Abstract
The Drosophila pair-rule gene even skipped (eve) is required for embryonic segmentation and later in specific cell lineages in both the nervous system and the mesoderm. We previously generated eve mesoderm-specific mutants by combining an eve null mutant with a rescuing transgene that includes the entire locus, but with the mesodermal enhancer removed. This allowed us to analyze in detail the defects that result from a precisely targeted elimination of mesodermal eve expression in the context of an otherwise normal embryo. Absence of mesodermal eve causes a highly selective loss of the entire eve-expressing lineage in this germ layer, including those progeny that do not continue to express eve, suggesting that mesodermal eve precursor specification is not implemented. Despite the resulting absence of a subset of muscles and pericardial cells, mesoderm-specific eve mutants survive to fertile adulthood, providing an opportunity to examine the effects of these developmental abnormalities on adult fitness and heart function. We find that in these mutants, flying ability, myocardial performance under normal and stressed conditions, and lifespan are severely reduced. These data imply a nonautonomous role of the affected pericardial cells and body wall muscles in developing and/or maintaining cardiac performance and possibly other functions contributing to normal lifespan. Given the similarities of molecular-genetic control between Drosophila and vertebrates, these findings suggest that peri/epicardial influences may well be important for proper myocardial function.
Keywords: cardiac development, heart rate, cardiac failure, aging, muscle
Although much effort has gone into defining developmental pathways, many of the mechanistic aspects of regulatory processes occurring during tissue specialization and organogenesis remain to be elucidated. The homeobox-containing gene even skipped (eve) is involved in several such aspects of tissue specialization. It was first identified in Drosophila as a segmentation gene.1 Later during development, eve is expressed in the nervous system and in dorsal muscle progenitors and pericardial cells.2 Regulatory elements for each of these aspects of the pattern were localized, 3,4 and a transgenic copy of the eve gene was shown to fully supply eve function.3 Specific repressor domains of Eve were identified in cultured cell assays5,6 and subsequently shown to interact functionally with corepressors.7–9
eve is expressed in, and required for the formation of, a subset of dorsal muscle and pericardial (PC) cells.10–12 In the developing mesoderm, early progenitors in the cardiogenic region express eve in segmentally repeated clusters that later differentiate into eve-positive pericardial cells (EPCs) and dorsal acute muscle 1 (DA1).2 A transgene was generated that fully rescues the phenotype of eve null mutants in all other tissues, but gives no detectable expression or function in the mesoderm, and resulting embryos were shown to develop mesodermal-specific defects that include disruption of EPC and DA1 muscle formation.13
Although much progress has been made in recent years in understanding the specification of cell types in the embryonic dorsal vessel,14 contributions of PC cells to the formation and function of the larval and adult heart are unexplored.15 Here, we focus on 4 key aspects of that function. First, we find that eve is required for the correct specification of cell types that arise from the eve-expressing lineage. Second, we find that eve is required for formation not only of the lineage in which it is continuously expressed, but also of a second lineage in which its expression is normally transient. Third, we find that the repressor function of Eve is necessary and sufficient, in the context of its homeodomain, to provide full rescue, implying that Eve acts exclusively as a repressor. Fourth, in the absence of eve-dependent PC cells, whereas the myocardium forms normally, the larval and adult heart rate is reduced, and the susceptibility of the heart to fail is dramatically increased, suggesting that PC cells play a pivotal role in normal heart function.
Materials and Methods
Transgenic Lines, Constructs, and Antibodies
A genomic region spanning 15 kb (Figure 1A) is sufficient to completely rescue the phenotype of eve null mutants.3 An additional 0.6 kb at the 3′ end may increase expression at some chromosomal locations, but at the insertion sites used here, no differences are seen in the degree of rescue with or without this region. Details of transgenes and antibodies are provided in the online Data Supplement available at http://circres.ahajournals.org.
Figure 1.
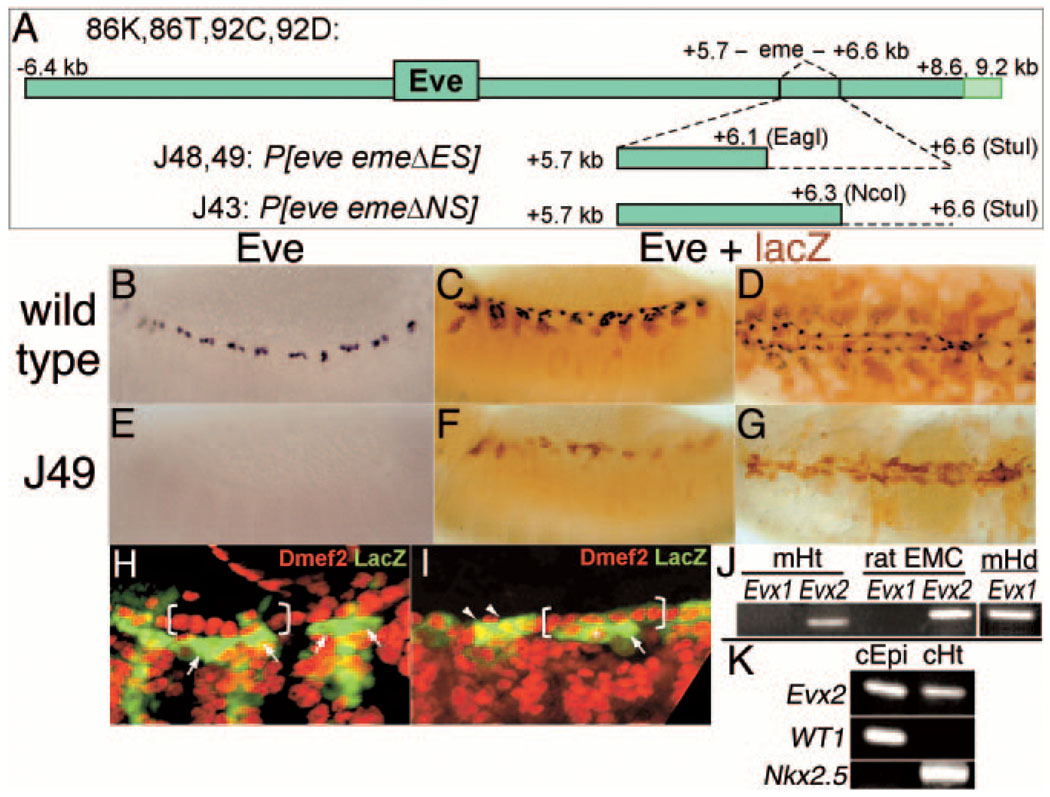
