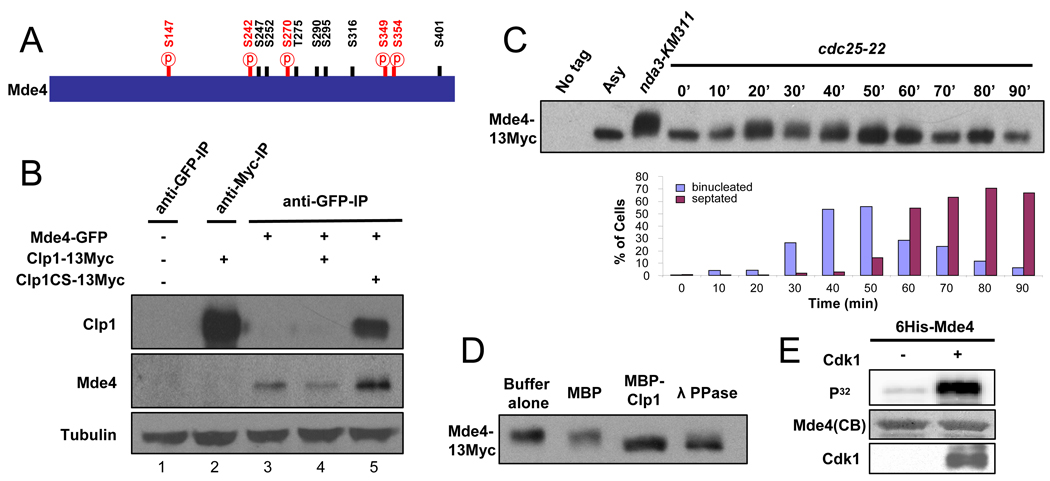
Previous studies found that clp1Δ mutants show chromosome segregation defects, implying that Clp1 substrates regulate chromosome segregation[15]. To identify such substrates, we purified protein complexes containing Clp1 and interacting proteins from mitotic cells using the substrate trapping mutant of Clp1, Clp1-C286S, which is catalytically inactive but binds more strongly to phosphorylated substrates than wild-type Clp1[15, 16]. Tandem affinity purification[17] was used to isolate Clp1-C286S protein complexes from metaphase-arrested cells. Protein complexes were then analyzed by tandem mass spectrometry. Numerous proteins that copurified with Clp1 were identified (data not shown) including the S. pombe monopolin proteins, Pcs1 and Mde4. Mde4 has twelve potential Cdk1 phosphorylation sites (SP/TP motifs), and five of them were identified as phosphorylated by mass spectrometry (Figure 1A). However, sequence coverage of Mde4 from the mass spectrometry analysis was only 30%, raising the possibility that the other 7 Cdk1 sites might be phosphorylated as well. Pcs1 has one potential Cdk1 site but it was not identified as phosphorylated in vivo (data not shown). To confirm the apparent interaction between monopolin and Clp1 identified using mass spectrometry, we carried out co-immunoprecipitation and western blotting experiments. Mde4-GFP co-immunoprecipitated with the substrate-trapping mutant, Clp1-C286S-13Myc, but not with wild-type Clp1-13Myc (Figure 1B), suggesting that the interaction between Clp1-C286S and Mde4 is mediated by phosphorylation on Mde4. Because Clp1 dephosphorylates sites phosphorylated by Cdk1, whose activity peaks in early mitosis, we investigated whether Mde4 is phosphorylated during mitosis. In synchronous cultures, slower migrating forms of Mde4-13Myc appeared during early mitosis, decreased during late mitosis, and then disappeared after mitotic exit (Figure 1C). To confirm that the shift was due to phosphorylation, Mde4-13Myc isolated from metaphase-arrested cells was treated with recombinant Clp1 purified from E. coli. Treatment with Clp1 eliminated the migration shift of Mde4-13Myc (Figure 1D), confirming that Mde4 is phosphorylated in vivo, and that Clp1 can dephosphorylate Mde4 in vitro. Moreover, Mde4 purified from E. coli could be efficiently phosphorylated by Cdk1 isolated from metaphase arrested cells, showing that Mde4 can be phosphorylated by Cdk1 in vitro (Figure 1E). To examine the contribution of Clp1 to Mde4 dephosphorylation in vivo we synchronized wild-type and clp1Δ cells in metaphase, then released them from the arrest and monitored Mde4 phosphorylation. Although Mde4 is more highly phosphorylated at the metaphase arrest point in clp1Δ cells (Supplemental Figure S1A), Mde4 still becomes dephosphorylated as these cells exit mitosis (Supplemental Figure S1B). Thus although Mde4 specifically purifies in a complex with the substrate trapping allele of Clp1, and can be dephosphorylated by it in vitro, our data indicate that other phosphatases are able to effect the dephosphorylation in vivo in the absence of Clp1. The identification of these phosphatases will be of considerable interest, and will be the subject of future studies. Together these data show that Mde4 is phosphorylated in early mitosis, and then becomes dephosphorylated in anaphase.
Figure 1. Mde4 is phosphorylated during mitosis and interacts with Clp1.
(A) Schematic representation of Mde4 depicting 12 consensus Cdk1 phosphorylation sites. 5 of the 12 predicted phosphorylation sites shown as red ℗ were identified by mass spectrometric analysis of Clp1-C286S-TAP purified from cells arrested at metaphase using the nda3-KM311 and mts3-1 mutants and from cells 60 min after release from a cdc25-22 arrest. (B) Interaction between Mde4 and Clp1 was determined by immunoprecipitation followed by western blotting using cell lysates from the following asynchronous cultures: wild-type (1), clp1-13Myc (2), mde4-GFP (3), mde4-GFP clp1-13Myc (4), and mde4-GFP clp1-C286S-13Myc (5). The lower panel shows the tubulin loading control from whole cell extracts prior to immunoprecipitation. (C) Cell cycle dependent changes in Mde4 phosphorylation was determined by analyzing the gel migration of Mde4-13Myc. cdc25-22 mde4-13Myc cells were arrested at the restrictive temperature of 36°C for 4 hours to synchronize them in G2 phase, then shifted to the permissive temperature of 25°C. Samples were taken at the indicated time points and the migration shift of Mde4-13Myc (upper panel) and cell cycle progression (lower panel) were determined by western blot analysis and microscopy respectively. Lysates from wild-type cells (No tag), asynchronous mde4-13Myc (Asy), and mde4-13Myc cells arrested in mitosis using the nda3-KM311 mutation (nda3-KM311) are also shown. (D) Immunoprecipitated Mde4-13Myc from mde4-13Myc clp1Δ cells arrested in metaphase using the nda3-KM311 mutation was treated with buffer alone, recombinant MBP (Maltose binding protein), MBP-Clp1, or lambda phosphatase (λ PPase) then analyzed using western blotting. (E) In vitro kinase assays were performed using Cdk1 immunoprecipitated, using Cdc13 (S. pombe cyclin B) antibodies, from metaphase arrested nda3-KM311 cells, and bacterially expressed 6His-Mde4 as substrate. Protein labeled by γ-32P was detected with a Phospho Imager (Molecular Dynamics), and the gel was stained with Coomassie Blue (CB) as a loading control. The level of Cdk1 was determined by western blotting.
Clp1 promotes loading of monopolin onto the spindle during anaphase
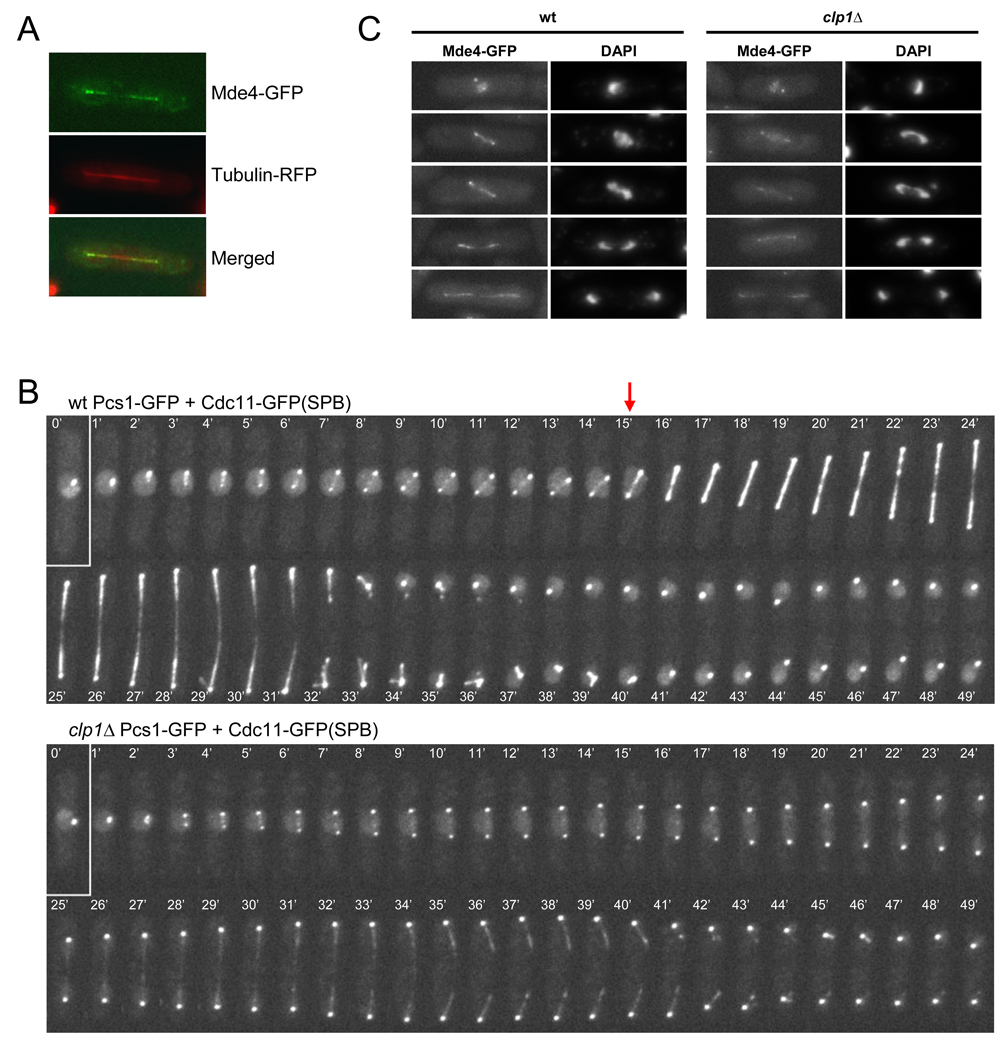
To test how phosphorylation affects Mde4 function, we examined whether changes in Mde4 phosphorylation state correlated with changes in Mde4 localization. Previous results showed that in interphase, when Mde4 is dephosphorylated, the Mde4-Pcs1 complex localizes to the nucleolus and to the kinetochores, which cluster at the nuclear periphery next to the spindle pole bodies (SPBs) [6, 8]. As cells enter mitosis and Mde4 becomes phosphorylated, Mde4-Pcs1 leave the nucleolus but remain at the kinetochores [6, 8]. Additionally we found that in anaphase, when Mde4 becomes dephosphorylated, Mde4 and Pcs1 localize to both ends of the anaphase spindle (Figure 2A–B). Because Mde4 is more highly phosphorylated in clp1Δ cells just prior to anaphase onset, we examined Pcs1 and Mde4 localization in c/p1Δ mutants as they progressed through mitosis. Pcs1-GFP localization was observed by time-lapse microscopy, using cells co-expressing the SPB marker Cdc11-GFP to monitor cell cycle progression (Figure 2B). In wild-type cells faint Pcs1-GFP spots, presumably corresponding to the kinetochores, could be observed between the two SPBs in early mitosis. At anaphase onset Pcs1-GFP localized to the spindle (Figure 2B, red arrow) and then to both ends of the spindle at late anaphase. In c/p1Δ cells, Pcs1-GFP spots at putative kinetochores were observed as in wild-type cells, however Pcs1-GFP was only observed faintly at both ends of the spindle at late anaphase (Figure 2B). As with Pcs1-GFP, Mde4-GFP localized faintly on the spindle in late, but not early, anaphase in c/p1Δ cells (Figure 2C). The reduced spindle localization of Mde4 and Pcs1 in c/p1Δ cells was not due to reduced protein levels since both proteins were present at or above wild-type levels in c/p1Δ cells (Figure S1A, and data not shown). In interphase, localization of Mde4 and Pcs1 to the nucleolus and kinetochores was not affected in c/p1Δ cells (Figure 2C, and data not shown). These results suggest that dephosphorylation of Mde4 may be important for monopolin localization to the spindle. The ability of monopolin to localize to late anaphase spindles in c/p1Δ mutants is consistent with our results showing that other phosphatases besides Clp1 can promote Mde4 dephosphorylation.
Figure 2. Clp1 promotes loading of the monopolin onto the spindle during anaphase.
(A) mde4-GFP cells expressing mRFP-α-tubulin were imaged by fluorescence microscopy. (B) pcs1-GFP cdc11-GFP (upper panel) and clp1Δ pcs1-GFP cdc11-GFP (lower panel) cells were analyzed using fluorescent time-lapse microscopy. Images were collected at one minute intervals, beginning immediately prior to entry into mitosis. The SPB marker Cdc11-GFP was used to monitor cell cycle progression. At time zero both cells are in interphase just prior to mitotic entry. The arrow indicates anaphase onset, when Pcs1 begins to localize to the spindle. (C) Mde4-GFP localization is shown in wild-type (left panels) and clp1Δ cells (right panels). Interphase cells are shown in the top row, and cells from early to late anaphase are shown in the lower panels. Cells were grown asynchronously, fixed, stained with DAPI, and imaged by fluorescence microscopy.