Generation of eve meso− flies and Evx2 expression in vertebrate heart. A, Transgenes used. Top, The entire rescue construct, showing the location of the Eve coding region, the mesodermal enhancer eme, and the alternative 3′ ends that each provide full rescue of eve null mutants. Middle and bottom, The dotted line indicates the region of eme deleted in the J48, J49, and J43 transgenes (restriction enzyme sites used are shown in parentheses). B, Expression of Eve (anti-Eve staining) in wild-type stage 13 embryos. C, Lateral view of the expression of Eve (black, anti-Eve staining) and a UAS-τLacZ transgene (brown, anti–β-gal staining) driven by eme-Gal4 in a wild-type stage 14 embryo. D, As in C, except late stage 16, dorsal view. E, As in B, but eve meso−, which is deficient for eve and carries 2 copies of the J49 transgene. Note the complete absence of Eve staining in the mesoderm (expression is normal elsewhere). F, As in C, but eve meso−. Note that there is still expression of the transgene in each segment, although the lack of proper differentiation in the eve lineages (see Results) reduces the number of cells stained. G, As in D, but eve meso−. H and I, Dmef2 (red) and eme-driven lacZ protein expression (green) of stage 14 embryos. H, Wild-type. Note cytoplasmic lacZ protein in 2 EPCs (arrows) adjacent to 6 myocardial cells per hemisegment (bracket). I, As in H, but eve meso−. Note the formation and normal alignment of 6 myocardial cells per hemisegment (bracket) and persistence of LacZ expression in myocardial (arrowheads) or pericardial cells (arrow). Ectopic Dmef2-positive nuclei coexpressing lacZ are also occasionally observed (asterisk). J, Reverse transcription polymerase chain reaction (RT-PCR) for Evx1 and Evx2 was performed using 800 ng of RNA isolated from 12.5dpc embryonic mouse heart (mHt) and an established rat epicardial cell line (ratEMC). Note the Evx2-specific band of 150bp in mouse (left) and of 194bp in rat (right). As a positive control for Evx1 amplification, RT-PCR was performed using RNA isolated from stage 11 embryonic mouse head (mHd), resulting in the amplification of a specific band of 194 bp. K, RT-PCR for Evx2 was performed with RNA obtained from primary epicardial monolayers from HH21 embryonic chick heart (cEpi).33 Note amplification of the chick Evx2-specific band of 462bp in these primary epicardial cells. RT-PCR for Evx2, WT1, and Nkx2.5 were also performed using RNA from the hearts (cHt) used to isolate the epicardium monolayers. Wilm’s tumor gene expression (WT1) was used as an epicardial-specific marker,34,35 and Nkx2.5 expression as a myocardial-specific marker. Note the lack of significant myocardial contamination in the epicardial sample, as evidenced by the lack of Nkx2.5 expression, indicating that the Evx2 signal in the epicardial sample does not come from myocardial contamination.
Analysis of Embryos
eve null backgrounds, either Df(2R)eve, Df(2R)eve/eve3, or eve3/eve3, all which gave indistinguishable results, were used in combination with mesodermal enhancer–deficient eve rescue transgenes and are referred to as eve “meso−” throughout the text. Embryos shown in Figure 1 are Df(2R)eve, whereas muscle preparations from larvae and adult are Df(2R)eve/eve3 or eve3/eve3.
Evx Expression, Pericardial Cell Counts, Cardiac and Lifespan Measurements, and Flight Assay
Details of assays for Evx expression, pericardial cell counts, heart rates, cardiac stress–induced failure rates, flight ability, and lifespan, are provided in the Data Supplement.
Results
Mesoderm-Specific eve Mutants
Removing the mesodermal enhancer eme in the context of a rescue transgene removes all detectable mesodermal eve expression without affecting any other aspect of expression (Figure 1). When combined with a null mutation at the endogenous locus, this results in an eve “mesoderm negative” (eve meso−) mutant. A separate transgene containing eme upstream of a reporter gene (lacZ) can be used to faithfully mark the cells that would normally express eve. In the absence of mesodermal eve, the body wall muscles no longer show reporter gene expression (Figure 1F and 1G). In cells associated with the heart, however, the stable reporter protein persists until the formation of a seemingly normal heart tube (Figure 1G; supplemental Figure I). Except for the lack of eve expression (Figure 1B through 1G), other heart cell types, particularly the myocardial cells, appear to be present in their normal numbers and positions (Figure 1H and 1I; supplemental Figure I). In the absence of eve, the cells in which the reporter persists appear to be naïve and able to adopt other (cardiac) fates (Figure 1H and 1I), consistent with eve acting as an essential factor for specification of their lineage. Interestingly, the vertebrate eve homolog, Evx2, is also expressed in the heart, in an epicardial cell line, and in cells from primary epicardial explants (Figure 1J and 1K). Thus, eve/Evx may play a role in heart development in both insects and mammals.