Loss of monopolin causes defects in microtubule attachment to kinetochores and activates the spindle checkpoint
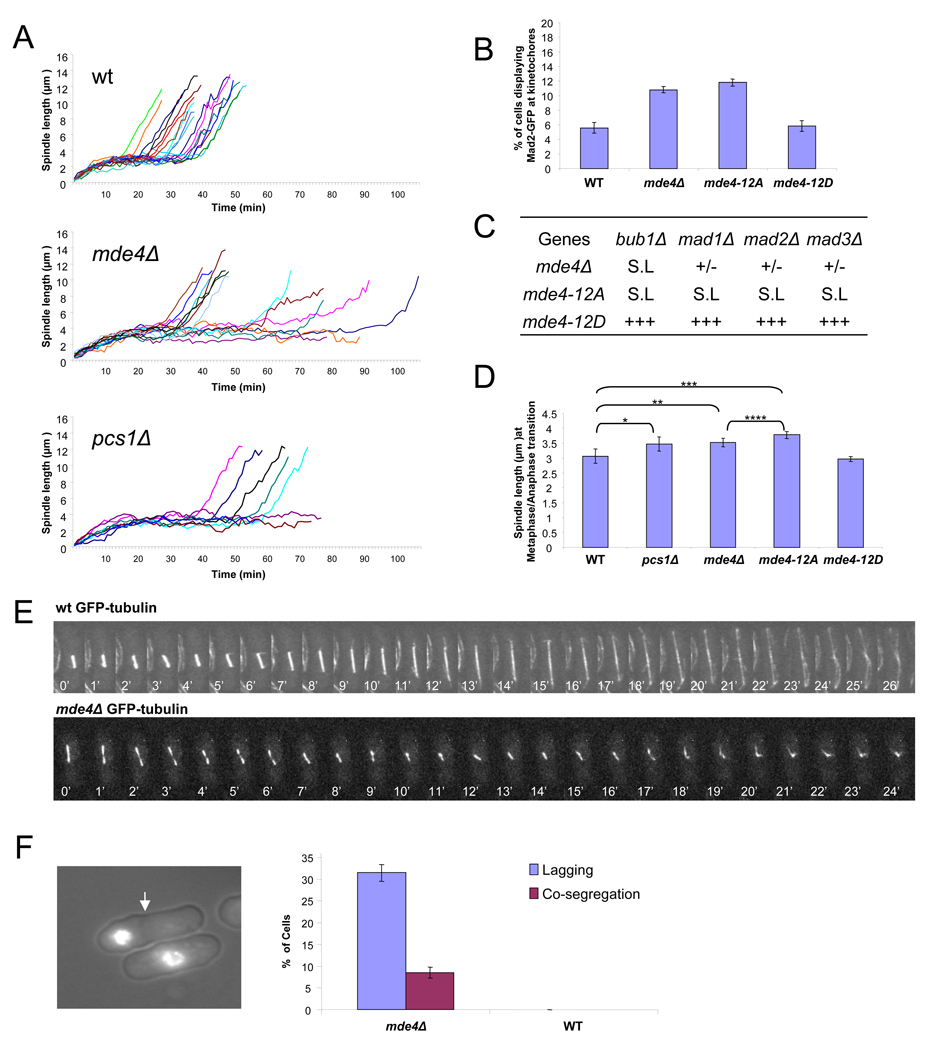
To test whether spindle localization of monopolin is important for spindle function, we examined spindle dynamics in monopolin mutants. Fission yeast spindle elongation can be divided into three distinct phases [18–20]. During phase I, which begins when cells enter mitosis, the spindle forms and grows to span the nucleus at a length of 2.5∼3µm. In phase II (metaphase/anaphase A), the spindle maintains the 2.5∼3µm length, and then elongates during phase III, which corresponds to anaphase B. Mitotic spindles were observed by time-lapse fluorescent microscopy in wild-type, mde4Δ, and pcs1Δ cells expressing GFP-α-tubulin. A comparison of the kinetics of spindle elongation in mde4Δ, pcs1Δ, and wild-type cells revealed that cells lacking monopolin spend significantly more time in phase II (Figure 3A). Wild-type cells stayed less than 20 min in phase II, but 8 of 14 mde4Δ cells, and 6 of 8 pcs1Δ cells showed a prolonged phase II suggesting a delay in metaphase. A metaphase delay is often observed in cells when microtubule attachments to kinetochores are defective, which triggers the spindle checkpoint to inhibit anaphase onset. Observation of asynchronous wild-type and mde4Δ cells showed that an elevated percent of mde4Δ cells displayed spindle checkpoint activation as judged by the presence of Mad2 at kinetochores (Figure 3B). We tested for genetic interactions between the mde4Δ mutant and the spindle checkpoint mutants, mad1Δ, mad2Δ, mad3Δ and bub1Δ. The mde4Δ mutation was synthetic lethal with bub1Δ since no double mutants were recovered. Double mutants between mde4Δ and mad1Δ, mad2Δ or mad3Δ showed strong synthetic growth defects (Figure 3C), consistent with the notion that mde4Δ cells have defects in attachment of microtubules to kinetochores, which trigger spindle checkpoint dependent delays.
Figure 3. Monopolin mutants have delays in anaphase onset and show occasional spindle collapse.
(A) Spindle elongation was monitored in wild-type and monopolin mutant cells. Spindle length were measured every one minute from time-lapse sequences of asynchronous GFP-atb2 (n=17), GFP-atb2 mde4Δ (n=14), and GFP-atb2 pcs1Δ (n=8) cells. (B) Asynchronous wild-type and mde4Δ cells expressing Mad2-GFP were scored for the presence of Mad2-GFP puncta at kinetochores. Error bars represent standard deviation. (C) Genetic interactions between the mde4Δ, mde4-12A, and mde4-12D mutations and spindle assembly checkpoint mutations. The mde4Δ, mde4-12A and mde4-12D mutants were crossed with the spindle assembly checkpoint mutants bub1Δ, mad1Δ, mad2Δ and mad3Δ. Genetic interactions are shown as synthetic lethality (S.L), strong growth defect (+/−) and normal growth (+++). (D) A comparison of spindle lengths of wild-type and monopolin mutant cells at the metaphase/anaphase transition. The mean lengths of the spindle were compared between wild-type and pcs1Δ (single asterisks, p<0.01), mde4Δ (double asterisk, p<0.0001), or mde4-12A (triple asterisk p<0.0001), as well as mde4Δ and mde4-12A (quadruple asterisk p<0.0001) by a t test. Error bars represent standard deviation. (E) Time-lapse analysis using GFP-tubulin expressing wild-type (upper panel) and mde4Δ (lower panel) mutant cells is shown. The images were collected at one minute intervals. (F) An example of the chromosome co-segregation phenotype observed in mde4Δ cells is shown (left panel, arrow indicates septum position). The frequency of anaphase cells with lagging chromosomes and septated cells with co-segregated chromosomes is shown for wild-type and mde4Δ cells (right panel). Error bars represent standard deviation.
We found another difference in spindle morphology in monopolin mutants. Spindle lengths at the phase II/III transition in mde4Δ and pcs1Δ mutants were longer, 3.52 ± 0.15 µm and 3.47 ± 0.23 µm respectively, than that of wild-type cells, 3.07 ± 0.23 µm (Figure 3D). Elongated phase II spindles have also been observed in mutants defective in either microtubule attachment to kinetochores [21], or cohesion between sister chromatids [22], which may suggest that poorly attached kinetochores or reduced cohesion in monopolin mutants causes an imbalance of pushing and pulling forces in the spindle leading a somewhat elongated metaphase spindle. Previous results showed that monopolin is involved in preventing merotelic attachments [6], which are not thought to be monitored by the spindle checkpoint. However our results suggest an additional role for monopolin in attachment of microtubules to kinetochores or cohesion, which is monitored by the spindle checkpoint.
Monopolin may stabilize anaphase spindle microtubules to prevent chromosome co-segregation
The analysis of spindle elongation in mde4Δ cells revealed a function for monopolin in promoting anaphase spindle stability. In 6.4% (3/47) of mde4Δ cells the spindles appeared to break and/or collapse (Figure 3E, Figure S2A, and Movie S1, Movie S2, and Movie S3). Consistent with this, we found that about 8.5 % of septated mde4Δ cells showed chromosome co-segregation, in which one daughter cell inherited all of the chromosomes (Figure 3F). This phenotype likely arises because the nuclear envelope does not break down during mitosis in S. pombe. As a consequence of this, when the spindle breaks prematurely the chromosomes collapse back into a single mass inside the nuclear envelope. Similar results have been observed when the mitotic spindle was cut using laser microsurgery [19, 23]. In those studies, when the spindle was cut near the middle, the mitotic spindle completely collapsed. However, when the mitotic spindle was cut near one SPB, the spindle continued to elongate unidirectionally, with the end of the spindle lacking an SPB pushing out a finger of nuclear envelope as it elongated. Once the broken end of the spindle reached the end of the cell, it pushed against the cell tip causing the other spindle pole to move to the opposite end of the cell. Of the 3 cells that showed spindle elongation defects, 2 of them showed spindle breakage and collapse (Figure 3E, and Movie S1 and Movie S2), and one showed unidirectional spindle elongation suggesting that the spindle may have broken near one pole (Movie S3 and Figure S2A). Another case of apparent spindle breakage followed by chromosome co-segregation was observed by time-lapse analysis of mde4Δ cells expressing histone H3-GFP (Figure S2B). Furthermore, thin protrusions of nuclear envelope possibly caused by broken spindle ends were also observed in some mde4Δ but not wild-type cells (Figure S2C). Taken together, our data suggest that monopolin may have a novel function in stabilization of anaphase spindles.
Characterization of non-phosphorylatable and phospho-mimetic mutants of Mde4
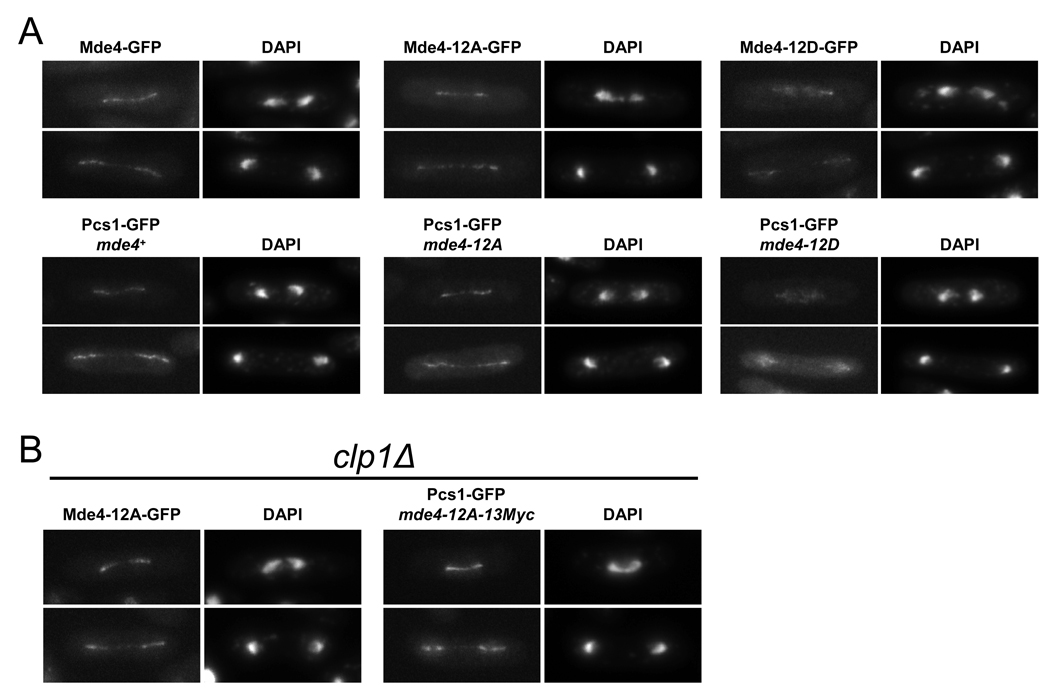
To more clearly define the role of phosphorylation of Mde4 in regulating the monopolin complex, we constructed non phosphorylatable and phospho-mimetic mutants. Since incomplete sequence coverage of Mde4 in our mass spectrometry did not allow us to identify all in vivo Cdk1 phosphorylation sites on Mde4, we mutated all twelve of the Cdk1 consensus sites to alanine (S/T to A) to prevent phosphorylation or to acidic residues (S to D and T to E) to create a phospho-mimetic mutant. We named them mde4-12A and mde4-12D respectively, and constructed strains where the endogenous mde4+ locus was replaced by mde4-12A and mde4-12D. As expected, the Mde4-12A mutant protein was no longer hyperphosphorylated in metaphase-arrested cells (Figure S3A), and Mde4-12A was not phosphorylated by Cdk1 in vitro (Figure S3B), indicating that the major sites of mitotic phosphorylation had been eliminated. Additionally, Mde4-12A did not interact with Clp1-C286S (Figure S3C), showing that phosphorylation is required for interaction with the Clp1-C286S substrate trapping allele. We do not think that the 12A and 12D mutations grossly perturbed the Mde4 protein structure because they did not affect Mde4-GFP localization to kinetochores and the nucleolus in interphase cells (Figure S3D). Additionally, kinetochore and nucleolar localization of Pcs1-GFP was abolished in the absence of Mde4 (Figure S3E), but was retained in mde4-12A and mde4-12D mutants (Figure S3E) suggesting that the Cdk1 site mutations do not disrupt formation of the monopolin complex.