Altered Expression of Identity Genes
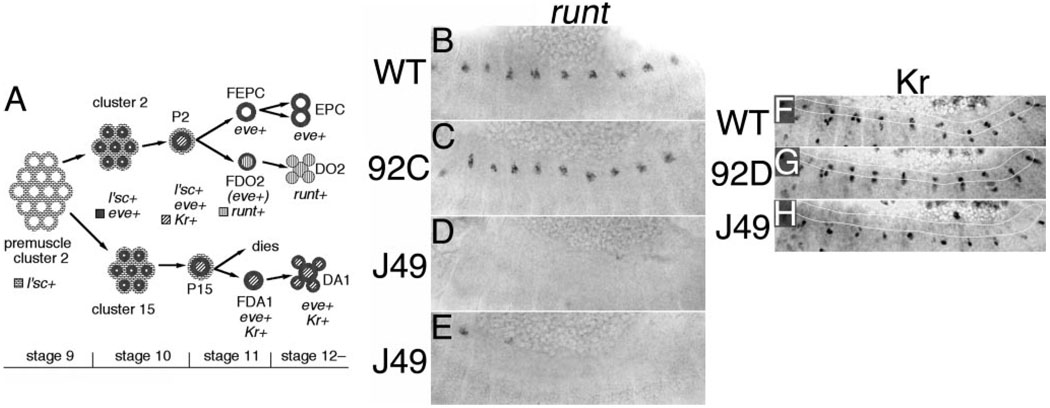
Mesodermal eve expression normally occurs in founder cells that give rise to a subset of pericardial cells and to 2 muscles per hemisegment, DO2 and DA1 (Figure 2A).16–18 A cell within 1 eve-expressing cluster (cluster 2) initiates expression of Krüppel (Kr; Figure 2F). This Kr- and eve-expressing cell (progenitor 2) divides to yield 2 founder cells that express runt (Figure 2B), 1 of which is the founder of DO2 and continues to express runt, and the other of which is the EPC founder and turns off runt. The DO2 founder turns off eve shortly after runt is activated, whereas the EPC founder and the resulting EPCs continue to express eve. A cell within a second eve-expressing cluster (cluster 15) activates Kr (Figure 2F, outlined in white), then divides to yield the DA1 founder and a second cell that is fated to die. The DA1 muscle maintains eve expression.
Figure 2.
Loss of Kr and runt expression due to the absence of mesodermal eve. A, Diagram of gene expression in the eve mesodermal lineage.35a Eve expression is initiated in 2 cell clusters per hemisegment during stage 102, cluster 2 appearing earlier than cluster 15, and each derived from premuscle cluster 2 that expresses lethal of scute (l’sc).36 One cell per cluster is selected through lateral inhibition to become a muscle progenitor,36–38 and initiates Kr expression.39,40 Progenitor 2 (P2) divides to form the founder of eve expressing pericardial cells (FEPC), which extinguishes l’sc and Kr expression and subsequently divides to form 2 EPCs and the founder of DO2 (FDO2), which extinguishes l’sc and Kr expression, then activates runt and loses eve expression, subsequently fusing with other myoblasts nearby to form DO2.17,40,41 Progenitor 15 (P15) gives rise to a cell that is fated to die and to the founder of DA1 (FDA1), which extinguishes l’sc but maintains Kr and eve expression, subsequently fusing with other myoblasts to form DA1.17,41–44 B through E, runt expression in stage 13 embryos, stained by in situ hybridization: Wild-type (B), eve null mutants rescued by the complete rescue transgene (92C) (C); eve meso− embryos (eve null mutants rescued by the eme-deleted J49 transgene) (D; note the absence of runt expression); and another example of the same line as in D (E; note the residual expression in some anterior parasegments). F through H, Kr expression in stage 12 embryos (anti-Kr staining); the region where the DA1 founders lie is outlined. F, Wild-type. G, eve null mutants rescued by the complete rescue transgene (92D); note the normal pattern of Kr expression. H, eve meso− embryos (eve null mutants rescued by J49); note the absence of the dorsally located set of Kr-expressing cells (also absent at stage 11), 1 in each parasegment, which are the DA1 founders (the more ventrally located Kr-expressing cells are derived from a separate lineage, and serve as an internal control).
Previous studies suggested that eve function is required for normal EPC differentiation12 and for the normal pattern of expression of ladybird.11,13 As a way to define the role of eve, we examined both runt and Kr expression in eve meso− embryos. In the absence of eve, both runt and Kr expression are either completely absent from or dramatically reduced in these muscle founder lineages (Figure 2). This strongly suggests that eve is required for these cells to adopt their normal fates. Thus, eve has either a direct or an indirect role (repression of a repressor) in activating Kr and runt. We distinguish between these possibilities below.
Muscle Defects in eve Meso− Larvae
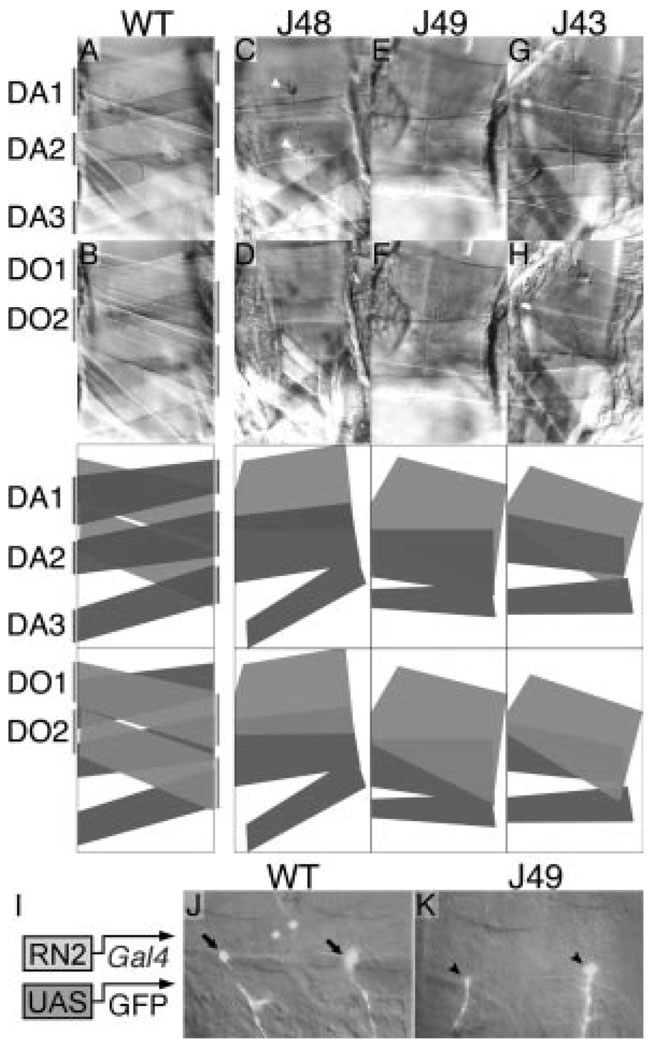
To determine the extent to which eve function is required for normal muscle formation, we examined the musculature in eve meso− third instar larvae. The normal arrangement of dorsal muscles within each segment is clearly altered (Figure 3A through 3H). Both DA1 and DO2 are severely defective or aberrant, and other muscles in the vicinity exhibit alterations in their placement and size. A simple interpretation of the effects seen in a majority of segments is that DA1 is missing, and a single, large muscle occupies the normal positions of DO1 and DO2. The muscle occupying the normal position of DA2 is also enlarged. These enlarged muscles suggest that myoblasts that normally fuse with the DA1 and DO2 founders may instead fuse with other muscles nearby, or, alternatively, that in the absence of DA1 and DO2, attachment sites for adjacent muscles expand.