Phosphorylation of Mde4 is required to prevent monopolin localization to the prometaphase/metaphase spindle
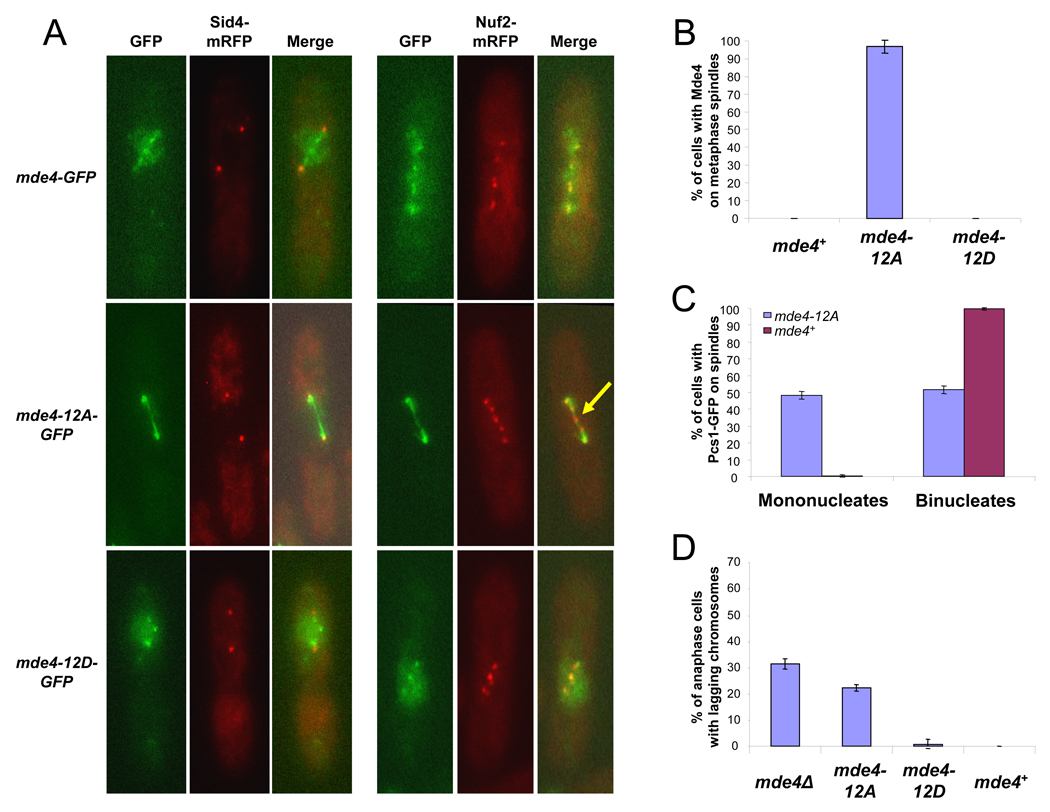
Unlike wild-type Mde4, Mde4-12A-GFP was often observed on the spindle in pre-anaphase cells (data not shown), suggesting that it may load onto the spindle prematurely. To examine this in greater detail, we arrested cells at metaphase by overexpressing the spindle assembly checkpoint protein Mad2, which prevents cyclin B and securin destruction by suppressing the activity of the APC/C-Cdc20 complex [24]. Under these conditions, Mde4-GFP and Mde4-12D-GFP were seen as several faint dots between the two spindle pole bodies, which were labeled with Sid4-mRFP (Figure 4A). These dots co-localized with a kinetochore protein, Nuf2-mRFP (Figure 4A). In contrast to Mde4-GFP, Mde4-12A-GFP localized to the metaphase spindle and could not be detected at kinetochores (Figure 4A, yellow arrow). Mde4-12A localized to the metaphase spindle in 97% (n=115) of metaphase cells, in contrast to Mde4 (n=126) and Mde4-12D (n=103), which did not localize on the spindle in metaphase cells (Figure 4B). To examine how the mde4-12A mutation affects Pcs1 localization, we counted cells showing spindle localization of Pcs1-GFP in asynchronous mde4+ and mde4-12A cells and classified them into mononucleate (early mitosis) and binucleate cells (anaphase). In wild-type cells, Pcs1-GFP localization on the spindle was observed almost exclusively in anaphase cells (307 of 308). In mde4-12A cells Pcs1-GFP localized normally to anaphase cells, but was also observed in pre-anaphase cell; specifically, 48% (n=350) of cells showing Pcs1-GFP localization to the spindle were mononucleates (Figure 4C). Thus, in each assay the non-phosphorylatable Mde4-12A mutant localizes the monopolin complex inappropriately to prometaphase/metaphase spindle, implying that Cdk1 phosphorylation acts to prevent premature localization of Mde4 to the mitotic spindle.
Figure 4. Mde4-12A prematurely localizes to the spindle instead of kinetochores before anaphase onset and displays lagging chromosomes in anaphase.
(A) Localization of Mde4, Mde4-12A, and Mde4-12D at metaphase was examined in the following strains: mde4-GFP sid4-mRFP, mde4-12A-GFP sid4-mRFP, mde4-12D-GFP sid4mRFP (left panel), mde4-GFP nuf2-mRFP, mde4-12A-GFP nuf2-mRFP, and mde4-12D-GFP nuf2-mRFP (right panel). Cells were arrested in metaphase by overexpression of Mad2 using the pREP3X-Mad2 plasmid for 19 hr in the absence of thiamine. Cells were fixed and imaged by fluorescence microscopy. The yellow arrow indicates a kinetochore that does not appear to label with Mde4-12A-GFP. (B) The frequency of Mde4 localization to the metaphase spindle was determined. mde4-GFP sid4-mRFP, mde4-12A-GFP sid4-mRFP, mde4-12D-GFP sid4-mRFP cells were arrested at metaphase by Mad2 overexpression for 19h. Cells showing spindle localization were counted among metaphase arrested cells exhibiting unseparated condensed chromosomes and separated spindle pole bodies. Error bars represent standard deviation. (C) Quantification of Pcs1 spindle localization in pre-anaphase, and post-anaphase mde4+ and mde4-12A cells. pcs1-GFP and pcs1-GFP mde4-12A cells were grown asynchronously at 30°C then fixed and stained with DAPI. Of cells that showed Pcs1 spindle localization, the frequency that were mononucleate or binucleate is shown. Error bars represent standard deviation. (D) Frequency of lagging chromosomes in mde4 mutants was determined by counting the percent of anaphase cells with lagging or unevenly segregated chromosomes from asynchronously growing mde4Δ, mde4-12A, mde4-12D, and wild-type cells. Error bars represent standard deviation.
Localization of monopolin at kinetochores has been proposed to prevent merotelic attachments, which cause lagging chromosomes [6, 8]. We observed that 22% of mde4-12A cells (n=304) showed lagging chromosomes in anaphase, which is comparable to the 31% observed in mde4Δ cells (n=327) (Figure 4D). In contrast, less than 1% of mde4-12D cells (n=311) in anaphase showed lagging chromosomes (Figure 4D). Our data shows that Cdk1 phosphorylation on Mde4 is important for its role in preventing merotelic attachment of kinetochores to spindle microtubules. One explanation for this could be that Cdk1 phosphorylation promotes kinetochore localization of Mde4 by inhibiting premature localization of Mde4 to microtubules in prometaphase/metaphase.
Mde4 Dephosphorylation promotes localization of monopolin to anaphase spindles
We used the mde4-12A and mde4 12D mutants to explore the role of Mde4 dephosphorylation in monopolin localization to the anaphase spindle (Figure 5A). Mde4-GFP and Mde4-12A-GFP localized similarly to the spindle in early and late anaphase. In contrast, Mde4-12D-GFP showed a diffuse nuclear signal in anaphase, with less than 10% of cells showing faint spindle localization. Pcs1-GFP localized in mde4+, mde4-12A, and mde4-12D cells the same as each Mde4 mutant protein (Figure 5A lower panel), consistent with the notion that the phosphorylation status of Mde4 regulates localization of the monopolin complex to the spindle. The poor localization of Mde4-12D to the spindle is similar to that observed for wild-type Mde4 in early anaphase clp1Δ cells, presumably because Mde4 is not dephosphorylated as rapidly in clp1Δ cells. If this is the case, then disruption of Mde4 phosphorylation should rescue the Mde4 spindle localization defect in clp1Δ cells. This proved to be the case, since Mde4-12A localized strongly to anaphase spindles in clp1Δ cells (Figure 5B). Additionally, expression of Mde4-12A was sufficient to restore Pcs1 association with the mitotic spindle (Figure 5B). Together these results show that Cdk1 phosphorylation on Mde4 must be removed to allow the monopolin complex to localize to anaphase spindles.
Figure 5. Mde4-12D localizes poorly to anaphase spindles.
(A) Early and late anaphase localization of wild-type Mde4-GFP, Mde4-12A-GFP, Mde4-12D-GFP (upper panel), and Pcs1-GFP in mde4+, mde4-12A, and mde4-12D cells (lower panel) is shown. (B) Early (top row) and late anaphase (bottom row) localization of Mde4-12A-GFP in clp1Δ and Pcs1-GFP in mde4-12A clp1Δ cells is shown. In both panels (A) and (B), cells were grown asynchronously, fixed, and imaged by fluorescence microscopy.
Both mde4-12A and mde4-12D cells display distinct defects in spindle elongation
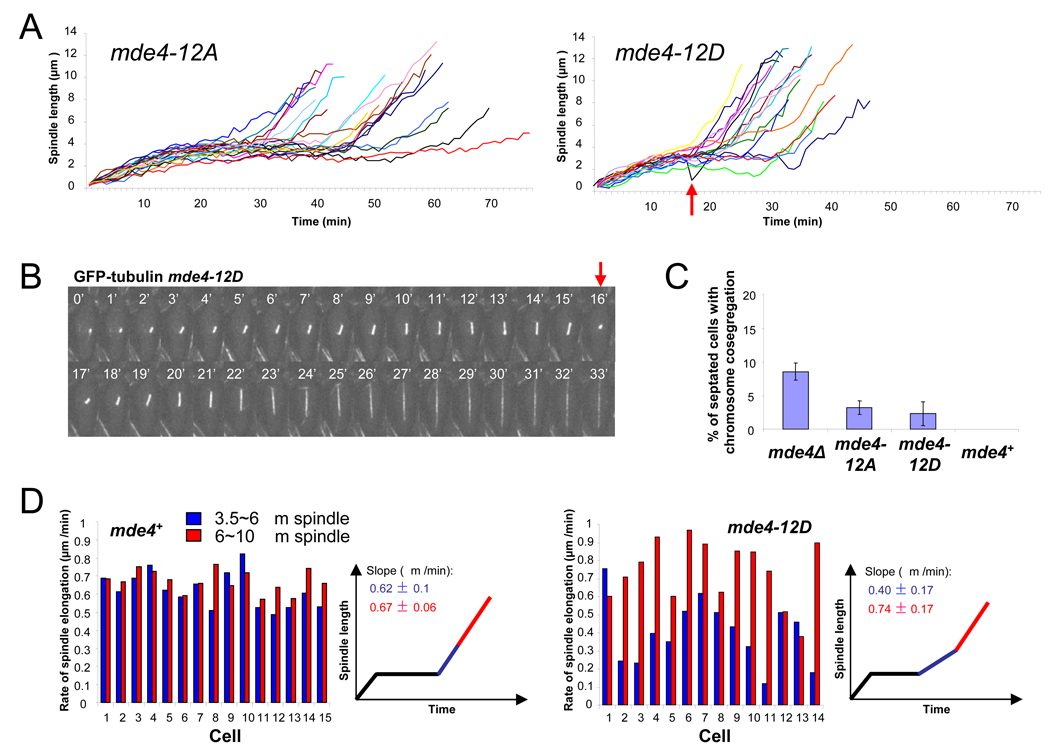
To better characterize spindle dynamics in mde4-12A and mde4-12D mutants, spindle elongation was observed by time-lapse microscopy in these cells. Spindle elongation in mde4-12A cells resembled mde4Δ cells (compare Figure 6A with Figure 3A), in that 50% (10 of 20) mde4-12A cells remained longer in phase II than mde4+ cells. However unlike mde4Δ cells, we did not observe spindle collapse or breakage in mde4-12A cells. The extended phase II in mde4-12A cells suggests that, like mde4Δ cells, they have microtubule attachment defects that trigger a spindle assembly checkpoint-dependent delay. To test this idea, we crossed mde4-12A to various spindle checkpoint mutants. The mde4-12A mutant was synthetically lethal with the mad1Δ, mad2A, mad3Δ and bub1Δ checkpoint mutants (Figure 3C), suggesting that the extended duration at phase II in mde4-12A cells is due to the activation of the spindle checkpoint. Notably, this interaction is even stronger than that observed for mde4Δ, since mde4Δ was only lethal with bub1Δ. Additionally, mde4-12A cells displayed significantly longer spindle lengths at phase II/III transition (3.78±0.12µm) than both wild-type and mde4Δ cells (Figure 3D). These results suggest that mde4-12A mutants may have greater defects in microtubule attachment to kinetochores than mde4Δ cells. This could be explained if the defects in mde4-12A cells are due not just to loss of Mde4 from the kinetochores in metaphase, but also to premature localization of Mde4 to the spindle.
Figure 6. Both mde4-12A and mde4-12D cells display distinct defects in spindle elongation.
(A) Spindle elongation was monitored in mde4-12A and mde4-12D cells. Spindle length was measured every one minute from time-lapse sequences of asynchronous GFP-atb2 mde4-12A (n=20) and GFP-atb2 mde4-12D (n=17) cells. Arrow indicates spindle collapse in the cell shown in part B. (B) Spindle collapse and re-elongation was observed in 1 of 17 the GFP-atb2 mde-12D cells shown part A. Images were collected every minute and spindle collapse was reconfirmed by examining a 3D view of the spindle from stacked images (data not shown) at the time point of spindle shortening (red arrow). (C) Frequency of chromosome co-segregation in mde4 mutants was determined by counting the percent of septated cells with a single DNA mass in only one daughter cell. Error bars represent standard deviation. (D) Comparison of spindle elongation rate between mde4+ (n=15) and mde4-12D (n=14) cells. Spindle elongation rates were obtained by measuring the slope of plots of spindle length over time using linear regression in individual mde4+ and mde4-12D anaphase cells. Slopes were calculated for early (spindle lengths of 3.5um to 6um, shown as a blue bar) and late (spindle lengths of 6 to 10 um, shown as a red bar) anaphase. Each pair of red and blue bars represents an individual cell.
Overall mde4-12D cells showed spindle behavior similar to wild-type cells but most cells (14 of 16) displayed slightly shorter phase II (Figure 6A and Figure 3A). Spindle lengths at the phase II/III transition in mde4-12D cells were 2.97 ± 0.09 µm, which is similar to that of wild-type cells (Figure 3D), and mde4-12D cells did not show negative genetic interactions with the spindle checkpoint mutants, mad1Δ, mad2Δ, mad3Δ and bub1Δ (Figure 3C), suggesting that microtubule attachment in mde4-12D cells is normal. A small fraction of mde4-12D cells exhibited spindle collapse followed by recovery (Figure 6B). Furthermore, 2.3% of mde4-12D cells (13 of 561) showed chromosome co-segregation (Figure 6C). While this represents a minority of mde4-12D cells, this was not observed in an equivalent number of wild-type cells (Figure 6C). These data are consistent with the view that reduced levels of Mde4 on the spindle may compromise its stability. Unexpectedly, we also found that mde4-12D cells display a reduced rate of spindle elongation in early anaphase B (Figure 6D). We measured the spindle elongation rate during early (3.5 to 6 µm length of the spindle) and late (6 to 10 µm length of the spindle) phase III. Although in wild-type cells the rate of spindle elongation (∼0.65 µm/min) was the same in early and late phase III, in mde4-12D cells the rate of spindle elongation was slower during early phase III (∼0.40 µm/min) but was similar to wild-type cells in late phase III (∼0.74 µm/min). Taken together, our data suggests that dephosphorylation of Mde4 at Cdk1 sites triggers spindle localization of monopolin, which promotes proper spindle elongation in early anaphase and enhances spindle stability.