Figure 3.
Muscle defects and innervation by eve-expressing neurons in eve meso− larvae. Either wild-type (A and B) or eve meso− (C through H) third instar larvae were dissected to reveal the dorsal musculature (anterior is to the left, dorsal up). Three meso− lines are shown, each null mutant for eve and rescued by the indicated eme-deleted transgene (J48 in C and D; J49 in E and F; J43 in G and H). Each panel in the second row shows the same embryo as that above it. The focal plane in the top row is that of the normal DA1, DA2, and DA3, whereas the focal plane in the second row is that of the normal DO1 and DO2. The muscle pattern in each pair of panels is diagrammed in the 2 panels directly below them, with dark gray representing muscles in the DA1,2,3 focal plane and light gray representing muscles in the DO1,2 focal plane. Note in the meso− embryos the absence of a muscle in the normal DA1 position, the wider-than-normal muscle in the DA2 position, and the single muscle occupying most of the normal DO1,2 region. A similar range of phenotypes as that shown, including the particular variations in muscle widths, is seen in each of the meso− lines. Note also the innervation of both of the abnormal muscles in C, as evidenced by the tree-like branching structures of boutons (white arrowheads; visualized by anti-fasciclin II staining). I, Transgenes used to label the eve-expressing neurons (RP2 and aCC) that innervate DA1 and DO1. The mesodermal regulatory element derived from the eve gene (RN2-Gal4)22 drives expression of the Gal4 activator in the RP2 and aCC motorneurons (as well as in the interneuron pCC), which in turn activates the UAS-GFP reporter transgene. J and K, GFP image of a 2 segment-wide swath of a late stage 16 embryo is overlayed with the corresponding visible light image. J, Wild type; note the bulbous synapse at a point where DA1 and DO1 intersect on their ventral side (arrows). K, J49 eve meso−; note the innervation (arrowheads) at a point of intersection of the two remaining muscles described above.
The DA1 and DO1 muscles are innervated in wild-type embryos by motorneurons that express eve.19–21 We used transgenes that express green fluorescent protein (GFP) in these neurons to label their axons in the muscle field.22 We found that in eve meso− embryos, one or both muscles in the normal positions of DA1/2 and DO1/2 are innervated by these eve-expressing neurons (Figure 3I through 3K). We also found that in eve meso− third instar larvae, both DA1/2 and DO1/2 are innervated (Figure 3C).
Eve Functions as a Repressor in the Mesoderm
The changes in gene expression observed in eve meso− embryos (Figure 2) suggest that eve acts, directly or indirectly, as an activator of Kr and runt. Previous analyses of eve function suggested that it acts as a repressor of transcription. 8,22–24 If this is true in the mesoderm, then at least 1 intermediary gene that is repressed by eve normally represses runt and Kr. To study the domain requirements of eve, we expressed transgenes in the eve meso− background that contained the eve mesodermal enhancer driving expression of modified Eve proteins. With the wild-type Eve coding region, the mesodermal defect is completely rescued (supplemental Figure IIA and IIB). In contrast, when the Eve homeodomain (HD)-containing region alone is so expressed, a very limited degree of rescue is observed (supplemental Figure IIC and IID). Importantly, when the heterologous Engrailed repressor domain is added to the HD construct, full rescuing ability is restored (supplemental Figure IIE), implying that eve acts exclusively as a repressor in the mesoderm.
We also examined the ability of Eve to act as a direct repressor in the mesoderm by targeting it to a reporter transgene using the Gal4 DNA binding domain. When a Gal4-Eve fusion protein containing both of the repressor domains of Eve is combined with an eve lineage–specific Gal4-UAS–containing reporter, the reporter is strongly repressed (supplemental Figure IIF through IIH).
eve Meso− Larvae Have Fewer Pericardial Cells
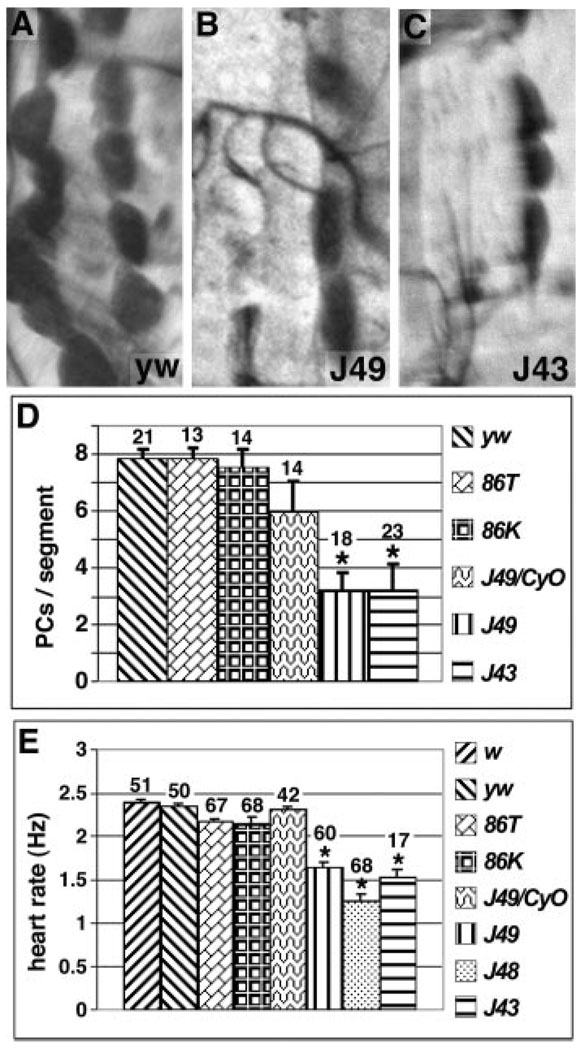
eve meso− embryos develop into viable adults, providing an opportunity to examine the role of PC cells in larval and adult heart function. The absence of mesodermal eve does not noticeably affect the assembly of the myocardial cells at the dorsal midline, which will give rise to the contractile part of the heart tube (data not shown). To examine the contribution of EPCs to larval heart development, eve meso− larvae were dissected and PC cells were counted. Wild-type and wild-type eve-rescued larvae show 7 to 8 PC cells per segement, and eve meso− heterozygotes show 6 (Figure 4A and 4D). In contrast, 2 independent eve meso− lines displayed a marked reduction, with an average of only 3 to 4 PC cells per segment (Figure 4B through 4D). Thus, eve is required to produce the normal complement of larval PC cells.