Discussion
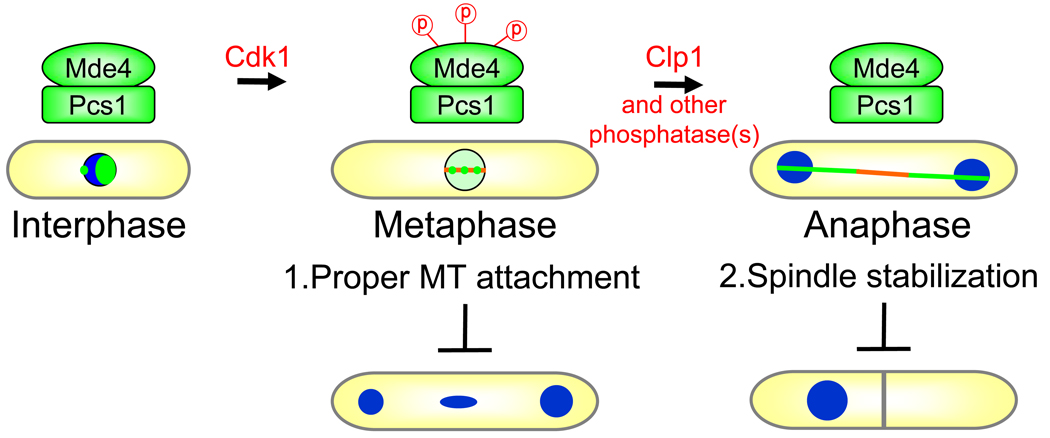
For faithful segregation of genetic material, chromosomes must be attached to microtubules in a bi-oriented manner at metaphase, and segregated to each daughter cell through spindle elongation at anaphase. Our studies show that regulated phosphorylation of the monopolin complex allows it to carry out discrete functions in each of these steps (Figure 7). When cells enter mitosis, Mde4-Pcs1 is released from the nucleolus and Mde4 becomes phosphorylated on Cdk1 sites. Mde4 phosphorylation does not appear to cause release of Mde4 from the nucleolus, since Mde4-12A is still released from the nucleolus in early mitosis. Instead, phosphorylation of Mde4 is required to prevent localization of the Mde4-Pcs1 complex to the metaphase spindle. Furthermore, the Mde4-12A protein not only localizes prematurely to the spindle, it also appears to localize poorly to the kinetochores. This could either be because monopolin localization to kinetochores requires Mde4 phosphorylation, or because the Mde4-12A protein has higher affinity for the spindle than for the kinetochores. We prefer the later model, since the Mde4-12A protein localizes normally to kinetochores in interphase when there is no spindle to compete with the kinetochores for binding to Mde4. Our data indicate that Mde4 phosphorylation on Cdk1 sites serves the dual function of (1) allowing monopolin to localize to kinetochores, where it can carry out its function in promoting proper chromosome attachment, and (2) phosphorylation blocks premature localization of monopolin to the spindle, which might interfere with metaphase spindle dynamics. After anaphase onset, Clp1 along with other phosphatases dephosphorylate Mde4 to allow the monopolin complex to load onto the anaphase spindle where it is required for both spindle stability and for proper spindle elongation.
Figure 7. Model of phosphorylation-dependent regulation of chromosome segregation by the monopolin complex.
Mde4-Pcs1 (green) localize to the nucleolus (green spot in nucleus) and the clustered kinetochores (smaller green spot at periphery of the nucleus) in interphase. In early mitosis Mde4 becomes phosphorylated (probably by Cdk1), which is required to maintain Pcs1-Mde4 at kinetochores (3 small green spots) to promote proper attachment of microtubules (MTs) to kinetochores to prevent lagging chromosomes in anaphase. In anaphase, Clp1 and other phosphatases dephosphorylate Mde4 to promote loading of monopolin onto the spindle (spindle shown in orange) to stabilize it and allow for proper elongation to prevent chromosome cosegregation.
Monopolin function at kinetochores
Previous studies in budding and fission yeast led to a model for monopolin function which proposed that the monopolin complex acts to clamp microtubule binding sites together at the kinetochore to ensure that they attach to microtubules from the same spindle pole [6–8, 10]. Further work in budding yeast showed that monopolin may also function to clamp the rDNA repeats together to keep them in register and prevent unequal crossing over [25]. Budding yeast monopolin requires separate adapters to target it to the meiosis I kinetochores and the rDNA repeats. These results suggest that monopolin can act as a crosslinker for different structures through the use of unique adapter molecules. Many crosslinking molecules such as the microtubule crosslinker Ase1 (PRC1 in humans) form multimeric complexes [26, 27]. Consistent with this, we have found that Mde4 and Pcs1 not only form heterodimers (Figure S4A), but may form either hetero-tetramers or higher order multimers since Mde4-GFP pulled down Mde4-13Myc from cell lysates from diploid cells expressing both tagged proteins (Figure S4B). Similar results were observed for cells expressing two different tagged versions of Pcs1 (Figure S4B). Thus it is possible that in S. pombe the Mde4-Pcs1 complex acts to cross-link microtubule binding sites, spindle microtubules, and possibly rDNA repeats. However, candidate adapter molecules for these locations have not been identified. TAP-purification of Mde4 and Pcs1 did not identify other kinetochore or microtubule binding proteins that could serve as adapters for Mde4-Pcs1 at these locations [6] (and our unpublished data).
In addition, our finding that monopolin mutants have extended metaphase delays and depend on the spindle checkpoint for normal growth suggests that monopolin may have an additional function to promote attachment of microtubules to kinetochores, since merotelic attachments are not thought to trigger the spindle checkpoint [2]. One way to reconcile this data with the earlier model would be to suppose that monopolin acts to clamp together and stabilize parallel microtubule ends at the kinetochore. Thus, monopolin could both stabilize attachments and favor kinetochore binding to microtubules from the same pole. Alternatively, monopolin could carry out separate functions in preventing merotelic attachments and promoting or stabilizing microtubule attachment to kinetochores.
Role of monopolin function in spindle elongation
Our work has uncovered a novel function for monopolin in promoting elongation and stability of the mitotic spindle. The spindle breakage phenotype we observed in mde4Δ cells indicates that Mde4 may have a structural role in maintaining spindle integrity. The mde4-12D mutant, which localizes poorly to the mitotic spindle, also has spindle integrity defects as judged by the chromosome cosegregation phenotype observed in fixed cells, and the occasional spindle collapse observed in time-lapse analysis. Both the exclusion of monopolin from anti-parallel microtubules in the spindle midzone, and the spindle breakage phenotype could be explained if monopolin prefers to bind to and crosslink only parallel microtubules. By crosslinking spindle microtubules, monopolin could promote spindle stability much like the spindle midzone crosslinker Ase1 [26, 27]. In an attempt to study the microtubule binding properties of monopolin in vitro, we were able to produce small amounts of Mde4-Pcs1 complex in bacteria, however the bacterially produced protein complex did not bind to microtubules in vitro (unpublished observations), suggesting that monopolin either does not bind to microtubules directly or that it requires other proteins or secondary modifications that are only present in vivo. In addition to the spindle collapse defect, we also found that mde4-12D mutant cells display slowed spindle elongation in early anaphase. We also observed a slowed rate of spindle elongation in mde4Δ cells (data not shown), however the phenotype in these cells may be complex since, unlike the mde4-12D mutant cells, mde4Δ cells have a high incidence of lagging chromosomes, which also slows spindle elongation [28]. The cause of slowed spindle elongation in mde4-12D cells is not known, but could be explained if monopolin has a role in loading and/or activation of microtubule motors on the spindle in early anaphase.