Figure 4.
Functional defects in eve meso− larvae. A through C, One segment is shown from the following genotypes of a dissected third instar larval heart stained with neutral red to label PC cells: A, yw: wild-type larvae contain 7 to 8 PCs per segment, on average; B and C, J49 and J43 meso−, respectively (see Figure 1A for transgene diagrams); only 3 PC cells are visible. D, Graph of the average number of PC cells per segment in larvae of the indicated genotypes. E, Graph of average heart rates (in beats/second) of white pre-pupae of the indicated genotypes. D and E, “J49/CyO” is heterozygous for the J49 eve meso− chromosome and the CyO balancer, which is wild-type for the eve gene. All other genotypes (except w and yw, which are considered wild-type) are eve null mutants, rescued with the indicated eve transgene. The number of segments counted (3 to 5 per dissected larva) is shown above each bar. Error bars represent standard deviation. Asterisk indicates significantly different from yw Student’s t test, P<0.01).
Functional Heart Defects
Because eve meso− animals are missing half or more of their PC cells, we examined the effect on heart function in pupae and adults. We found that neither wild-type eve-rescued pupae nor those heterozygous for eve and carrying one copy of a meso− rescue transgene exhibited heart rates that differed significantly from controls (Figure 4E). However, all 3 meso− lines, which carry independent transgene insertions, exhibited a significant reduction in heart rate (30% to 50%, Figure 4E).
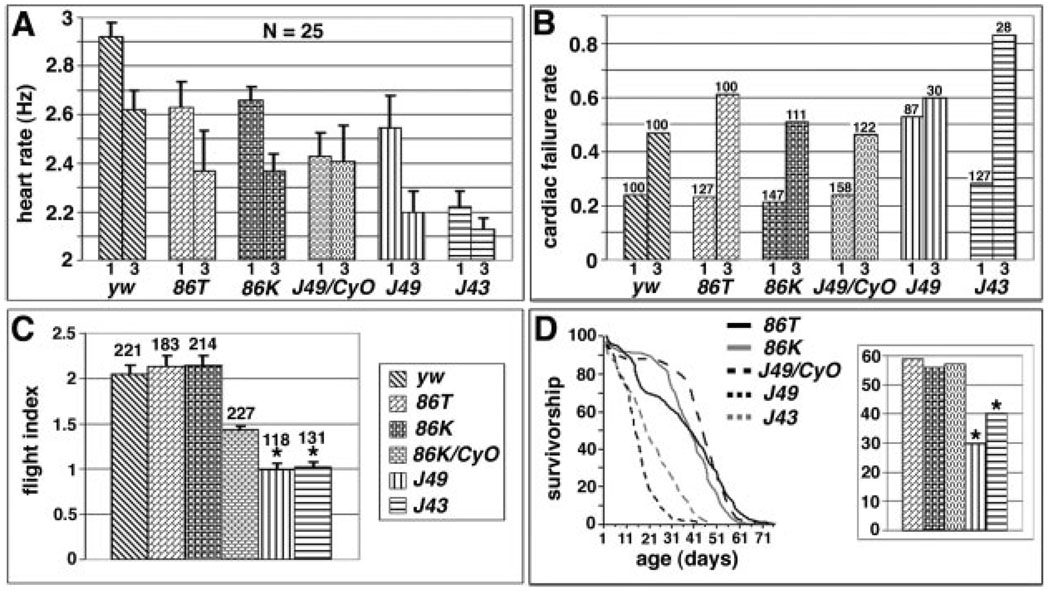
We examined adults of the same genotypes to assess whether defective functions persist through partial remodeling of the heart during metamorphosis.25 The wild-type heart rate is 2.9 beats/second (Hz) in 1-week-old adults and 2.6 Hz in 3-week-old adults (Figure 5A).26 Compared with wild type, both the eve null background rescued by a wild-type rescue transgene and the eve meso− heterozygotes (J49/CyO) exhibit a reduced heart rate at both ages, probably attributable to a genetic background effect inherent to these eve rescue lines that serve as controls. eve meso− flies, however, develop a dramatically lower heart rate with age, and one of the meso− lines also shows a severely reduced heart rate at an early age (Figure 5A).
Figure 5.