Reversal of Cdk1 phosphorylation and anaphase spindle function
Upon entry into mitosis, Cdk1 phosphorylation, either directly or indirectly, drives disassembly of the interphase microtubule cytoskeleton and assembly of the mitotic spindle. However, cells are able to maintain a mitotic spindle even when Cdk1 activity drops in anaphase. This and other studies have shown that anaphase spindle stability is accomplished at least in part by the loading of certain proteins onto the spindle specifically in anaphase [29–31]. Tightly coupling the localization and/or activity of these proteins to the level of Cdk1 activity seems to be a common strategy, as a number of spindle-associated proteins are inhibited in early mitosis by Cdk1 activity and then dephosphorylated in anaphase. Examples include in budding yeast Sli15 (human INCENP), Ase1, and Fin1 [29–31]. Dephosphorylation by Cdc14 promotes spindle localization of Sli15 and Fin1 [30, 31], and promotes the activity of Ase1 [29]. Similar to our findings with non-phosphorylatable Mde4, non-phosphorylatable variants of Sli15 and Fin1 localize prematurely to the mitotic spindle, which results in chromosome segregation defects [30, 31 and this study]. Premature localization of monopolin to the metaphase spindle also seems to cause additional defects in spindle dynamics or microtubule attachment to kinetochores since double mutants between mde4-12A and spindle checkpoint mutants such as mad2Δ were lethal, in contrast to mde4Δ mad2Δ mutants which were sick but viable.
Conservation of monopolin function?
The large number of microtubule binding sites at each mammalian kinetochore suggests that they should be particularly vulnerable to merotelic attachments. Consistent with this, time-lapse analysis of cultured mammalian cells showed that merotelic attachments are the major cause of chromosome mis-segregation [3]. Therefore we expect that mammalian cells have mechanisms to clamp together microtubule binding sites on each kinetochore to ensure that they become attached to microtubules from the same spindle pole. It will be interesting in future studies to identify structural or functional homologs of monopolin that act to prevent merotelic attachments in mammalian cells.
Supplementary Material
Acknowledgements
We thank Dr. Mitsuhiro Yanagida, Dr Kevin Hardwick, Dr Kim Nasmyth, Dr Fred Chang, Dr. Mohan Balasubramanian, Dr. Jean-Paul Javerzat, Dr. Phong Tran, and Dr. Kathleen Gould, for yeast strains, plasmids and antibodies. We are grateful to Dr. Peter Pryciak for helpful discussions. This work was supported by NIH grant R01GM068786 to D.M. V.S. is supported by ISREC and the Swiss National Science Foundation, and M-P.P-G was supported by the Marie Heim-Vogtlin fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 2.Cimini D. Detection and correction of merotelic kinetochore orientation by Aurora B and its partners. Cell Cycle. 2007;6:1558–1564. doi: 10.4161/cc.6.13.4452. [DOI] [PubMed] [Google Scholar]
- 3.Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregan J, Riedel CG, Pidoux AL, Katou Y, Rumpf C, Schleiffer A, Kearsey SE, Shirahige K, Allshire RC, Nasmyth K. The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr Biol. 2007;17:1190–1200. doi: 10.1016/j.cub.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 8.Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, Tanaka TU, Nasmyth K. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell. 2003;4:535–548. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- 9.Toth A, Rabitsch KP, Galova M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis i. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- 10.Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clifford DM, Wolfe BA, Roberts-Galbraith RH, McDonald WH, Yates JR, 3rd, Gould KL. The Clp1/Cdc14 phosphatase contributes to the robustness of cytokinesis by association with anillin-related Mid1. J Cell Biol. 2008;181:79–88. doi: 10.1083/jcb.200709060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cueille N, Salimova E, Esteban V, Blanco M, Moreno S, Bueno A, Simanis V. Flp1, a fission yeast orthologue of the s. cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J Cell Sci. 2001;114:2649–2664. doi: 10.1242/jcs.114.14.2649. [DOI] [PubMed] [Google Scholar]
- 13.Trautmann S, Wolfe BA, Jorgensen P, Tyers M, Gould KL, McCollum D. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr Biol. 2001;11:931–940. doi: 10.1016/s0960-9822(01)00268-8. [DOI] [PubMed] [Google Scholar]
- 14.Wolfe BA, McDonald WH, Yates JR, 3rd, Gould KL. Phospho-regulation of the Cdc14/Clp1 phosphatase delays late mitotic events in S. pombe. Dev Cell. 2006;11:423–430. doi: 10.1016/j.devcel.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 15.Trautmann S, Rajagopalan S, McCollum D. The S. pombe Cdc14-like phosphatase Clp1p regulates chromosome biorientation and interacts with Aurora kinase. Dev Cell. 2004;7:755–762. doi: 10.1016/j.devcel.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Wolfe BA, Gould KL. Fission yeast Clp1p phosphatase affects G2/M transition and mitotic exit through Cdc25p inactivation. Embo J. 2004;23:919–929. doi: 10.1038/sj.emboj.7600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tasto JJ, Carnahan RH, McDonald WH, Gould KL. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast. 2001;18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]
- 18.Hagan IM. The fission yeast microtubule cytoskeleton. J Cell Sci. 1998;111(Pt 12):1603–1612. doi: 10.1242/jcs.111.12.1603. [DOI] [PubMed] [Google Scholar]
- 19.Khodjakov A, La Terra S, Chang F. Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr Biol. 2004;14:1330–1340. doi: 10.1016/j.cub.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 20.Nabeshima K, Nakagawa T, Straight AF, Murray A, Chikashige Y, Yamashita YM, Hiraoka Y, Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toyoda Y, Furuya K, Goshima G, Nagao K, Takahashi K, Yanagida M. Requirement of chromatid cohesion proteins rad21/scc1 and mis4/scc2 for normal spindle-kinetochore interaction in fission yeast. Curr Biol. 2002;12:347–358. doi: 10.1016/s0960-9822(02)00692-9. [DOI] [PubMed] [Google Scholar]
- 23.Tolic-Norrelykke IM, Sacconi L, Thon G, Pavone FS. Positioning and elongation of the fission yeast spindle by microtubule-based pushing. Curr Biol. 2004;14:1181–1186. doi: 10.1016/j.cub.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 24.He X, Patterson TE, Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci U S A. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J, Brito IL, Villen J, Gygi SP, Amon A, Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loiodice I, Staub J, Setty TG, Nguyen NP, Paoletti A, Tran PT. Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol Biol Cell. 2005;16:1756–1768. doi: 10.1091/mbc.E04-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuyler SC, Liu JY, Pellman D. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J Cell Biol. 2003;160:517–528. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pidoux AL, Uzawa S, Perry PE, Cande WZ, Allshire RC. Live analysis of lagging chromosomes during anaphase and their effect on spindle elongation rate in fission yeast. J Cell Sci. 2000;113(Pt 23):4177–4191. doi: 10.1242/jcs.113.23.4177. [DOI] [PubMed] [Google Scholar]
- 29.Khmelinskii A, Lawrence C, Roostalu J, Schiebel E. Cdc14-regulated midzone assembly controls anaphase B. J Cell Biol. 2007;177:981–993. doi: 10.1083/jcb.200702145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pereira G, Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- 31.Woodbury EL, Morgan DO. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat Cell Biol. 2007;9:106–112. doi: 10.1038/ncb1523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.