Functional defects in eve meso− adults. A, Adult heart rates. For each of the indicated genotypes (see Figure 1A), the average heart rate (in Hz, ie, cycles/second) is graphed at both 1 week of age (left bar) and 3 weeks of age (right bar). Error bars represent standard deviation. Twenty-five flies were tested for each genotype. Note that J49 meso− flies (J49) have a significantly lower heart rate at 3 weeks than all controls (Student t test, P<0.01), whereas J43 meso− (J43) flies have a significantly lower heart rate than all controls at both 1 and 3 weeks (Student t test, P<0.01). B, Cardiac failure rates in response to external electrical pacing. For each of the indicated genotypes, the average stress-induced cardiac failure rate is shown at both 1 week of age (left bar) and 3 weeks of age (right bar). The number of flies tested is shown above each bar. Note that J49 flies have a failure rate at 1 week that is significantly higher than all controls (binomial confidence interval, P<0.01), whereas J43 flies have a failure rate at 3 weeks significantly higher than all controls (binomial confidence interval, P<0.01). C, Quantification of flight ability of the indicated genotypes. A flight index on a scale of 0 to 4 was used to assess each line, with 0 meaning no flying ability and 4 being the highest possible score (see Materials and Methods). The number of flies tested is shown above each bar. Error bars represent the standard error of the mean. Note that 86K/CyO has a significantly lower flight ability than wild type (Student t test, *P<0.01), whereas both J49 and J43 scored significantly lower than even 86K/CyO. D, Lifespan measurements for the indicated genotypes, graphed as percentage of survival versus time (in days). Both J49 and J43 flies had a significantly shorter lifespan than all wild-type rescue controls (Student t test, *P<0.01). Right panel, Histogram depicting mean lifespan from the same experimental series. The same fills indicate the genotypes as in A through C.
Heart function can also be assayed by quantifying stress tolerance, using an external current to briefly pace the heart to about twice the normal rate, then charting the percentage that undergo either fibrillation or cardiac arrest26 (termed heart failure). In wild-type flies, the ability of the heart to withstand such stress is highly age-dependent, with stress-induced failure rates increasing dramatically (2- to 3-fold) between 1 and 5 weeks of age.26,27 Because eve meso− flies seldom reach 5 weeks of age, we examined flies at 1 and 3 weeks of age. Neither heterozygous eve meso− nor eve rescued flies differed from wild type (Figure 5B). In contrast, one eve meso− line showed a significantly increased failure rate at 1 week of age, whereas the other showed a disastrously high failure rate at 3 weeks of age (Figure 5B). Although dorsal somatic muscle defects might also conceivably affect heart function, these results suggest that the reduction in PC cell number causes a slowed heartbeat and reduces cardiac stress resistance.
Effects on Lifespan
To further assess the importance of fully functional heart and muscle activity and potentially of other results of mesodermal eve expression (see Figure 5C, and “Other functional defects” in the Data Supplement), we examined the life spans of eve meso− flies. Such flies display a significantly reduced mean and maximal lifespan (Figure 5D), suggesting that the presence of mesodermal Eve is required not only for normal activity levels but also for a normal lifespan.
Discussion
Functional Requirements in All eve-Expressing Mesodermal Lineages
By constructing a mesoderm-specific eve mutant, we established that eve is required not only in the eve-expressing lineages in which it is maintained during terminal differentiation (the eve-expressing pericardial cells and the DA1 muscle), but also in the lineage in which it is expressed in the progenitor but turned off in the muscle founder cell and the resulting DO2 muscle. Thus, both DA1 and DO2 require mesodermal eve to be specified, and without this specification, the pattern and size control of some remaining muscles are compromised (Figure 2 and Figure 3). Importantly for heart function (discussed below), in the absence of mesodermal eve expression, the majority of the large larval PC cells are missing (Figure 4).
It is intriguing that even though ectopic Eve expression can interfere with the DO2 fate,11 and eve is normally turned off as runt is activated in the lineage (Figure 2A), eve function is nonetheless required for DO2 formation, apparently because of a requirement in the progenitor before the lineage divisions.
Eve Protein Function in the Mesoderm
When normal eve expression in the mesoderm is replaced by expression of the eve HD (with repressor domains deleted), a similar but less severe muscle deficiency is observed compared with the complete absence of mesodermal eve (supplemental Figure II). In particular, a muscle in the DO2 position is usually formed, whereas DA1 is still absent. Additionally, there is occasionally an extra muscle ventral to DO2, as if the DO2 founder was duplicated. Importantly, however, when the heterologous Engrailed repressor domain is added to the HD construct, full rescuing ability is restored (supplemental Figure II). This suggests that Eve functions in the mesoderm primarily or exclusively as a repressor, and in turn that eve acts indirectly to activate Kr and runt in the mesoderm. Good candidates for intermediary repressors are ladybird and the muscle identity gene msh.11,13
Eve and PC Cells Influence Larval and Adult Heart Function
A reduced number of larval PC cells (and dorsal somatic muscles) caused by a lack of mesodermal eve expression results in severely compromised heart function and is likely to contribute to a shortened lifespan. A less drastic effect on cardiac performance and lifespan is observed when manipulating insulin signaling exclusively in the heart.27 The functional role of pericardial cells in insect hearts is not well understood,15 but they may contribute to heart function by secreting hormones or by gathering such peptides from circulating hemolymph and “presenting” them to the myocardium. It has been suggested that pericardial cells may function as nephrocytes,15 and at this point we cannot rule out that a potential accumulation of toxic agents, as a consequence of fewer pericardial cells, contributes to the observed phenotypes.
As in insects, the developmental and functional interactions between the vertebrate epicardium and the myocardium are not well understood. Recent studies have suggested that the loss of epicardial function results in impaired growth of the myocardium at mid-gestation.28–30 The epicardium is thought to be a source of signals and secreted factors that affect myocardial proliferation and differentiation, as well as influencing formation of the conduction system.31 Even though it cannot yet be decided whether the mammalian epicardium has a developmental program in common with a fly’s pericardial cells, they both depend on GATA factors for formation.30,32 In addition, the Evx2 homolog of eve is indeed expressed in the mammalian heart, including in epicardial tissue (Figure 1J and 1K). Our findings are consistent with pericardial cells in Drosophila functioning as a source of signals that affect the myocardium. Possibly because the myocardium, which is maintained by proliferation in vertebrates, does not proliferate in flies after it is developmentally specified,25 pericardial deficiency does not appear to result in morphological heart defects (supplemental Figure I). Rather, defects manifest themselves as functional deficits. This provides an opportunity to study the influence of these heart-associated cell types on cardiac physiology in the absence of myocardial defects. Epicardial lineages in vertebrates may contribute analogously to normal cardiac physiology and performance.
Supplementary Material
Acknowledgments
M.Z. is a recipient of a fellowship from the Spanish Ministry of Science and Education. P.R-L. is funded by the National Institutes of Health (National Heart, Lung, and Blood Institute). J.B.J. received support for this work from the National Institutes of Health and the National Science Foundation (award 0416760), and R.B. from the National Heart, Lung, and Blood Institute and the National Institute on Aging of the National Institutes of Health. We thank Manfred Frasch, Corey Goodman, Steve Small, the Developmental Studies Hybridoma Bank at the University of Iowa, and the Bloomington Stock Center for reagents. We thank Jian Zhou and Erin Fitzgerald for excellent technical assistance.
Footnotes
Reprints: Information about reprints can be found online at http://www.lww.com/static/html/reprints.html
References
- 1.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 2.Frasch M, Hoey T, Rushlow C, Doyle H, Levine M. Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 1987;6:749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development. 1999;126:2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sackerson C, Fujioka M, Goto T. The even-skipped locus is contained in a 16-kb chromatin domain. Dev Biol. 1999;211:39–52. doi: 10.1006/dbio.1999.9301. [DOI] [PubMed] [Google Scholar]
- 5.Han K, Manley JL. Transcriptional repression by the Drosophila even-skipped protein: definition of a minimal repression domain. Genes Dev. 1993;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- 6.Jaynes JB, O’Farrell PH. Active repression of transcription by the engrailed homeodomain protein. EMBO J. 1991;10:1427–1433. doi: 10.1002/j.1460-2075.1991.tb07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erkner A, Roure A, Charroux B, Delaage M, Holway N, Core N, Vola C, Angelats C, Pages F, Fasano L, Kerridge S. Grunge, related to human Atrophin-like proteins, has multiple functions in Drosophila development. Development. 2002;129:1119–1129. doi: 10.1242/dev.129.5.1119. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi M, Goldstein RE, Fujioka M, Paroush Z, Jaynes JB. Groucho augments the repression of multiple even-skipped target genes in establishing parasegment boundaries. Development. 2001;128:1805–1815. doi: 10.1242/dev.128.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang S, Xu L, Lee J, Xu T. Drosophila Atrophin homolog functions as a transcriptional corepressor in multiple developmental processes. Cell. 2002;108:45–56. doi: 10.1016/s0092-8674(01)00630-4. [DOI] [PubMed] [Google Scholar]
- 10.Abmayr SM, Erickson MS, Bour BA. Embryonic development of the larval body wall musculature of Drosophila melanogaster. Trends Genet. 1995;11:153–159. doi: 10.1016/s0168-9525(00)89030-7. [DOI] [PubMed] [Google Scholar]
- 11.Jagla T, Bidet Y, Da Ponte JP, Dastugue B, Jagla K. Cross-repressive interactions of identity genes are essential for proper specification of cardiac and muscular fates in Drosophila. Development. 2002;129:1037–1047. doi: 10.1242/dev.129.4.1037. [DOI] [PubMed] [Google Scholar]
- 12.Su MT, Fujioka M, Goto T, Bodmer R. The Drosophila homeobox genes zfh-1 and even-skipped are required for cardiac-specific differentiation of a numb-dependent lineage decision. Development. 1999;126:3241–3251. doi: 10.1242/dev.126.14.3241. [DOI] [PubMed] [Google Scholar]
- 13.Han Z, Fujioka M, Su M, Liu M, Jaynes JB, Bodmer R. Transcriptional integration of competence modulated by mutual repression generates cell-type specificity within the cardiogenic mesoderm. Dev Biol. 2002;252:225–240. doi: 10.1006/dbio.2002.0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bodmer R, Wessells RJ, Johnson EC, Dowse H. Heart development and function. In: Gilbert LI, Iatrou K, Gill S, editors. Comprehensive Molecular Insect Science, Volumes 1–7. Vol. 2. Oxford, UK: Elsevier; 2004. pp. 199–250. [Google Scholar]
- 15.Rizki T. The circulatory system and associated cells and tissues. In: Ashburner M, Wright TRF, editors. The Genetics and Biology of Drosophila. London, UK: Academic Press; 1978. pp. 1839–1845. [Google Scholar]
- 16.Han Z, Bodmer R. Myogenic cells fates are antagonized by Notch only in asymmetric lineages of the Drosophila heart, with or without cell division. Development. 2003;130:3039–3051. doi: 10.1242/dev.00484. [DOI] [PubMed] [Google Scholar]
- 17.Carmena A, Gisselbrecht S, Harrison J, Jimenez F, Michelson AM. Combinatorial signaling codes for the progressive determination of cell fates in the Drosophila embryonic mesoderm. Genes Dev. 1998;12:3910–3922. doi: 10.1101/gad.12.24.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alvarez AD, Shi W, Wilson BA, Skeath JB. Pannier and pointedP2 act sequentially to regulate Drosophila heart development. Development. 2003;130:3015–3026. doi: 10.1242/dev.00488. [DOI] [PubMed] [Google Scholar]
- 19.Landgraf M, Bossing T, Technau GM, Bate M. The origin, location, and projections of the embryonic abdominal motorneurons of Drosophila. J Neuroscience. 1997;17:9642–9655. doi: 10.1523/JNEUROSCI.17-24-09642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sink H, Whitington PM. Location and connectivity of abdominal motoneurons in the embryo and larva of Drosophila melanogaster. J Neurobiology. 1991;22:298–311. doi: 10.1002/neu.480220309. [DOI] [PubMed] [Google Scholar]
- 21.Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126:4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- 22.Fujioka M, Lear BC, Landgraf M, Yusibova GL, Zhou J, Riley KM, Patel NH, Jaynes JB. Even-skipped, acting as a repressor, regulates axonal projections in Drosophila. Development. 2003;130:5385–5400. doi: 10.1242/dev.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujioka M, Yusibova GL, Patel NH, Brown SJ, Jaynes JB. The repressor activity of even-skipped is highly conserved, and is sufficient to activate engrailed and to regulate both the spacing and stability of parasegment boundaries. Development. 2002;129:4411–4421. doi: 10.1242/dev.129.19.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujioka M, Jaynes JB, Goto T. Early even-skipped stripes act as morphogenetic gradients at the single cell level to establish engrailed expression. Development. 1995;121:4371–4382. doi: 10.1242/dev.121.12.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molina MR, Cripps RM. Ostia, the inflow tracts of the Drosophila heart, develop from a genetically distinct subset of cardial cells. Mech Dev. 2001;109:51–59. doi: 10.1016/s0925-4773(01)00509-3. [DOI] [PubMed] [Google Scholar]
- 26.Wessells RJ, Bodmer R. Screening assays for heart function mutants in Drosophila. Biotechniques. 2004;37:58–64. doi: 10.2144/04371ST01. [DOI] [PubMed] [Google Scholar]
- 27.Wessells RJ, Fitzgerald E, Cypser JR, Tatar M, Bodmer R. Insulin regulation of heart function in aging fruit flies. Nat Genet. 2004;36:1275–1281. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- 28.Chen TH, Chang TC, Kang JO, Choudhary B, Makita T, Tran CM, Burch JB, Eid H, Sucov HM. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acid-inducible trophic factor. Dev Biol. 2002;250:198–207. doi: 10.1006/dbio.2002.0796. [DOI] [PubMed] [Google Scholar]
- 29.Stuckmann I, Evans S, Lassar AB. Erythropoietin and retinoic acid, secreted from the epicardium, are required for cardiac myocyte proliferation. Dev Biol. 2003;255:334–349. doi: 10.1016/s0012-1606(02)00078-7. [DOI] [PubMed] [Google Scholar]
- 30.Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci U S A. 1257;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gittenberger-de Groot AC, Blom NM, Aoyama N, Sucov H, Wenink AC, Poelmann RE. The role of neural crest and epicardium-derived cells in conduction system formation. Novartis Found Symp. 2003;250:125–134. doi: 10.1002/0470868066.ch8. discussion 134–141, 276–279. [DOI] [PubMed] [Google Scholar]
- 32.Klinedinst SL, Bodmer R. Gata factor pannier is required to establish competence for heart progenitor formation. Development. 2003;130:3027–3038. doi: 10.1242/dev.00517. [DOI] [PubMed] [Google Scholar]
- 33.Dettman RW, Denetclaw W, Jr, Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- 34.Moore AW, McInnes L, Kreidberg J, Hastie ND, Schedl A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development. 1999;126:1845–1857. doi: 10.1242/dev.126.9.1845. [DOI] [PubMed] [Google Scholar]
- 35.Perez-Pomares JM, Phelps A, Sedmerova M, Carmona R, Gonzalez-Iriarte M, Munoz-Chapuli R, Wessels A. Experimental studies on the spatiotemporal expression of WT1 and RALDH2 in the embryonic avian heart: a model for the regulation of myocardial and valvuloseptal development by epicardially derived cells (EPDCs) Dev Biol. 2002;247:307–326. doi: 10.1006/dbio.2002.0706. [DOI] [PubMed] [Google Scholar]
- 35a.Carmena A, Buff E, Halfon MS, Gisselbrecht S, Jiménez F, Baylies MK, Michelson AM. Reciprocal regulatory interactions between the Notch and Ras signaling pathways in the Drosophila embryonic mesoderm. Dev Biol. 2002;244:226–242. doi: 10.1006/dbio.2002.0606. [DOI] [PubMed] [Google Scholar]
- 36.Carmena A, Bate M, Jimenez F. Lethal of scute, a proneural gene, participates in the specification of muscle progenitors during Drosophila embryogenesis. Genes Dev. 1995;9:2373–2383. doi: 10.1101/gad.9.19.2373. [DOI] [PubMed] [Google Scholar]
- 37.Bate M, Rushton E, Frasch M. A dual requirement for neurogenic genes in Drosophila myogenesis. Development Suppl. 1993:149–161. [PubMed] [Google Scholar]
- 38.Corbin V, Michelson AM, Abmayr SM, Neel V, Alcamo E, Maniatis T, Young MW. A role for the Drosophila neurogenic genes in mesoderm differentiation. Cell. 1991;67:311–323. doi: 10.1016/0092-8674(91)90183-y. [DOI] [PubMed] [Google Scholar]
- 39.Ruiz-Gomez M, Romani S, Hartmann C, Jackle H, Bate M. Specific muscle identities are regulated by Krüppel during Drosophila embryogenesis. Development. 1997;124:3407–3414. doi: 10.1242/dev.124.17.3407. [DOI] [PubMed] [Google Scholar]
- 40.Carmena A, Murugasu-Oei B, Menon D, Jimenez F, Chia W. inscuteable and numb mediate asymmetric muscle progenitor cell divisions during Drosophila myogenesis. Genes Dev. 1998;12:304–315. doi: 10.1101/gad.12.3.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halfon MS, Carmena A, Gisselbrecht S, Sackerson CM, Jimenez F, Baylies MK, Michelson AM. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103:63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- 42.Buff E, Carmena A, Gisselbrecht S, Jimenez F, Michelson AM. Signalling by the Drosophila epidermal growth factor receptor is required for the specification and diversification of embryonic muscle progenitors. Development. 1998;125:2075–2086. doi: 10.1242/dev.125.11.2075. [DOI] [PubMed] [Google Scholar]
- 43.Michelson AM, Gisselbrecht S, Buff E, Skeath JB. Heartbroken is a specific downstream mediator of FGF receptor signalling in Drosophila. Development. 1998;125:4379–4389. doi: 10.1242/dev.125.22.4379. [DOI] [PubMed] [Google Scholar]
- 44.Michelson AM, Gisselbrecht S, Zhou Y, Baek KH, Buff EM. Dual functions of the heartless fibroblast growth factor receptor in development of the Drosophila embryonic mesoderm. Dev Genet. 1998;22:212–229. doi: 10.1002/(SICI)1520-6408(1998)22:3<212::AID-DVG4>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.